Abstract
The YAP transcription coactivator has been implicated as an oncogene and is amplified in human cancers. Recent studies have established that YAP is phosphorylated and inhibited by the Hippo tumor suppressor pathway. Here we demonstrate that the TEAD family transcription factors are essential in mediating YAP-dependent gene expression. TEAD is also required for YAP-induced cell growth, oncogenic transformation, and epithelial–mesenchymal transition. CTGF is identified as a direct YAP target gene important for cell growth. Moreover, the functional relationship between YAP and TEAD is conserved in Drosophila Yki (the YAP homolog) and Scalloped (the TEAD homolog). Our study reveals TEAD as a new component in the Hippo pathway playing essential roles in mediating biological functions of YAP.
Keywords: TEAD, YAP, CTGF, Hippo, transcription, cancer
Recent genetic studies in Drosophila have identified a novel tumor suppressor pathway, the Hippo pathway (Harvey et al. 2003; Wu et al. 2003; Lai et al. 2005; Edgar 2006; Hariharan and Bilder 2006; Harvey and Tapon 2007). Genetic experiments demonstrated that the Yki transcription coactivator is inhibited by the Hippo pathway (Huang et al. 2005). Consistently, biochemical studies showed that Yki is directly phosphorylated and inhibited by the Wts protein kinase, which is phosphorylated and activated by the Hippo (Hpo) protein kinase (Dong et al. 2007). Yki induces expression of genes like cyclin E and Diap1, and therefore promotes proliferation and inhibits apoptosis (Udan et al. 2003; Huang et al. 2005). However, Yki does not have a DNA-binding domain, and therefore must interact with a DNA-binding transcription factor(s) to regulate gene expression. Scalloped (Sd), a transcription factor in Drosophila, has been reported recently to act downstream from Yki (Goulev et al. 2008; Wu et al. 2008; Zhang et al. 2008).
Components of the Hippo pathway are highly conserved, and recent studies from us and other groups have demonstrated the function of the Hippo pathway in mammalian cell growth (Hao et al. 2007; Zhao et al. 2007). YAP, the human homolog of Yki, is phosphorylated by the Lats tumor suppressor, which is a homolog of Drosophila Wts. Phosphorylation of YAP by Lats results in cytoplasmic translocation and, therefore, inactivation of YAP. This mechanism of YAP regulation is involved in cell contact inhibition and tissue growth control (Zhao et al. 2007).
The importance of the Hippo pathway in human cancer was gradually uncovered. Mutation of the Hippo pathway components, such as the NF2 tumor suppressor, is known to contribute to human tumorigenesis (McClatchey and Giovannini 2005). More importantly, YAP is the candidate oncogene in the human chromosome 11q22 amplicon, which is evident in several human cancers (Overholtzer et al. 2006; Zender et al. 2006). YAP overexpression stimulates proliferation and increases saturation cell density in monolayer culture of NIH-3T3 cells (Zhao et al. 2007). Furthermore, YAP overexpression in MCF10A cells induces epithelial– mesenchymal transition (EMT), which is a hallmark of tumorigenic transformation (Overholtzer et al. 2006). Moreover, elevated YAP protein levels and increased nuclear localization have been observed in multiple human cancer tissues (Zhao et al. 2007). Interestingly, YAP overexpression causes a dramatic increase in liver size and eventually leads to tumor growth (Camargo et al. 2007; Dong et al. 2007). These observations have established the importance of the Hippo pathway in human cancer.
Several transcription factors, including ErbB4, Runx2, TEAD, and p73, have been reported to interact with YAP (Yagi et al. 1999; Vassilev et al. 2001; Basu et al. 2003; Komuro et al. 2003). However, the significance of these transcription factors in mediating the biological functions of YAP, especially in promoting cell growth, has not been demonstrated. In this study, we identified TEAD as the most potent YAP target from a transcription activity-based screen. By means of dominant-negative or RNAi, we further showed that TEAD is required for YAP to stimulate gene expression, cell growth, anchorage-independent growth, and EMT. We identified the connective tissue growth factor (CTGF) as a direct target gene of YAP and TEAD. Interestingly, knockdown of CTGF blocks YAP-stimulated cell growth and significantly reduces YAP-induced colony formation in soft agar. Furthermore, experiments in Drosophila demonstrated that Sd and Yki genetically interact to enhance tissue growth and organ size. Together, our observations establish TEAD as the key transcription factor in the Hippo pathway acting downstream from YAP.
