Abstract
Oxidative tissues such as heart undergo a dramatic perinatal mitochondrial biogenesis to meet the high-energy demands after birth. PPARγ coactivator-1 (PGC-1) α and β have been implicated in the transcriptional control of cellular energy metabolism. Mice with combined deficiency of PGC-1α and PGC-1β (PGC-1αβ−/− mice) were generated to investigate the convergence of their functions in vivo. The phenotype of PGC-1β−/− mice was minimal under nonstressed conditions, including normal heart function, similar to that of PGC-1α−/− mice generated previously. In striking contrast to the singly deficient PGC-1 lines, PGC-1αβ−/− mice died shortly after birth with small hearts, bradycardia, intermittent heart block, and a markedly reduced cardiac output. Cardiac-specific ablation of the PGC-1β gene on a PGC-1α-deficient background phenocopied the generalized PGC-1αβ−/− mice. The hearts of the PGC-1αβ−/− mice exhibited signatures of a maturational defect including reduced growth, a late fetal arrest in mitochondrial biogenesis, and persistence of a fetal pattern of gene expression. Brown adipose tissue (BAT) of PGC-1αβ−/− mice also exhibited a severe abnormality in function and mitochondrial density. We conclude that PGC-1α and PGC-1β share roles that collectively are necessary for the postnatal metabolic and functional maturation of heart and BAT.
Keywords: Transcriptional regulation, heart development, mitochondria, energy metabolism
The transcriptional coactivator peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (Ppargc1a, or commonly called PGC-1α) was discovered based on its functional interaction with the nuclear receptor PPARγ in brown adipocytes (Puigserver et al. 1998). Thereafter, two related transcriptional coactivators, PGC-1β (Ppargc1b) and PGC-1-related coactivator (Pprc1, or commonly called PRC), were identified (Andersson and Scarpulla 2001; Kressler et al. 2002; Lin et al. 2002a). PGC-1α and PGC-1β exhibit the greatest degree of homology among the PGC-1 family members and are preferentially expressed in tissues with high-capacity mitochondrial function such as heart, slow-twitch skeletal muscle, and brown adipose tissue (BAT). PGC-1 coactivators dock to specific target transcription factors, providing a platform for the recruitment of protein complexes that exert powerful effects on target gene transcription by remodeling chromatin and enabling access by the RNA polymerase II machinery (Puigserver et al. 1999; Wallberg et al. 2003; Sano et al. 2007). Studies focused largely on PGC-1α have shown that it exerts its biologic actions by coactivating a variety of nuclear receptor (e.g., PPARγ, PPARα, estrogen-related receptor [ERR] α), and non-nuclear receptor (e.g., nuclear respiratory factors [NRFs], FOXO1) transcription-factor targets (Wu et al. 1999; Vega et al. 2000; Huss et al. 2002; Puigserver et al. 2003; Schreiber et al. 2003).
As opposed to the majority of known transcriptional coactivators, the expression of PGC-1α and, to a lesser extent PGC-1β, is highly inducible in response to developmental stage-specific and physiological cues (Puigserver et al. 1998; Wu et al. 1999; Lehman et al. 2000; Finck and Kelly 2006; Handschin and Spiegelman 2006). PGC-1α expression is induced by cold exposure, exercise, and fasting in tissue-specific patterns. Studies of murine heart have shown that PGC-1α is induced at birth and during postnatal stages coincident with a dramatic energy metabolic maturation that involves a robust mitochondrial biogenic response and switch to reliance on fatty acids as the chief fuel (Lehman et al. 2000; Lehman and Kelly 2002; Taha and Lopaschuk 2007; Buroker et al. 2008). Overexpression studies in mammalian cells in culture or in transgenic mice have shown that both PGC-1α and PGC-1β are capable of activating the expression of a cascade of genes involved in mitochondrial biogenesis and respiratory function in adipocytes, cardiac myocytes, and myogenic cells (Wu et al. 1999; Lehman et al. 2000; Lin et al. 2002a; St-Pierre et al. 2003). In addition, gain-of-function studies have shown that PGC-1α activates gene regulatory programs involved in hepatic gluconeogenesis, (Herzig et al. 2001; Yoon et al. 2001; Puigserver and Spiegelman 2003; Koo et al. 2004), muscle glucose uptake (Michael et al. 2001; Wende et al. 2007), and slow-twitch muscle fiber type determination (Lin et al. 2002b).
More recently, loss-of-function studies have been conducted in mice in an attempt to define specific biological roles for the PGC-1 coactivators and to determine the necessity of these molecules in the regulation of biologic processes in vivo. Two independent mouse lines with generalized inactivation of the PGC-1α gene have been generated (Lin et al. 2004; Leone et al. 2005). PGC-1α−/− mice exhibit a surprisingly minimal phenotype under basal physiologic conditions, indicating that it is dispensable for the fundamental process of mitochondrial biogenesis or fetal development. However, physiological conditions that impose increased energy demands such as cold exposure, fasting, or exercise, precipitate phenotypes in the PGC-1α-deficient mice (Lin et al. 2004; Leone et al. 2005). The baseline cardiac phenotype of PGC-1α-deficient mice is remarkably minimal given the importance of a high-capacity mitochondrial system for this organ. However, PGC-1α-deficient mice develop ventricular dysfunction after prolonged pressure overload (Arany et al. 2006).
Recently, three independent lines of generalized PGC-1β-deficient mice were generated. PGC-1β−/− mice exhibit stress-induced phenotypes that are generally milder but very similar to PGC-1α−/− mice (Lelliott et al. 2006; Vianna et al. 2006; Sonoda et al. 2007). These results suggest the possibility that PGC-1α and PGC-1β control a subset of overlapping targets and are, therefore, capable of compensating for the loss of the other factor in the PGC-1 loss-of-function mice. To address this question and to learn more about the role of PGC-1 coactivators in vivo, we generated mice that are doubly deficient in PGC-1α and PGC-1β (PGC-1αβ−/−) by targeting all four alleles. As described by others, we found that our independent line of PGC-1β-deficient mice exhibits a mild basal phenotype similar to that of PGC-1α-deficient mice. In striking contrast, PGC-1αβ−/− mice die shortly after birth as a result of heart failure related to a perinatal developmental arrest in cardiac maturation including a block in mitochondrial biogenesis.
Results
Generation and general characterization of PGC-1β-null mice
Previously, three research groups have reported and characterized PGC-1β-deficient mice (Lelliott et al. 2006; Vianna et al. 2006; Sonoda et al. 2007). We generated an independent line of PGC-1β-deficient mice. Briefly, a cre-lox strategy was used to delete exons 4–6 of the murine PGC-1β gene (Supplemental Fig. 1A). The targeted deletion introduced a predicted amino acid frameshift, resulting in a premature stop codon in exon 7. The efficacy of the gene targeting event and generation of the mutant transcript was confirmed by PCR, Southern blotting, RNA blotting, and immunoblotting studies (Supplemental Fig. 1B–D).
Heterozygous mice (PGC-1β+/−) were bred to generate PGC-1β−/− offspring. As described previously (Sonoda et al. 2007), survival rates were modestly, but significantly reduced (17% observed, 25% expected; Supplemental Table 1). The surviving mice appeared normal. Given that PGC-1α is necessary for adaptive physiological responses to stressors that demand increased mitochondrial oxidative capacity (Lin et al. 2004; Leone et al. 2005), the PGC-1β−/− mice were subjected to short-term cold exposure and exercise. Six-week-old PGC-1β−/− mice were subjected for 4 h to a cold environment (4°C) without food. The PGC-1β−/− mice were unable to maintain core body temperature to the same degree as sex- and weight-matched wild-type controls (Supplemental Fig. 2A).
Electron microscopic analysis of the BAT of PGC-1β−/− mice did not reveal any significant abnormalities in mitochondrial volume density or ultrastructure (data not shown). Exercise performance was assessed using a low-intensity, run-to-exhaustion exercise protocol on a motorized treadmill. The mean running duration for the PGC-1β−/− mice was less than that of PGC-1β+/+ mice (164 ± 14 vs. 202 ± 10 min, P < 0.05) (Supplemental Fig. 2B). Histologic analyses and electron microscopic studies of soleus muscles did not reveal any overt cellular or ultrastructural abnormalities in the PGC-1β−/− mice (data not shown). However, mean state 3 respiration rates of mitochondria isolated from hindlimb muscle were modestly but significantly decreased in PGC-1β−/− mice compared with PGC-1β+/+ controls (73.93 ± 6.12 vs. 102.58 ± 3.22 nmol O2 per minute per milligram of protein, P < 0.05) (Supplemental Fig. 2C). This finding of reduced muscle state 3 respiratory rates in the PGC-1β−/− mice is consistent with the results of recent studies demonstrating that the expression of genes involved in mitochondrial oxidative phosphorylation (OXPHOS) are down-regulated in skeletal muscle (Lelliott et al. 2006; Vianna et al. 2006; Sonoda et al. 2007) and BAT (Lelliott et al. 2006; Sonoda et al. 2007) in PGC-1β-deficient mouse lines. Taken together, these results demonstrate that the muscle and BAT phenotypes of the PGC-1β−/− mice are similar to that of PGC-1α-deficient mice (Lin et al. 2004; Leone et al. 2005) indicating that both PGC-1α and PGC-1β are necessary for adaptive thermogenesis to cold and exertional exercise.
To determine the necessity of PGC-1β for cardiac function, echocardiographic studies were performed on age-matched PGC-1β−/− and PGC-1β+/+ control mice at 2 mo of age. Mean left ventricular (LV) mass (64.87 ± 3.52 vs. 73.33 ± 4.13 mg) and cardiac systolic function (% LV fractional shortening, 64.96 ± 3.53 vs. 58.32 ± 0.99) were not different between the groups. To assess the cardiac response to stress, exercise echocardiography was performed. The post-exercise heart rate response and LV fractional shortening was similar between the groups (data not shown). Tissue histologic studies of the PGC-1β−/− ventricle did not reveal any significant fibrosis or cellular abnormalities (data not shown). Electron microscopic analyses of PGC-1β−/− papillary muscle revealed normal sarcomeric architecture, mitochondrial morphology, and mitochondrial volume density (Supplemental Fig. 3). Collectively, these results indicate that, as we reported previously for PGC-1α (Leone et al. 2005), PGC-1β is dispensable for normal cardiac development, structure, and function.
PGC-1α compensates for the loss of PGC-1β
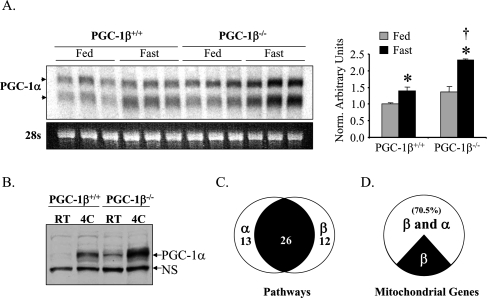
The lack of a cardiac phenotype in PGC-1β−/− mice under nonstressed conditions strongly suggested that compensatory mechanisms are activated, possibly through the actions of the related coactivator, PGC-1α. As an initial step to explore this possibility, levels of PGC-1α gene expression were determined in several relevant PGC-1β−/− tissues. Under basal conditions, PGC-1α mRNA levels in PGC-1β−/− heart were not significantly different from that of littermate PGC-1β+/+ controls (Fig. 1A). However, after short-term starvation, a condition that increases demands on mitochondrial oxidation of fatty acids and ketones in heart, PGC-1α mRNA levels were induced to higher levels in PGC-1β−/− hearts compared with controls (Fig. 1A). In addition, mean levels of BAT PGC-1α protein were higher in PGC-1β−/− mice compared with PGC-1β+/+ mice under basal conditions and following exposure to cold (Fig. 1B). Thus, PGC-1α gene expression is induced in the heart and BAT of PGC-1β−/− mice, particularly in the context of physiological stressors that increase requirements for mitochondrial ATP production.
Figure 1.
PGC-1α and PGC-1β drive a significant subset of overlapping gene regulatory programs. (A, left) Representative autoradiograph of a Northern blot using RNA isolated from heart of PGC-1β+/+ and PGC-1β−/− mice on standard chow (Fed) or post 36 h fast (Fast) is shown using a full-length PGC-1α cDNA as a probe. PGC-1α transcripts are denoted by the arrows. Ethidium bromide staining of 28s ribosomal RNA is shown at the bottom as a loading control. (Right) Quantitative RT–PCR (TaqMan) of total RNA from heart was used to characterize the level of PGC-1α gene expression in fed (gray bar) and fasted (black bars) PGC-1β+/+ and PGC-1β−/− hearts. The mRNA levels were normalized to 36Β4 mRNA content, and are shown relative to the PGC-1β+/+ fed values (=1.0). (*) P < 0.05 compared with the fed group of the same genotype; (†) P < 0.05 compared with the fasted group of the PGC-1β+/+. (B) Western blot analysis of whole-cellprotein extracts preparedfromBAT of PGC-1β+/+ and PGC-1β−/− mice at room temperature or at 4°C. The top arrow designates the full-length PGC-1α protein. A nonspecific band (NS) is shown as a loading control. (C–D) NRCM in culture were infected with Ad-GFP, Ad-PGC-1α, or Ad-PGC-1β, and gene expression array analysis was performed using the Affymetrix Rat Expression Set 230 chip. (C) Venn diagram showing the Gene Ontology pathways that were up-regulated by either PGC-1α or PGC-1β compared with Ad-GFP-infected cells. The left circle represents the pathways up-regulated by PGC-1α, and right circle represents those up-regulated by PGC-1β with overlapping portion (black) representing pathways up-regulated by both. (D) The pie chart represents genes in the Gene Ontology category “mitochondrion” that were up-regulated at least 1.5-fold by PGC-1β compared with Ad-GFP-infected cells (P < 0.05). The white portion represents those that were also up-regulated by PGC-1α by at least 1.5-fold (P < 0.05).
We next sought to determine whether the PGC-1 coactivators share gene targets in cardiac myocytes given the minimal cardiac phenotype of the PGC-1β−/− and PGC-1α−/− mice. To this end, gene expression profiling was conducted using RNA isolated from neonatal rat cardiac myocytes (NRCM) infected with adenovirus overexpressing PGC-1α, PGC-1β, or GFP alone (adenoviral backbone control). NRCM were used because, in culture conditions, the myocytes assume a late fetal metabolic phenotype including minimal expression of PGC-1 coactivators and targets (Lehman et al. 2000). Pathway analysis revealed that 38 pathways were up-regulated by PGC-1β, 26 of which were also regulated by PGC-1α (Fig. 1C). As expected, mitochondrial-associated pathways were predominantly targeted by both coactivators (Supplemental Table 2). Mitochondrial targets shared by the coactivators include genes involved in fatty acid oxidation (FAO), the TCA cycle, OXPHOS, and glucose oxidation (Supplemental Table 3). Notably, 70.5% of genes involved in mitochondrial metabolism up-regulated by PGC-1β were also induced by PGC-1α (Fig. 1D). These results suggest that there is significant overlap in the cardiac metabolic gene targets regulated by PGC-1α and PGC-1β.
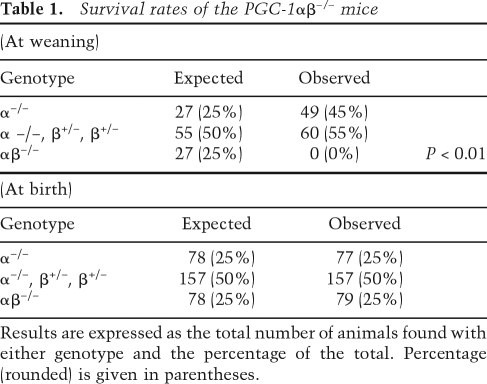
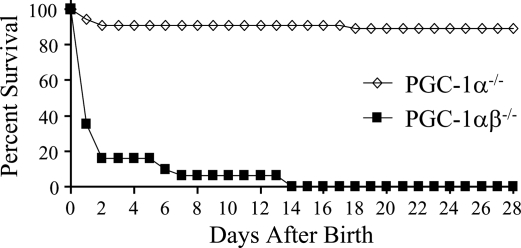
We next sought to evaluate the functional redundancy of the two coactivators by generating mice with targeted deactivation of all four PGC-1α and PGC-1β alleles (PGC-1αβ−/− mice). For these studies, PGC-1β+/− mice on a PGC-1α-null background (PGC-1α−/−β+/−) were generated and intercrossed to generate double-deficient offspring (PGC-1αβ−/−). Of 109 pups generated from the PGC-1α−/−β+/− × PGC-1α−/−β+/− crosses, no double homozygous null mice (PGC-1αβ−/−) were found at weaning. Furthermore, the PGC-1α−/−β+/− mice that survived until weaning were not found in the expected 2:1 ratio with PGC-1α−/− mice (Table 1, top), indicating that the PGC-1αβ−/− genotype is lethal. To determine the age of death in the PGC-1αβ−/− group, timed breedings were performed. Inspection of birth sacs during the late fetal period did not reveal any evidence of embryonic lethality. At birth, all of the PGC-1αβ−/− mice were viable and genotyping revealed the expected Mendelian ratio (Table 1, bottom), although their birth weights were less than the other genotypes (Supplemental Fig. 4). However, the majority (∼70%) of PGC-1αβ−/− pups died within 24 h of birth and all died within 14 d (Fig. 2). These results demonstrate the importance of having at least one PGC-1α or PGC-1β allele for survival following birth.
Table 1.
Survival rates of the PGC-1αβ−/− mice
Results are expressed as the total number of animals found with either genotype and the percentage of the total. Percentage (rounded) is given in parentheses.
Figure 2.