Results
TEAD mediates YAP-dependent gene induction
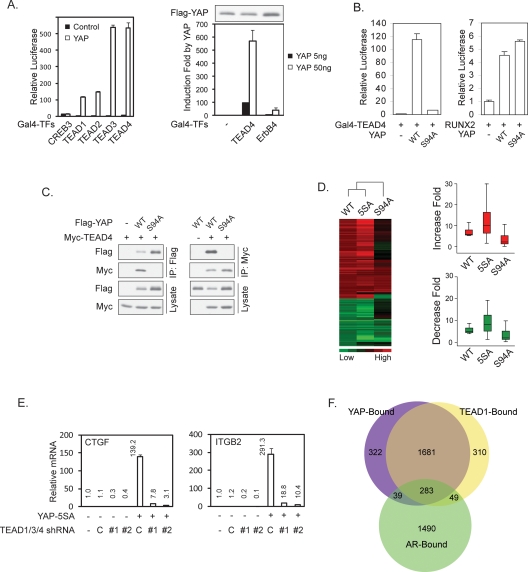
To identify YAP target transcription factors, we screened a human transcription factor library in which the known or putative transcription factors were fused to Gal4 DNA-binding domain. Clones of the Gal4-TF library (a total of 1100) (J.D. Lin, unpubl.) were individually cotransfected with a 5× UAS-luciferase reporter, which is driven by five Gal4-binding elements, in the presence or absence of YAP cotransfection. This unbiased strategy identified TEAD2, TEAD3, and TEAD4 as the strongest positives based on the transcription reporter assay. The human genome contains four TEAD transcription factors. TEAD1 was not present in our Gal4-TF library, but it could also be potently activated by YAP (Fig. 1A). Several other transcription factors, including ErbB4 and RUNX2, have been reported to interact with YAP (Yagi et al. 1999; Komuro et al. 2003). However, the activation of ErbB4 by YAP is much weaker than that of TEAD (Fig. 1A). Furthermore, YAP showed a strong physical interaction with TEAD but little interaction with RUNX2 (data not shown). These data indicate that the TEADs may represent the major target transcription factors of YAP.
Figure 1.
TEAD is required for YAP-induced gene expression. (A) YAP potently activates TEAD family transcription factors. The indicated Gal4-fused transcription factors were cotransfected with a 5× UAS-Luc reporter and a CMV-β-gal construct into 293T cells in the presence or absence of YAP. The β-galactosidase activity normalized luciferase activity in the absence of YAP (Gal4-TEAD1 in the absence of YAP in the left panel) was set to 1. Flag-YAP Western blot shows that the YAP expression level was not decreased by ErbB4. (B) YAP-S94A cannot activate TEAD4. The indicated plasmids were cotransfected with a 5× UAS-luciferase reporter for Gal4-TEAD4 or a 6× OSE2-luciferase reporter for RUNX2 into 293T cells. Luciferase activity was measured and normalized to cotransfected β-galactosidase. (C) Serine 94 of YAP is required for its interaction with TEAD4. The indicated plasmids were transfected into HEK293 cells. Flag-YAP (left panel) or Myc-TEAD4 (right panel) was immunoprecipitated, and the immunoprecipitates were probed as indicated. D. YAP-S94A is defective in gene expression regulation. The left panel shows cluster analysis of gene expression profiles in YAP-WT, 5SA, or S94A-overexpressing MCF10A cells. The group of genes presented was chosen by the following standard: a P call in all samples and up-regulated more than fivefold or down-regulated more than fourfold by YAP-wild-type overexpression. Cluster analysis was done with Eisen Lab Cluster software using average linkage clustering. (Right panel) The same data sets were drawn into boxplots using the R program. Red and green indicate up-regulated and down-regulated genes, respectively. (E) TEAD is required for YAP-induced expression of CTGF and ITGB2. The indicated shRNAs were infected into native or YAP-5SA-expressing MCF10A cells. Expression of CTGF and ITGB2 was determined by quantitative RT–PCR and compared to vector control cells. (C) Scramble shRNA control; (#1 and #2) two different shRNAs targeting TEAD1/3/4. (F) YAP and TEAD1 occupy common promoters. ChIP-on-chip was performed with YAP or TEAD1 antibody against endogenous proteins in MCF10A cells. Genome-wide location analysis was performed. AR ChIP was included as a negative control.