Deficiency of both PGC-1α and PGC-1β is lethal. Mortality curve depicting the percent survival of male and female PGC-1α−/− (diamonds, n = 55) and PGC-1αβ-deficient (PGC-1αβ−/−, squares, n = 31) pups 28 d after birth.
The PGC-1αβ−/− pups exhibited a labored breathing pattern during the first hours after birth, suggestive of a metabolic or cardiopulmonary crisis. Blood glucose values were not significantly different among the four genotypes with the exception that glucose levels in PGC-1αβ−/− mice were modestly but significantly lower than PGC-1αβ+/+ controls, yet similar to that of PGC-1α−/− and PGC-1β−/− mice (Supplemental Fig. 5A). In addition, blood lactate levels were not different across all four genotypes following a 4-h fast (Supplemental Fig. 5B). Postmortem histologic analyses of the PGC-1αβ−/− lungs revealed evidence of alveolar collapse, whereas sections taken before death were normal, suggesting that the abnormalities were secondary, possibly to congestive heart failure (Supplemental Fig. 6A). In addition, postmortem gross and histologic analyses of the PGC-1αβ−/− mice did not reveal overt abnormalities in brain, liver, and kidney (Supplemental Fig. 6B,C; data not shown). The PGC-1αβ−/− hearts were significantly smaller than the hearts from the other genotypes, but did not exhibit any gross morphologic abnormalities; all four chambers were present, the great vessels were intact and in the proper orientation, and the ductus arteriosus was appropriately closed post-birth (data not shown).
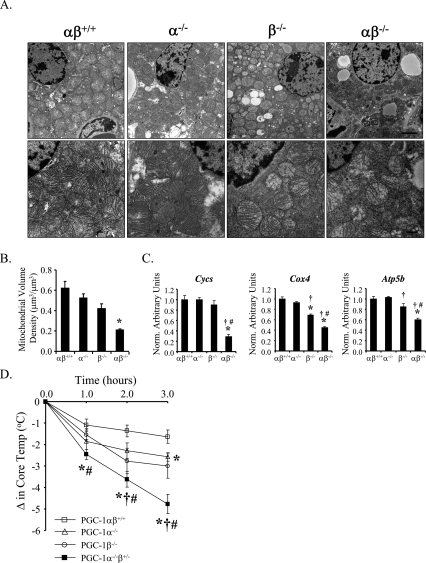
PGC-1 loss-of-function studies conducted in isolated adipocytes have strongly suggested that the presence of either PGC-1α or PGC-1β is necessary for the differentiation and mitochondrial maturation of brown adipocytes in vitro (Uldry et al. 2006). Accordingly, we characterized BAT structure and function in the doubly mutant mice. Lipid droplet size and density appeared greater in brown adipocytes of PGC-1α−/−, PGC-1β−/−, and PGC-1αβ−/− mice compared with the wild-type control, with the PGC-1αβ−/− group being slightly more abnormal (Supplemental Fig. 7A). Triglyceride content in BAT from all four genotypes paralleled the histologic results (Supplemental Fig. 7B). Electron microscopy coupled with quantitative morphometry demonstrated that whereas BAT mitochondrial volume density was not different in PGC-1α−/− and PGC-1β−/− BAT compared with wild-type control, it was significantly lower in PGC-1αβ−/− mice (Fig. 3A,B). The mitochondria of PGC-1αβ−/− BAT also exhibited reduced cristae density (Fig. 3A). Consistent with the ultrastructural findings, BAT expression of key PGC-1 mitochondrial gene targets (cytochrome c, somatic, Cycs; cytochrome oxidase 4, Cox4; and ATP synthase, H+ transporting, mitochondrial F1 complex, β polypeptide, Atp5b) was significantly reduced in PGC-1αβ−/− mice compared with the other genotypes (Fig. 3C). To assess functional correlates of the BAT histologic and gene expression results, cold exposure studies were conducted with 5- to 6-wk-old PGC-1α−/−β+/− mice. Triple allele mutants were used because the PGC-1αβ−/− mice did not survive. After 2 h, the core temperature of PGC-1α−/−β+/− mice dropped significantly lower than age- and sex-matched wild-type, PGC-1α−/−, or PGC-1β−/− mice (Fig. 3D). Taken together, these results suggest that all four alleles are necessary for a normal adaptive thermogenic response, but that some functional overlap exists between the two PGC-1 coactivator proteins for mitochondrial biogenesis.
Figure 3.
BAT phenotype in PGC-1αβ-deficient mice. (A) Representative electron micrographs of BAT at PD0.5 from wild-type (αβ+/+), PGC-1α−/− (α−/−), PGC-1β−/− (β−/−), and PGC-1αβ−/− (αβ−/−) mice at two different magnifications. Each genotype label denotes the vertical column below it. Bars: top row, 2 μm; bottom row, 500 nm. (B) Quantitative morphometric measurements of the cellular volume density for the mitochondrial fraction based on analysis of electron micrographs. Bars represent mean ± SEM. (*) P < 0.05 compared with αβ+/+. (C) Quantitative real-time RT–PCR analysis of RNA extracted from hearts of PD0.5 wild-type, PGC-1α−/−, PGC-1β−/−, and PGC-1αβ−/− mice for the following: oxidative phosphorylation-cytochrome c, somatic (Cycs), cytochrome oxidase 4 (Cox4), ATP synthase, H+ transporting, mitochondrial F1 complex, β polypeptide (Atp5b). The mRNA levels were normalized to 18s rRNA content, and expressed relative to PGC-1αβ+/+ values. Bars represent mean ± SEM. (*) P < 0.05 compared with αβ+/+; (†) P < 0.05 compared with α−/−; (#) P < 0.05 compared with β−/−. (D) Thirty-six-day-old to 42-d-old PGC-1αβ+/+ (open squares, n = 11), PGC-1α−/− (open triangle, n = 7), PGC-1β−/− (open circle, n = 13), and PGC-1α−/−β+/− mice (black squares, n = 13) were subjected to cold (4°C). The change in core temperature ± SEM is shown in the graph as a function of time. (*) P < 0.05, compared with αβ+/+; (†) P < 0.05 compared with α−/−; (#) P < 0.05 compared with β−/−.
PGC-1αβ−/− mice die of heart failure
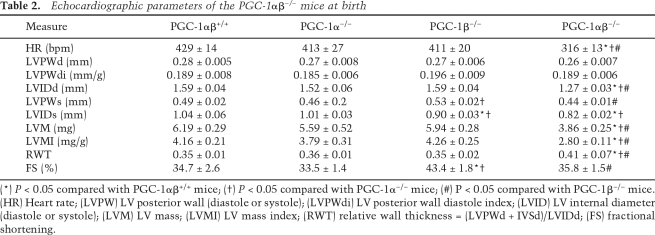
To assess cardiac structure and function in the PGC-1αβ−/− mice, echocardiographic and Doppler studies were performed at ∼12 h after birth. Both absolute LV mass (LVM) and LVM corrected for body weight (LVMI) were significantly reduced in PGC-1αβ−/− mice compared with wild-type, PGC-1α−/−, and PGC-1β−/− groups consistent with a growth defect (Table 2). PGC-1αβ−/− hearts also exhibited a significant reduction in mean LV internal diameter (LVID) during diastole consistent with reduced heart size (Table 2). Mean heart rate was significantly lower in the PGC-1αβ−/− mice compared with the other genotypes (Table 2). Careful inspection of the heart rate and Doppler velocity waveforms representing the passive (E wave) and atrial contraction (A wave) components of diastolic LV filling revealed that they were not always coupled to a systolic LV outflow jet, consistent with intermittent second-degree heart block, which occurred in a variety of A:V conduction patterns including 2:1, 3:1, 4:1, 6:1, and 8:1 (Supplemental Fig. 8).
Table 2.
Echocardiographic parameters of the PGC-1αβ−/− mice at birth
(*) P < 0.05 compared with PGC-1αβ+/+ mice; (†) P < 0.05 compared with PGC-1α−/− mice; (#) P < 0.05 compared with PGC-1β−/− mice. (HR) Heart rate; (LVPW) LV posterior wall (diastole or systole); (LVPWdi) LV posterior wall diastole index; (LVID) LV internal diameter (diastole or systole); (LVM) LV mass; (LVMI) LV mass index; (RWT) relative wall thickness = (LVPWd + IVSd)/LVIDd; (FS) fractional shortening.
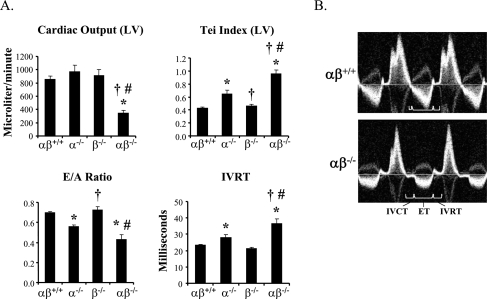
Cardiac output measurements were made in the neonatal mice by measuring aortic diameter and blood flow velocity via Doppler in the proximal portion of the descending aorta. Cardiac output was markedly reduced in the PGC-1αβ−/− mice compared with the other genotypes (Fig. 4A,B), despite preservation of LV fractional shortening (Table 2). To further analyze cardiac performance, the Tei index, a noninvasively derived parameter of combined systolic and diastolic function, based on the timing of events during the cardiac cycle, was determined using the pulse wave Doppler spectra of the trans-mitral and LV outflow tract velocities (Tei et al. 1995). To account for the potential confounding effect of heart rate differences among the groups, the heart rate of wild-type mice was lowered to that measured in the PGC-1αβ−/− mice by the sinus node inhibitor, Zatebradine. The mean Tei index was abnormal in the PGC-1αβ−/− mice compared with the other genotypes (Fig. 4A), reflecting a significant abnormality in each of the three components of the index (increased isovolumic contraction time and isovolumic relaxation time [IVRT], and decreased ejection time, depicted in Fig. 4B). Interestingly, the PGC-1α-deficient heart exhibited a significant decrease in performance compared with the PGC-1αβ+/+ heart, albeit not as severe as that of the PGC-1αβ−/− mice. Lastly, Doppler-derived parameters of diastolic filling showed decreased E/A ratio and prolonged IVRT in PGC-1αβ−/− mice, consistent with impaired ventricular diastolic relaxation (Fig. 4A). Taken together, the cardiac function results indicate that the PGC-1αβ−/− mice have markedly reduced postnatal cardiac output, likely due to an inappropriately low heart rate combined with reduced contractile and diastolic function.
Figure 4.
Evidence for cardiac failure in PGC-1αβ-deficient mice. To evaluate cardiac function noninvasively in all four genotypes, high-resolution echocardiography was performed within a few hours after birth. (A) Bar graphs show representative indices of systolic (cardiac output), diastolic (E/A ratio, IVRT), and combined (Tei index) left ventricular performance. (B) Representative images of the trans-mitral/left ventricular outflow tract (LVOT) Doppler spectra from wild-type (αβ+/+) and PGC-1αβ−/− (αβ−/−) mice demonstrate markedly altered cardiac time intervals and reduced LVOT velocities in the double null mice. (IVRT) Isovolumic relaxation time; (IVCT) isovolumic contraction time; (ET) ejection time.
Despite our findings of a significant cardiac phenotype in the PGC-1αβ−/− mice, it was possible that extracardiac effects including, but not limited to, abnormal thermogenesis contributed to the early postnatal lethality. To address this, mice were generated in which the PGC-1β gene was deleted specifically in heart on a generalized PGC-1α-deficient background (PGC-1α−/−βf/f/MHC-Cre) via Cre-recombinase-mediated excision of exons 4–6 using the same parent targeting vector used to generate the generalized PGC-1β-null mice (strategy shown in Supplemental Fig. 9A). Cardiac specificity of the PGC-1β gene deletion was achieved through the use of αMHC-Cre mice (Agah et al. 1997), which expresses Cre recombinase specifically in cardiac myocytes driven by the cardiac α myosin heavy chain promoter (Supplemental Fig. 9B; data not shown). Combined PGC-1α−/− and cardiac-specific PGC-1β-deficient mice (PGC-1α−/−βf/f/MHC-Cre) were generated by breeding. Among the four genotypes expected in the offspring, three were viable and produced with the expected 1:1:1 ratio (Supplemental Table 4). Similar to the generalized PGC-1αβ−/− mice, PGC-1α−/−βf/f/MHC-Cre mice were born alive, 67% of the pups died within 24 h of birth, and all were dead within 7 d (Supplemental Fig. 10). The mean echocardiographic-derived cardiac output measurements with the neonatal PGC-1α−/−βf/f/MHC-Cre mice (controls, 920 ± 40 vs. PGC-1α−/−βf/f/MHC-Cre, 479 ± 28 μL per minute) were strikingly similar to that of the PGC-1αβ−/− mice (αβ+/+, 852 ± 52 vs. αβ−/−, 342 ± 47 μL per minute). These results indicate that the PGC-1α−/−βf/f/MHC-Cre mice phenocopy the generalized PGC-1αβ-deficient mice, providing further support for the conclusion that the PGC-1αβ−/− mice die of heart failure.
Evidence for a mitochondrial maturational arrest in the hearts of PGC-1αβ−/− mice
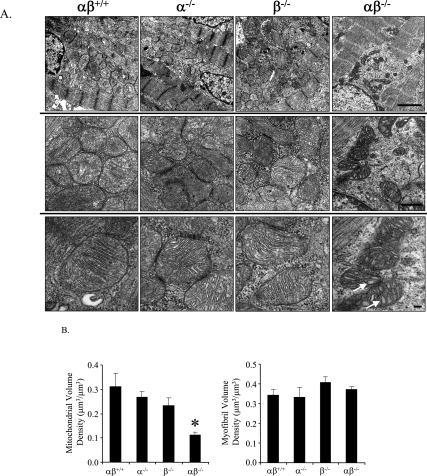
We next sought to investigate the basis for the cardiac failure in the PGC-1αβ−/− mice. Evidence for apoptosis or fibrotic changes was absent based on TUNEL, caspase, and trichrome staining (data not shown). However, electron microscopic studies demonstrated dramatic mitochondrial abnormalities in the hearts of PGC-1αβ−/− mice, most prominent of which was a significant diminution in mitochondrial number and size, consistent with a defect in mitochondrial biogenesis (Fig. 5A). Cardiac mitochondria of the PGC-1αβ−/− mice also exhibited a variety of ultrastructural abnormalities including vacuoles and reduced cristae density, suggesting a defect in biogenesis or swelling (Fig. 5A). Quantitative morphometry confirmed a significant reduction in mean cellular mitochondrial volume density despite normal myofibrillar volume density in the cardiac myocytes of the PGC-1αβ−/− mice (Fig. 5B). Importantly, the myocyte mitochondrial volume density of PGC-1α−/− and PGC-1β−/− hearts was not reduced. This conclusion was also supported by the results of mitochondrial DNA quantification (Supplemental Fig. 11).
Figure 5.
Abnormal mitochondrial density and structure in hearts of PGC-1αβ−/− mice. (A) Representative electron micrographs of cardiac muscle (LV free wall) at PD0.5 from wild-type (αβ+/+), PGC-1α−/− (α−/−), PGC-1β−/− (β−/−), and PGC-1αβ−/− (αβ−/−) mice at three different magnifications. Each genotype label denotes the vertical column below it. Bars: top row, 2 μm; middle row, 500 nm; bottom row, 100 nm. Arrows indicate vacuolar abnormalities within mitochondria of the PGC-1αβ−/− mice. (B) Quantitative morphometric measurements of the cellular volume density for the mitochondrial (left) and myofibrilar (right) fractions based on analysis of electron micrographs. Bars represent mean ± SEM. (*) P < 0.05 compared with αβ+/+.
A variety of cellular ultrastructural abnormalities were also noted in the cardiac ventricles of PGC-1αβ−/− mice. A subset of myocytes that were largely or partially devoid of mitochondria, sarcomeres, and other cellular organelles was noted (Supplemental Fig. 12A,B). Similar cellular and mitochondrial derangements, including a marked reduction in mitochondrial number and size, were also seen in the postnatal hearts of the PGC-1α−/−βf/f/MHC-Cre mice (data not shown).
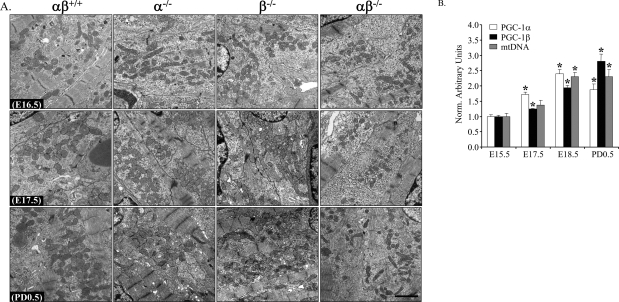
The mitochondrial abnormalities in hearts of PGC-1αβ−/− mice suggested a mitochondrial maturation arrest. A major surge in cardiac mitochondrial biogenesis occurs in heart during the late fetal period and continues through the early postnatal stages (Hallman 1971; Smolich et al. 1989; Marin-Garcia et al. 2000). To determine whether this perinatal mitochondrial biogenic response was defective in the PGC-1αβ−/− mice, electron microscopy studies were conducted on heart samples from mice at embryonic days 16.5 and 17.5 (E16.5 and E17.5) compared with that of postnatal day 0.5 (PD0.5) across all four genotypes. At E16.5, mitochondria were small and sparse compared with postnatal ventricular sections in all four genotypes (Fig. 6A). In striking contrast, at PD0.5 a marked cardiac biogenic response occurred in PGC-1αβ+/+, PGC-1α−/−, and PGC-1β−/− mice, but not the PGC-1αβ−/− group (Fig. 6A). At E17.5, a modest increase in mitochondrial density occurs in all genotypes except the PGC-1αβ−/− group, indicative of a block in this late prenatal surge of mitochondrial biogenesis in the doubly mutant mice. Mitochondrial DNA measurements also revealed the same pattern (Supplemental Fig. 13). Expression of the PGC-1α and PGC-1β genes is coordinately induced in wild-type murine heart from E15.5 to PD0.5, in parallel with the observed mitochondrial biogenic response (Fig. 6B) providing additional evidence supporting a role for these coactivators in the perinatal mitochondrial biogenic surge.