By point mutation scanning, we found that the YAP Ser 94 to alanine (S94A) mutant was defective in TEAD4 activation (Fig. 1B) as well as other TEADs activation (data not shown). However, YAP-S94A retains full potential to activate RUNX2 (Fig. 1B) and ErbB4 (data not shown). This indicates that mutation of YAP S94 selectively abolishes its ability to activate TEAD but does not impair its general transcriptional activity. Consistently, we observed that YAP-S94A lost its ability to physically interact with TEAD4 (Fig. 1C) and other TEADs (data not shown).
To assess the importance of TEAD interaction in YAP-induced gene expression, we established MCF10A stable pools with expression of YAP, constitutively active YAP-5SA (Zhao et al. 2007), and YAP-S94A. Gene expression profiles were determined by microarray (Supplemental Table S1). Our data showed that YAP-5SA caused a stronger induction of YAP-inducible genes than the wild-type YAP (Fig. 1D). Interestingly, YAP-S94A was severely compromised in gene regulation (both induction and repression) (Fig. 1D; Supplemental Table S1). We reported previously that YAP regulates gene expression in NIH-3T3 cells (Zhao et al. 2007). Comparing the data from NIH-3T3 and MCF10A cells by Gene Set Enrichment Analysis (GSEA) (Subramanian et al. 2005), we found a significant overlap of gene profiles between the two cell lines (Supplemental Fig. S1A). The majority of genes that are affected by YAP expression are similarly regulated (either up or down) in both NIH-3T3 and MCF10A cells (Supplemental Table S2), while a subset of genes is oppositely regulated in NIH-3T3 and MCF10A cells (Supplemental Table S2).
Among the confirmed YAP-inducible genes in MCF10A were CTGF and ITGB2 (integrin β 2). They were strongly induced by YAP-5SA but not by YAP-S94A (Supplemental Fig. S1B). Furthermore, coexpression of the dominant-negative TEAD1-ΔC, which has a deletion of the C-terminal YAP-binding domain, blocked the induction of both CTGF and ITGB2 (Supplemental Fig. S1B). The four TEAD family members are all expressed in MCF10A cells, while TEAD1 has the highest expression (data not shown). We generated lentiviral constructs with shRNAs designed in a region identical in TEAD1, TEAD3, and TEAD4. Indeed, these shRNAs were able to knock down TEAD1, TEAD3, and TEAD4 concurrently but not TEAD2 (Supplemental Fig. S1C). Nevertheless, these TEAD1/3/4 shRNAs strongly blocked the induction of CTGF and ITGB2 by YAP-5SA expression (Fig. 1E). These data demonstrate that in MCF10A cells, the TEAD1/3/4 transcription factors play a critical role in the expression of YAP-dependent genes.
If TEAD plays a major role in YAP-regulated gene expression, they should occupy a similar set of gene promoters. We performed genome-wide location analysis of YAP and TEAD1 occupancy in MCF10A cells by chromatin immunoprecipitation (ChIP)-on-chip experiments. Interestingly, our results demonstrated that YAP and TEAD1 co-occupy >80% of the promoters pulled down by either of them (Fig. 1F; raw data in Supplemental Table S3). The Androgen Receptor (AR)-associated genes were included as a control, which showed a much lesser degree of overlap with those occupied by YAP compared with TEAD1 (odds ratio = 34.6, P < 0.00001). This observation further supports that the overlap between YAP and TEAD1 targets is not a random event. Gene Set Enrichment Analysis (GSEA) demonstrated that a significant (P < 0.001) portion of YAP-bound genes are differentially expressed upon YAP overexpression in MCF10A cells. Since YAP does not have DNA-binding activity, these data strongly indicate that TEAD plays a major role in mediating the binding of YAP to gene promoters.
TEAD binding is required for YAP-induced cell growth and EMT
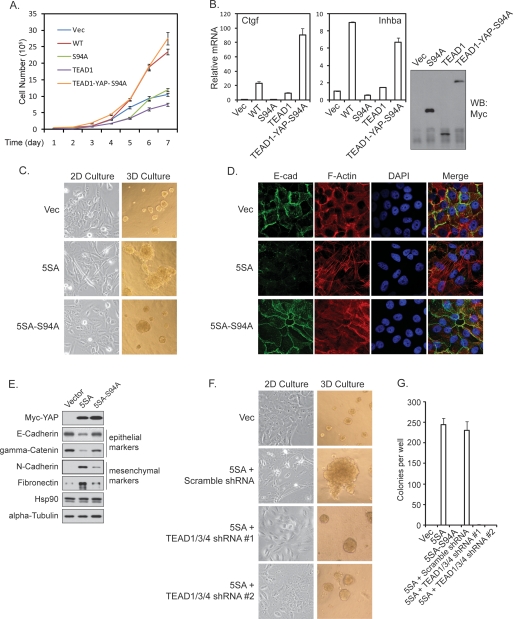
We reported that YAP expression in NIH-3T3 cells enhances cell growth (Zhao et al. 2007). NIH-3T3 stable pools with expression of YAP and YAP-S94A were established, and cell growth was determined. We found that YAP-S94A was much less potent than the wild-type YAP to stimulate NIH-3T3 cell growth (Fig. 2A). Furthermore, in MCF10A cells, wild-type YAP induced cell proliferation even when cells reached confluency, while the YAP-S94A mutant was largely inactive as determined by the staining of proliferation marker Ki67 (Supplemental Fig. S2A). To confirm that the loss of growth-promoting activity in YAP-S94A is due to the loss of its interaction with TEAD, we generated a TEAD1-YAP-S94A fusion protein. Interestingly, this fusion protein stimulated NIH-3T3 cell growth as effectively as the wild-type YAP, while neither TEAD1 nor YAP-S94A stimulated cell growth (Fig. 2A). Furthermore, the TEAD1-YAP-S94A fusion also rescued the expression of Ctgf and Inhba, two YAP target genes, in NIH-3T3 cells (Fig. 2B). We also examined the effect of S94A mutation in the constitutively active YAP-5SA background in MCF10A cells. Expression of YAP-5SA resulted in the formation of much larger acini in three-dimensional (3D) culture compared with vector control. Importantly, this effect was largely reduced if an S94A mutation was introduced into YAP-5SA (Fig. 2C). These results indicate that S94, hence TEAD binding, is required for YAP-induced cell proliferation.
Figure 2.
TEAD is required for YAP activity in growth promotion and EMT. (A) YAP-S94A is defective in promoting cell growth. The growth curve of NIH-3T3 stable cells with expression of Vector, YAP, YAP-S94A, TEAD1, or TEAD1-YAP-S94A was determined. (B) Fusion of YAP-S94A with TEAD1 rescued YAP target gene expression. (Right panel) NIH-3T3 stable cells with expression of YAP-S94A, TEAD1, and TEAD1-YAP-S94A fusion protein were generated, and the expression of these proteins was shown by anti-Myc-tag Western blot. The expression of Ctgf and Inhba, two YAP target genes in NIH-3T3 cells, was measured by quantitative PCR. The induction of these two genes by YAP-WT was also shown for comparison. (C) YAP-5SA-S94A is compromised in eliciting EMT-like morphology. Indicated MCF10A stable cells were cultured in monolayer or in 3D on reconstituted basement membrane for 16 d before pictures were taken. (D) YAP-5SA-S94A is defective in reducing membrane E-cadherin and cortical actin. The indicated MCF10A stable cells were stained by anti-E-cadherin (green), rhodamine-phalloidin (red), and DAPI (blue). (E) The TEAD-binding-defective YAP is compromised in altering EMT marker expression. Western blot of epithelial and mesenchymal markers was performed using lysates from indicated MCF10A stable cells. (F) TEAD1/3/4 shRNAs blocked YAP induced EMT-like morphology and acinar overgrowth. YAP-5SA-expressing MCF10A cells were infected with indicated shRNA lentiviruses. The morphology in 2D and 3D culture was documented as in C. (G) TEAD1/3/4 shRNAs blocked YAP-induced anchorage-independent growth in soft agar. The indicated MCF10A stable cells were plated in soft agar and allowed to grow for 3 wk, after which colonies were stained with crystal violet and counted.