Figure 6.
Perinatal mitochondrial biogenesis is blocked in PGC-1αβ−/− hearts. (A) Representative electron micrographs of cardiac muscle sections (LV free wall) at E16.5 (top panels), E17.5 (middle panels), and PD0.5 (bottom panels) from wild-type (αβ+/+), PGC-1α−/− (α−/−), PGC-1β−/− (β−/−), and PGC-1αβ−/− (αβ−/−) mice. Bar, 2 μm. (B) Quantitative real-time RT–PCR analysis of RNA extracted from hearts of E15.5, E17.5, E18.5, and PD0.5 C57BL6/J mice for the expression of PGC-1α (white bars) and PGC-1β (black bars) genes. The mRNA levels were normalized to 36B4 mRNA levels, and expressed relative to E15.5 values (=1.0). Quantitative PCR of total DNA from heart was performed to quantify mitochondrial DNA (gray bars) using primers for NADH dehydrogenase (ND1) and genomic DNA using primers for lipoprotein lipase (LPL). The ND1 levels were normalized to LPL DNA content, and expressed relative to E15.5 values (=1.0). Bars represent mean ± SEM. (*) P < 0.05 compared with E15.5.
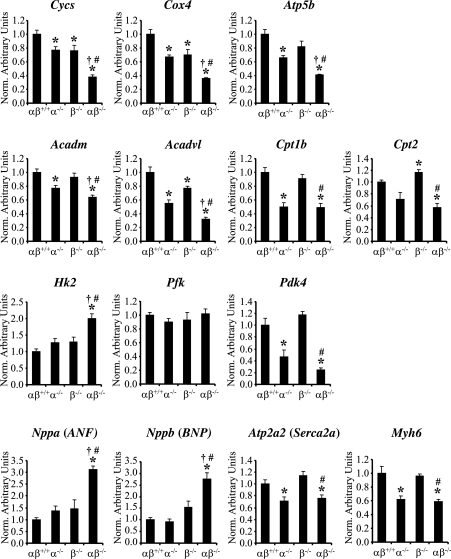
The small heart size and mitochondrial biogenic arrest noted in PGC-1αβ−/− mice strongly suggested a generalized defect in cardiac maturation. To further explore this possibility, gene regulatory signatures of postnatal cardiac myocyte maturation were assessed in the PGC-1αβ−/− mice and compared with the other genotypes. Known metabolic markers of terminal maturation include PGC-1 target genes involved in mitochondrial oxidative pathways such as FAO and OXPHOS. Quantitative RT–PCR analyses revealed that the expression of genes involved in FAO (acetyl-Coenzyme A dehydrogenase, medium chain [Acadm], acyl-Coenzyme A dehydrogenase, very long chain [Acadvl], carnitine palmitoyltransferase 1b [Cpt1b], carnitine palmitoyltransferase 2 [Cpt2]), and OXPHOS (Cycs, Cox4, and Atp5b) were significantly reduced in the PGC-1αβ−/− hearts compared with wild-type controls (Fig. 7). Glucose metabolic markers were also analyzed. Hexokinase 2 (Hk2) was increased and pyruvate dehydrogenase kinase 4 (Pdk4) was significantly decreased in the PGC-1αβ−/− hearts, suggesting that the programs directing the switch from reliance on glucose during the fetal period to fatty acids as the preferred fuel after birth was blocked. In addition, expression of several fetal cardiac gene markers not directly involved in cellular energy metabolism, including atrial natriuretic factor (Nppa) and brain natriuretic peptide (Nppb), remained elevated in the PGC-1αβ−/− cardiac ventricles (Fig. 7), whereas expression of α myosin heavy chain (Mhy6), an adult sarcomeric isoform, was reduced in PGC-1αβ−/− as well as PGC-1α−/− hearts. These gene marker results are consistent with a general arrest in cardiac maturation and suggest a regulatory link between metabolic pathways and the broad program of myocyte maturation.
Figure 7.
Cardiac gene expression markers are consistent with a block in fetal–adult transition. Quantitative real-time RT–PCR analysis of RNA extracted from hearts of PD0.5 wild-type (αβ+/+), PGC-1α−/− (α−/−), PGC-1β−/− (β−/−), and PGC-1αβ−/− (αβ−/−) mice for the following: oxidative phosphorylation-cytochrome c, somatic (Cycs), cytochrome oxidase 4 (Cox4), ATP synthase, H+ transporting, mitochondrial F1 complex, β polypeptide (Atp5b); fatty acid oxidation-acetyl-Coenzyme A dehydrogenase, medium chain (Acadm), acyl-Coenzyme A dehydrogenase, very long chain (Acadvl), carnitine palmitoyltransferase 1b (Cpt1b), carnitine palmitoyltransferase 2 (Cpt2); Glycolysis/Glucose oxidation-hexokinase 2 (Hk2), phosphofructokinase (Pfk), pyruvate dehydrogenase kinase 4 (Pdk4); General adult cardiac gene markers-atrial natriuretic factor (ANF), brain natriuretic peptide (BNP), ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2 (Serca2a), and α-myosin heavy chain (Myh6). The mRNA levels were normalized to β-actin mRNA content, and expressed relative to PGC-1αβ+/+ values. Bars represent mean ± SEM. (*) P < 0.05 compared with αβ+/+; (†) P < 0.05 compared with α−/−; (#) P < 0.05 compared with β−/−.
Discussion
The mammalian heart functions as a constant pump throughout the life of the organism. Following birth, the myocardium burns tremendous amounts of ATP daily to meet the energy demands of postnatal life. During the fetal period, the heart uses mainly glucose and lactate to generate ATP (Fisher et al. 1980, 1981; Girard et al. 1992). The enormous energy demands of the adult heart are met, in large part, by the oxidation of fatty acids in mitochondria (Bing 1955; Itoi and Lopaschuk 1993; Taha and Lopaschuk 2007). Accordingly, to meet the rigors of postnatal life, the heart undergoes a perinatal metabolic maturation that involves a fuel “switch” concordant with a dramatic increase in mitochondrial functional capacity (Marin-Garcia et al. 2000; Taha and Lopaschuk 2007). Herein, we show that the collective actions of PGC-1α and PGC-1β comprise a critical component of the molecular circuitry that drives the perinatal mitochondrial biogenesis necessary for metabolic and functional maturation of the murine heart and BAT.
Our results indicate that key late fetal and perinatal cardiac developmental events are not activated in the PGC-1αβ−/− mice. The heart chambers and great vessels of the PGC-1αβ−/− mice were overtly normal, suggesting that major early fetal developmental events were unimpaired. However, the hearts of the double mutant animals are small and the dramatic mitochondrial biogenic response known to occur at the time of birth was found to be completely absent in the PGC-1αβ−/− mice. The doubly deficient animals also exhibited evidence of an immature conduction system, including bradycardia and heart block. Consistent with this conclusion, mice deficient for ERRγ, a known target of PGC-1-mediated coactivation, were recently shown to exhibit gene regulatory signatures consistent with a block in the cardiac perinatal switch from relying on glucose to oxidative metabolism (Alaynick et al. 2007). Taken together, these results indicate that whereas PGC-1α and PGC-1β are not required for early formation of mitochondria, they are necessary for programs directing late fetal and postnatal cardiac maturation. It is possible that the related factor PRC is compensating for early fetal development processes. In addition, given that our original PGC-1α gene ablation strategy did not exclude the possible production of a smaller mutant PGC-1α protein (Leone et al. 2005), it is theoretically possible that a small amount of residual PGC-1α activity compensates during early developmental stages in the PGC-1αβ−/− mice.