It has been reported that YAP induces EMT in MCF10A cells (Overholtzer et al. 2006). Indeed, expression of the active YAP-5SA induced EMT-like morphological change in monolayer culture (Fig. 2C). However, YAP-5SA-S94A was not effective in eliciting EMT morphology. Furthermore, in 3D culture, YAP-5SA-S94A failed to induce complex-shaped large acini with spike-like projections and rough surface, which were obvious in YAP-5SA-expressing cultures (Fig. 2C). As another hallmark of EMT, YAP-5SA-expressing cells also displayed disorganized adherens junctions, as shown by the loss of cell–cell junction localized E-cadherin, and the switch from cortical actin to stress fibers (Fig. 2D). However, these phenotypes were not seen in YAP-5SA-S94A-expressing cells. YAP-5SA expression also changed the expression pattern of epithelial and mesenchymal markers, which was not induced by YAP-5SA-S94A expression (Fig. 2E). These results indicate that S94 of YAP, presumably by mediating TEAD interaction, is at least partially responsible for YAP function in inducing EMT.
To further confirm the function of TEAD, we used shRNAs to knock down TEAD1/3/4 in YAP-5SA-expressing cells. TEAD1/3/4 knockdown not only reversed the EMT-like morphology in monolayer and 3D cultures, but also rescued the expression of epithelial markers (Fig. 2F; Supplemental Fig. S2B). Knockdown of TEAD1/3/4 also significantly shrank the aberrantly enlarged acini caused by YAP-5SA expression, further supporting a role of TEAD in YAP-induced growth. A YAP-dependent function of TEAD in cell growth is also implicated in Sveinsson’s chorioretinal atrophy, a rare genetic disease caused by TEAD1 mutation and characterized by atrophic lesions involving retina and choroids (Fossdal et al. 2004; Kitagawa 2007). The mutated tyrosine Y406 is highly conserved in TEAD family members (Supplemental Fig. S2C), and is located within the YAP-binding domain (Supplemental Fig. S2D). Interestingly, mutation of this tyrosine residue in TEADs abolished their interaction with and their activation by YAP (Supplemental Fig. S2E–G), which may explain the atrophic phenotype caused by this mutation.
Anchorage-independent growth is a hallmark of oncogenic transformation. YAP overexpression is reported to induce anchorage-independent growth of MCF10A cells (Overholtzer et al. 2006). We observed that YAP-5SA potently induced MCF10A colony formation in soft agar. In contrast, YAP-5SA-S94A was unable to induce anchorage-independent growth of MCF10A cells (Fig. 2G; Supplemental Fig. S2H). Similarly, almost no colony was formed if TEAD1/3/4 were down-regulated in the YAP-5SA expressing cells (Fig. 2G; Supplemental Fig. S2H). These data indicate the requirement of at least one of TEAD1/3/4 for the YAP-induced anchorage-independent growth. Together, the above observations support a model in which TEAD is essential for the function of YAP in cell proliferation, EMT, and oncogenic transformation.