Our results strongly suggest that PGC-1α and PGC-1β share a subset of key gene targets and functions, at least in heart and BAT. This conclusion is supported by the following lines of evidence: (1) PGC-1α gene expression is induced in BAT and heart of PGC-1β-deficient mice, suggesting a compensatory response in this context; (2) the severity of the cold intolerance phenotype and BAT mitochondrial derangements increases in parallel with the number of deleted PGC-1α and PGC-1β alleles, results that are consistent with the findings of a previous study conducted in isolated brown adipocytes (Uldry et al. 2006); (3) gene expression profiling in cardiac myocytes demonstrated a high degree of overlap in PGC-1α and PGC-1β target pathways, especially those involved in mitochondrial metabolism; and (4) whereas the PGC-1β-deficient mice and PGC-1α-deficient mice survive with a minimal cardiac phenotype, combined deficiency results in 100% postnatal lethality due to heart failure. Further analysis of our gene expression array results also provided some idea about which of the downstream transcription factors may be involved in the shared PGC-1 functions. A significant subset of genes activated by both coactivators (Supplemental Tables 2, 3) are also direct targets for ERRα and ERRγ, known PGC-1 coactivating targets in cardiac myocytes based on gene expression profiling and ChIP-chip promoter occupation assays (Dufour et al. 2007). This comparative analysis reveals that a significant number of ERR target genes involved in mitochondrial FAO, respiration, and ATP synthesis were also shown to be activated by PGC-1α and PGC-1β in this study. These results suggest that the profound cardiac mitochondrial phenotype that occurs in the PGC-1αβ−/− mice is related to deactivation of the ERR gene regulatory pathway.
Although these results indicate significant gene target and functional redundancy, evidence for complementary PGC-1 coactivator-specific roles also exist. For example, the stress-induced phenotypes of the single PGC-1-null mice indicate that both coactivators are necessary to meet the full range of physiological demands imposed on postnatal life. Cold exposure, treadmill exercise, or fasting precipitate phenotypes in the single PGC-1 gene deletion mice (Lin et al. 2004; Leone et al. 2005; Lelliott et al. 2006; Vianna et al. 2006; Sonoda et al. 2007). In addition, the results of studies focused on noncardiac tissues have suggested that PGC-1β drives a subset of programs distinct from that of PGC-1α, including hepatic lipogenesis and cholesterol metabolism (Lin et al. 2005) and skeletal muscle IIx fiber type determination (Mortensen et al. 2006; Arany et al. 2007). Single and combined PGC-1α and PGC-1β loss-of-function studies conducted in brown adipocytes in culture have also revealed complementary effects on mitochondrial function (Uldry et al. 2006). It is likely that the two coactivators are regulated by distinct upstream circuits (Lin et al. 2002a) providing for the regulation of common and distinct gene targets in coactivator-specific patterns based on the specific physiological stimulus among tissues or cell types.
Extensive phenotyping revealed that the early postnatal death of the PGC-1αβ−/− mice is due to severe cardiac dysfunction. The PGC-1αβ−/− hearts exhibited intermittent second-degree AV block and generated a very low cardiac output. The basis for the low cardiac output likely relates to the relatively small size of the heart, bradycardia, and reduced contractile function. The contractile dysfunction is likely due, at least in part, to reduced capacity for mitochondrial ATP production related to immature mitochondria and a block in the postnatal induction of genes involved in FAO, the chief source of energy in the postnatal period. In support of this latter conclusion, inborn errors in mitochondrial FAO enzymes are an important cause of inherited cardiomyopathy in children (Kelly and Strauss 1994). In addition, mice with targeted ablation of the gene encoding ERRγ, a known target of PGC-1α and β (Huss et al. 2002; Kamei et al. 2003; Schreiber et al. 2003), die early after birth with a small heart and reduced expression of genes involved in myocardial FAO (Alaynick et al. 2007). Lastly, mice with cardiac-specific deficiency of PGC-1β in a generalized PGC-1α-deficient background (PGC-1α−/−βf/f/MHC-Cre mice) phenocopy the generalized PGC-1αβ−/− mice, providing additional support for the conclusion that the lethal phenotype is caused by cardiac derangements. However, given the severe abnormalities found in the BAT of the PGC-1αβ−/− mice, it is possible that this phenotype is also incompatible with postnatal survival.
Materials and methods
Generation of generalized PGC-1β-null and cardiac-specific PGC-1β-null mice
Sv129 genomic DNA was used as a template to create three amplicons using PCR, which were subsequently inserted into pGKNeo-p1339 (GenBank Accession #AF335420). The 3′ amplicon also contained an engineered LoxP site that was used to excise exons 4, 5, and 6 via Cre recombinase. The construct was linearized with XhoI and electroporated into SCC10 ES cells (derived from RW4) using G418 selection. The clones were screened by Southern blotting and PCR. One clone out of 216 screened was positive for the recombination event. This clone was injected into a C57BL6/J blastocyst. Germ-line transmission was confirmed by coat color as well as PCR analysis of tail DNA. The female mice containing the targeted allele were bred with the male EIIa-Cre mice to generate both complete and conditional knockout mice. The offspring were screened by Southern blotting and PCR. Mice that contained the recombination allele with the exon 4–6 cassette as well as the neomycin cassette removed were chosen for generation of the generalized “knockout” (PGC-1β−/−). Mice that had only the neomycin cassette removed were chosen to create the conditional knockout. Mice were maintained in the hybrid background, C57BL6/J × sv129. Littermates were used whenever possible (as indicated in the text) to control for strain effects.
Generation of PGC-1aβ double-deficient mice
Mice with a single gene deletion of either PGC-1α (Leone et al. 2005) or PGC-1β were bred to generate compound heterozygous (PGC-1α+/−β+/−) mice, which in turn were crossed to generate mice deficient for PGC-1α and heterozygous for PGC-1β (PGC-1α−/−β+/−). These mice were used as breeders to generate the offspring used in this study. Mice were maintained in a hybrid background (C57BL6/J × sv129) and littermates for PGC-1α−/− and PGC-1αβ−/− were used for comparative analysis to control for genetic background effects. PGC-1β−/− and PGC-1αβ+/+ mice were generated with separate breeding pairs.
Animal phenotyping studies
All animal experiments and euthanasia protocols were conducted in strict accordance with the National Institutes of Health guidelines for humane treatment of animals and were reviewed and approved by the Institutional Animal Care and Use Committee of Washington University School of Medicine.
Animals were weighed at different time points between 2 and 8 wk of age and compared directly to their sex-matched littermates. For cold exposure experiments, male and female PGC-1β+/+ and PGC-1β−/− mice were singly housed and placed for 3–4 h at 4°C without food. Core body temperatures were monitored by rectal probe at baseline and every hour thereafter. Mice were monitored at least every 30 min to check for lethargy. At the end of 4 h, mice were sacrificed and tissues harvested for RNA and protein extraction. For fasting studies, animals were singly housed and given water ad libitum. Food was removed from cages in the morning and tissues harvested at 36 h for RNA and histology. For prenatal and mortality curve studies, timed breedings were performed, and pregnancy was determined by detection of a vaginal plug (E0.5). The time of birth was closely monitored, and newborns were counted and genotyped within 12 h after birth for χ2 analysis. Postnatal survival was followed daily for 28 d. Blood glucose in newborns was measured with a One-Touch Ultra glucometer (LifeScan, Inc.). Blood lactate in newborns was measured with a Lactate Pro blood lactate test meter (ARKRAY, Inc.).
For the low-intensity exercise studies, 9-wk-old female PGC-1β+/+ (n = 6) and PGC-1β−/− (n = 6) mice were run to exhaustion using a motorized, speed-controlled modular treadmill system (Columbus Instruments). The treadmill was equipped with an electric shock stimulus and an adjustable inclination angle. Running velocity was set at 10 m/min for an hour, and increased by 2 m/min increments every 15 min until exhaustion was achieved.
For echocardiographic studies performed on adult mice, 2-mo-old female PGC-1β+/+ (n = 5) and PGC-1β−/− (n = 5) mice were lightly anesthetized with an intraperitoneal injection of 3% avertin (tribromoethanol, 0.01 mL/g). Cardiac ultrasound studies were performed as described previously (Rogers et al. 1999). Exercise echocardiography was performed on a motorized treadmill as described previously at a duration tolerated by the PGC-1β−/− mice (1–1.5 min) (Leone et al. 2005).
For neonatal echocardiography, male and female pups were imaged within 12 h after birth using a Vevo 770 ultrasound system (Visual Sonics, Inc.). Unanesthetized mice were placed on an imaging table under a heating lamp, and were lightly restrained in a left lateral decubitus position. Parasternal long- and short-axis images of the heart were obtained from standard echocardiographic views. Semi-apical long-axis views of the LV were used to interrogate the combined trans-mitral and LV outflow tract blood flow velocities. Basal short-axis views were used to image the pulmonary artery and the proximal portion of the descending thoracic aorta. Pulse wave Doppler sample volume was placed parallel with the direction of blood flow, and aortic diameter was measured at the same level. Cardiac output was measured as the product of aortic area and velocity time integral of the Doppler tracing. The Tei index was calculated as (IVCT + IVRT)/LVET, where IVCT is isovolumic contraction time (period between mitral valve closure and aortic valve opening), IVRT is isovolumic relaxation time (period between aortic valve closure and mitral valve opening), and LVET is the LV ejection period (time between aortic valve opening and closure).
RNA, DNA, protein, and tissue triglyceride analyses
Total RNA was isolated from various mouse tissues using the RNAzol method (Tel-Test). Northern blotting (Kelly et al. 1989) and quantitative real-time RT–PCR were performed as described (Huss et al. 2004). In brief, total RNA was isolated and reverse transcribed with Taqman reverse transcription reagents (Applied Biosystems). PCR reactions were performed in triplicate in a 96-well format using a Prism 7500 Sequence Detector (Applied Biosystems). The mouse-specific primer-probe sets used to detect specific gene expression can be found in Supplemental Table 5. Either β-actin (Applied Biosystems), 36B4, or 18s primer-probe sets (Supplemental Table 5) was included in a separate well (in triplicate) and used to normalize the gene expression data as noted in the figure legends.
Genomic/mitochondrial DNA was isolated using RNAzol, followed by back extraction with 4 M guanidine thiocyanate, 50 mM sodium citrate, and 1 M tris, and an alcohol precipitation. Mitochondrial DNA content was determined by SYBR green analysis (Applied Biosystems). To this end, the levels of NADH dehydrogenase subunit 1 (mitochondrial DNA) were normalized to the levels of lipoprotein lipase (genomic DNA). The primer sequences are noted in Supplemental Table 5.
For Southern blot analysis, 5 μg of genomic DNA was digested with SpeI, electrophoresed on a 0.8% TAE gel, and transferred to Gene Screen (Perkin Elmer) membrane for hybridization. Western blotting was performed as described (Cresci et al. 1996) using Super Signal West Dura Extended Duration Substrate (Pierce) for detection. The polyclonal PGC-1β antibody was a generous gift provided by Dr Anastasia Kralli. The PGC-1α antibody has been previously described (Lehman et al. 2000).
Total tissue triglyceride analysis was performed by the Animal Model Research Core at Washington University School of Medicine (supported by the CNRU) from frozen tissue using a modified Bligh and Dyer technique as described previously (Bligh and Dyer 1959).
Mitochondrial respiration studies
Mitochondrial respiration was assessed in isolated mitochondria from the hindlimb muscle with pyruvate as substrate as described previously (Bhattacharya et al. 1991). In brief, 3-mo-old male mice were euthanized by CO2 inhalation. The entire hindlimb was dissected from the bone and minced well, followed by a 5-min incubation in Ionic Medium (IM) plus Nagarse (100 mM sucrose, 10 mM EDTA, 100 mM Tris-HCl, 46 mM Kcl at pH 7.4, 10 mg nagarse). Samples were homogenized using an Eberbach homogenizer, centrifuged at 500g for 10 min at 4°C, and supernatant transferred to a clean tube. The supernatant was centrifuged at 12,000g and the pellet was resuspended in IM + 0.5% BSA. The sample was spun again and the pellet resuspended in Suspension Buffer (230 mM mannitol, 70 mM sucrose, 0.02 mM EDTA, 20 mM Tris-HCl, 5 mM K2HPO4 at pH 7.4). Total protein was quantified by a BCA assay (Pierce) and respiration was performed at 25°C using an optical probe (Oxygen FOXY Probe, Ocean Optics). Following measurement of basal respiration, maximal (ADP-stimulated) respiration was determined by exposing the mitochondria to 1 mM ADP. Uncoupled respiration was evaluated following addition of oligomycin (1 μg/mL). The solubility of oxygen in the respiration buffer at 25°C was taken as 246.87 nmol O2 per milliliter. Respiration rates were expressed as “nmol O2 ⋅ min−1 ⋅ mg protein−1.”
Histology and electron microscopy
Adult mice were anesthetized and perfused with Karnovsky’s fixative (2% glutaraldehyde, 1% paraformaldehyde, and 0.08% sodium cacodylate) to avoid artifact. Neonates were euthanized and hearts were fixed in Karnovsky’s fixative. Cardiac papillary muscle (adult), LV free wall (neonates), BAT, soleus, and EDL muscle were dissected and postfixed in 1% osmium tetroxide, dehydrated in graded ethanol, embedded in Poly Bed plastic resin, and sectioned for electron microscopy. Mitochondrial and myofibrillar volume densities were determined from electron micrographs as described previously (Russell et al. 2004). For each animal, three different fields at the magnification of 7500× were quantified in blinded fashion. Data were expressed as mean volume density of mitochondria or myofibrils in each field.
H&E staining was performed by the Morphology Core at Washington University School of Medicine (DDRCC). The tissues were fixed with 10% buffered formalin overnight, dehydrated in graded concentrations of alcohol, and embedded in paraffin from which 5-μm sections were prepared.
Gene expression profiling
Total RNA from cultured NRCM post adenoviral infection with either Ad-GFP, Ad-PGC-1α, or Ad-PGC-1β was reverse transcribed using Superscript II (Invitrogen Corp.) and primed with T7 promoter-polyA primer (T7T24), followed by second-strand synthesis according to the manufacturer’s protocol. Biotin-labeled cRNA was synthesized using T7-coupled ENZO BioArray High Yield RNA Transcript Labeling Kit (ENZO Diagnostics, Inc.). The Alvin J. Siteman Cancer Center’s Bioinformatics Core at Washington University School of Medicine performed hybridization to Affymetrix Rat Expression Set 230 chip. Affymetrix MAS 5.0 software was used for initial analysis and background normalization, and Z score calculation and subsequent data analysis were performed using Spotfire DecisionSite for Functional Genomics 9.0. Probe sets called “absent” by MAS 5.0 in all Ad-GFP control, Ad-PGC-1α, and Ad-PGC-1β were excluded. Two independent trials were performed. Student’s t-test was performed and a P-value <0.05 was used to determine genes significantly up-regulated. Signal intensity ratios were averaged from both trials and were calculated as either Ad-PGC-1α/Ad-GFP or Ad-PGC-1β/Ad-GFP to determine changes due to exogenous expression of either PGC-1α or PGC-1β. A gene with a calculated fold change ≥1.5 was considered an up-regulated gene target in cultured NRCM. For pathway analysis, the filtered data sets were uploaded into GenMAPP software to review the biopathways using the Gene Ontology database. GenMAPP produced a ranked list of Gene Ontology biological categories based on the following criteria: (1) at least five regulated genes in selected GO terms; (2) >50% of genes regulated in selected pathways (>50 “percent changed”). The “percent changed” was calculated by “the genes meeting the criteria/genes measured * 100.” The Z score was calculated by subtracting the expected number of genes meeting the criteria from the observed number, and then dividing by the standard deviation of the observed number of genes.
Statistical analysis
Data were analyzed using t-tests or ANOVA where appropriate. The level of significance was set at P < 0.05 in all cases. Data are reported as mean values ± the standard error of the mean, unless otherwise noted.
Acknowledgments
We thank Dr. Anastasia Kralli (Scripps) for providing the PGC-1β antibody; Juliet Fong, Michelle Trusgnich, and Alicia Wallis for help with mouse husbandry; William Kraft for expert technical assistance with electron microscopy; Mike Courtois for assistance with echocardiography; Dr. Suellen Greco, Dr. Erica Crouch, Dr. Feng Chen, and Dr. Robert Schmidt for careful analysis of the histopathology; Dr. Jeffrey Saffitz (Beth Israel Deaconess Medical Center) for consultative advice on electron microscopy; Dr. Patrick Jay for help with assessing cardiac morphology; and Mary Wingate for assistance with manuscript preparation. L.L. is supported by the AHA Fellowship award (0525743Z). C.Z. is a recipient of the Deutsche Forschungsgemeinschaft research Fellowship ZE 796/2-1. This work was supported by NIH grants RO1 DK045416, RO1 HL058427, Digestive Diseases Research Core Center (P30 DK052574), Diabetes Research and Training Center (P60 DK020579), Alvin J. Siteman Cancer Center Bioinformatics Core and Embryonic Stem Cell Core (P30 CA91842), and Clinical Nutrition Research Unit Core Center (P30 DK56341).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1661708.
References
- Agah R., Frenkel P.A., French B.A., Michael L.H., Overbeek P.A., Schneider M.D. Gene recombination in postmitotic cells. Targeted expression of cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaynick W.A., Kondo R.P., Xie W., He W., Dufour C.R., Downes M., Jonker J.W., Giles W., Naviaux R.K., Giguère V., et al. ERRγ directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Andersson U., Scarpulla R.C. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor-1-dependent transcription in mammalian cells. Mol. Cell. Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z., Novikov M., Chin S., Ma Y., Rosenzweig A., Spiegelman B.M. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPARγ coactivator 1α. Proc. Natl. Acad. Sci. 2006;103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z., Lebrasseur N., Morris C., Smith E., Yang W., Ma Y., Chin S., Spiegelman B.M. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S.K., Thakar J.H., Johnson P.L., Shanklin D.R. Isolation of skeletal muscle mitochonria using an ionic medium containing ethylenediaminetetraacetic acid and nagarse. Anal. Biochem. 1991;192:344–349. doi: 10.1016/0003-2697(91)90546-6. [DOI] [PubMed] [Google Scholar]
- Bing R.J. The metabolism of the heart. Harvey Lect. 1955;50:27–70. [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Buroker N.E., Ning X.-H., Portman M. Cardiac PPARα protein expression is constant as alternate nuclear receptors and PGC-1 coordinately increase during the postnatal metabolic transition. PPAR Research. 2008;2008:279531. doi: 10.1155/2008/279531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresci S., Wright L.D., Spratt J.A., Briggs F.N., Kelly D.P. Activation of a novel metabolic gene regulatory pathway by chronic stimulation of skeletal muscle. Am. J. Physiol. 1996;270:C1413–C1420. doi: 10.1152/ajpcell.1996.270.5.C1413. [DOI] [PubMed] [Google Scholar]
- Dufour C.R., Wilson B.J., Huss J.M., Kelly D.P., Alaynick W.A., Downes M., Evans R.M., Blanchette M., Giguère V. Genome-wide orchestration of cardiac functions by orphan nuclear receptors ERRα and γ. Cell Metabolism. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Finck B.N., Kelly D.P. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D.J., Heymann M.A., Rudolph A.M. Myocardial oxygen and carbohydrate consumption in fetal lambs in utero and in adult sheep. Am. J. Phys. 1980;238:H399–H405. doi: 10.1152/ajpheart.1980.238.3.H399. [DOI] [PubMed] [Google Scholar]
- Fisher D.J., Heymann M.A., Rudolph A.M. Myocardial consumption of oxygen and carbohydrates in newborn sheep. Pediatr. Res. 1981;15:843–846. doi: 10.1203/00006450-198105000-00003. [DOI] [PubMed] [Google Scholar]
- Girard J., Ferré P., Pégorier J.P., Duée P.H. Adaptations of glucose and fatty acid metabolism during perinatal period and suckling–weaning transition. Physiol. Rev. 1992;72:507–562. doi: 10.1152/physrev.1992.72.2.507. [DOI] [PubMed] [Google Scholar]
- Hallman M. Changes in mitochondrial respiratory chain proteins during perinatal development. Evidence of the importance of environmental oxygen tension. Biochim. Biophys. Acta. 1971;253:360–372. doi: 10.1016/0005-2728(71)90040-5. [DOI] [PubMed] [Google Scholar]
- Handschin C., Spiegelman B.M. Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- Herzig S., Long F., Jhala U.S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Huss J.M., Kopp R.P., Kelly D.P. PGC-1α coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. J. Biol. Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- Huss J.M., Pinéda Torra I., Staels B., Giguère V., Kelly D.P. ERRα directs PPARα signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoi T., Lopaschuk G.D. The contribution of glycolysis, glucose oxidation, lactate oxidation, and fatty acid oxidation to ATP production in isolated biventricular working hearts from 2-week-old rabbits. Pediatr. Res. 1993;34:735–741. doi: 10.1203/00006450-199312000-00008. [DOI] [PubMed] [Google Scholar]
- Kamei Y., Ohizumi H., Fujitani Y., Nemoto T., Tanaka T., Takahashi N., Kawada T., Miyoshi M., Ezaki O., Kakizuka A. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D.P., Strauss A.W. Inherited cardiomyopathies. N. Engl. J. Med. 1994;330:913–919. doi: 10.1056/NEJM199403313301308. [DOI] [PubMed] [Google Scholar]
- Kelly D.P., Gordon J.I., Alpers R., Strauss A.W. The tissue-specific expression and developmental regulation of the two nuclear genes encoding rat mitochondrial proteins: Medium-chain acyl-CoA dehydrogenase and mitochondrial malate dehydrogenase. J. Biol. Chem. 1989;264:18921–18925. [PubMed] [Google Scholar]
- Koo S.H., Satoh H., Herzig S., Lee C.H., Hedrick S., Kulkarni R., Evans R.M., Olefsky J., Montminy M. PGC-1 promotes insulin resistance in liver through PPARα-dependent induction of TRB-3. Nat. Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- Kressler D., Schreiber S.N., Knutti D., Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor α. J. Biol. Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- Lehman J.J., Kelly D.P. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin. Exp. Pharmacol. Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- Lehman J.J., Barger P.M., Kovacs A., Saffitz J.E., Medeiros D., Kelly D.P. PPARγ coactivator-1 (PGC-1) promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelliott C.J., Medina-Gomez G., Petrovic N., Kis A., Feldmann H.M., Bjursell M., Parker N., Curtis K., Campbell M., Hu P., et al. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:2042–2056. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone T.C., Lehman J.J., Finck B.N., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., et al. PGC-1α deficient mice exhibit multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control, and hepatic steatosis. PLoS Biol. 2005;3:672–687. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B.M. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002a;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotanni E., Olson E.N., et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibers. Nature. 2002b;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lin J., Wu P.-H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.-Y., Mootha V.K., Jãger S., Vianna C.R., Reznick R.M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lin J., Yang R., Tarr P.T., Wu P.-H., Handschin C., Li S., Yang W., Pei L., Uldry M., Tontonoz P., et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Marin-Garcia J., Ananthakrishnan R., Goldenthal M.J. Heart mitochondrial DNA and enzyme changes during early human development. Mol. Cell. Biochem. 2000;210:47–52. doi: 10.1023/a:1007031919298. [DOI] [PubMed] [Google Scholar]
- Michael L.F., Wu Z., Cheatham R.B., Puigserver P., Adelmant G., Lehman J.J., Kelly D.P., Spiegelman B.M. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen O.H., Frandsen L., Schjerling P., Nishimura E., Grunnet N. PGC-1α and PGC-1β have both similar and distinct effects on myofiber switching toward an oxidative phenotype. Am. J. Phys. Endocrinol. Metab. 2006;4:E807–E816. doi: 10.1152/ajpendo.00591.2005. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Spiegelman B.M. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): Transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Adelmant G., Wu Z., Fan M., Xu J., O'Malley B., Spiegelman B.M. Activation of PPARγ coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Rhee J., Donovan J., Walkey C.J., Yoon J.C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Rogers J.H., Tamirisa P., Kovacs A., Weinheimer C., Courtois M., Blumer K.J., Kelly D.P., Muslin A.J. RGS4 causes increased mortality and reduced cardiac hypertrophy in response to pressure overload. J. Clin. Invest. 1999;104:567–576. doi: 10.1172/JCI6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L.K., Mansfield C.M., Lehman J.J., Kovacs A., Courtois M., Saffitz J.E., Medeiros D.M., Valencik M.L., McDonald J.A., Kelly D.P. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ. Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- Sano M., Tokudome S., Shimizu N., Yoshikawa N., Ogawa C., Shirakawa K., Endo J., Katayama T., Yuasa S., Ieda M., et al. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor γ coactivator 1α. J. Biol. Chem. 2007;282:25970–25980. doi: 10.1074/jbc.M703634200. [DOI] [PubMed] [Google Scholar]
- Schreiber S.N., Knutti D., Brogli K., Uhlmann T., Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα) J. Biol. Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- Smolich J.J., Walker A.M., Campbell G.R., Adamson T.M. Left and right ventricular myocardial morphometry in fetal, neonatal, and adult sheep. Am. J. Phys. 1989;257:1–9. doi: 10.1152/ajpheart.1989.257.1.H1. [DOI] [PubMed] [Google Scholar]
- Sonoda J., Mehl I.R., Chong L.W., Nofsinger R.R., Evans R.M. PGC-1β controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc. Natl. Acad. Sci. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J., Lin J., Krauss S., Tarr P.T., Yang R., Newgard C.B., Spiegelman B.M. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- Taha M., Lopaschuk G.D. Alterations in energy metabolism in cardiomyopathies. Ann. Med. 2007;39:594–607. doi: 10.1080/07853890701618305. [DOI] [PubMed] [Google Scholar]
- Tei C., Ling L.H., Hodge D.O., Bailey K.R., Oh J.K., Rodeheffer R.J., Tajik A.J., Seward J.B. New index of combined systolic and diastolic myocardial performance: A simple and reproducible measure of cardiac function-a study in normals and dilated cardiomyopathy. J. Cardiol. 1995;26:357–366. [PubMed] [Google Scholar]
- Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Vega R.B., Huss J.M., Kelly D.P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianna C.R., Huntgeburth M., Coppari R., Choi C.S., Lin J., Krauss S., Barbatelli G., Tzameli I., Kim Y.B., Cinti S., et al. Hypomorphic mutation of PGC-1β causes mitochondrial dysfunction and liver insulin resistance. Cell Metab. 2006;4:453–464. doi: 10.1016/j.cmet.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A.E., Yamamura S., Malik S., Spiegelman B.M., Roeder R.G. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol. Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- Wende A.R., Schaeffer P.J., Parker G.J., Zechner C., Han D.H., Chen M.M., Hancock C.R., Lehman J.J., Huss J.M., McClain D.A., et al. A role for the transcriptional coactivator PGC-1α in muscle refueling. J. Biol. Chem. 2007;282:36642–36651. doi: 10.1074/jbc.M707006200. [DOI] [PubMed] [Google Scholar]
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Yoon J.C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C.R., Granner D.K., et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]