CTGF is a direct YAP-TEAD target gene required for cell growth
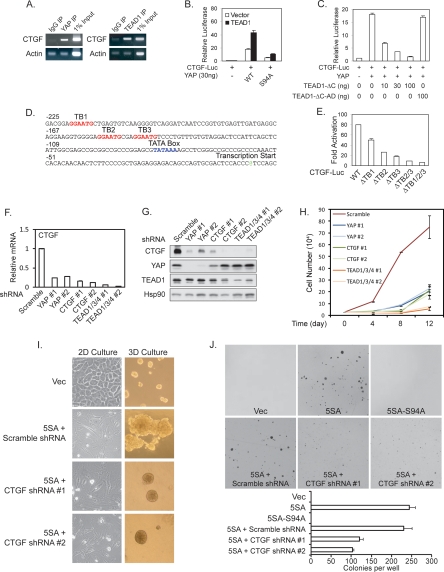
YAP expression affected many cell proliferation-related genes (Supplemental Table S1). However, cyclin E and IAP, the key Yki-inducible genes in Drosophila, were not significantly induced by YAP in either NIH-3T3 or MCF10A cells (Supplemental Table S1). This indicates that there might be different genes in mammalian cells to mediate YAP function. CTGF is highly induced by YAP expression in both NIH-3T3 and MCF10A cells, and its promoter is co-occupied by YAP and TEAD1, as shown by ChIP (Fig. 3A); therefore, it might be a direct YAP target gene. We cloned the CTGF promoter into a basic luciferase reporter and found that it was potently activated by YAP but not by YAP-S94A, and the activation was further enhanced by TEAD1 coexpression (Fig. 3B). Expression of the dominant-negative TEAD1-ΔC, but not the TEAD1-ΔC-AD (in which the C-terminal YAP-binding domain was replaced by the YAP transactivation domain), blocked the activation of CTGF reporter by YAP (Fig. 3C). These results indicate that YAP activates the CTGF promoter through TEAD. Examination of the CTGF promoter region revealed three putative TEAD-binding sites (Fig. 3D; Anbanandam et al. 2006). Individual or combinatory mutation of the putative TEAD-binding sites indicated that TB2 and TB3 were more important for CTGF promoter activity while TB1 was also involved (Fig. 3E).
Figure 3.
CTGF is a direct target of YAP and TEAD. (A) Both YAP and TEAD1 bind to the CTGF promoter. ChIP from MCF10A cells was performed with control IgG, YAP, or TEAD1 antibody as indicated. The presence of CTGF promoter was detected by PCR. (B) Activation of CTGF reporter by YAP and TEAD1. A luciferase reporter driven by CTGF promoter was cotransfected with YAP wild type or S94A mutant as indicated with or without TEAD1 cotransfection. Luciferase activity was measured and normalized to cotransfected β-galactosidase. (C) Dominant-negative TEAD1 blocks the YAP stimulation of the CTGF reporter. The indicated plasmids were cotransfected, and luciferase activity was determined as in B. (D) The human CTGF promoter region contains three putative TEAD-binding sites. The putative TEAD-binding sites (TB1–TB3) are shown in red. (E) The putative TEAD-binding sites are important for CTGF promoter activity. The putative TEAD-binding sites (TB) were mutated individually or in combination. The luciferase activity of each reporter was measured in the presence or absence of YAP and TEAD1. The activation folds by YAP and TEAD1 are shown. (F) YAP and TEAD are required for CTGF expression. ACHN cells were infected with the indicated shRNA lentiviruses, and CTGF mRNA levels were determined by quantitative RT–PCR. (G) Knockdown of YAP or TEAD1/3/4 decreases CTGF protein levels. Experiments were similar to F except Western blotting was performed with the indicated antibodies. (H) YAP, TEAD, and CTGF are important for the AHCN cell growth. YAP, TEAD1/3/4, and CTGF were knocked down by shRNAs. Cell growth rate was determined. (I) CTGF is required for YAP-induced growth and morphological change in 3D culture. MCF10A cells expressing YAP-5SA were infected with indicated shRNA lentiviruses. The morphology in 2D and 3D culture was documented as in Figure 2C. (J) CTGF knockdown attenuates YAP induced anchorage-independent growth in soft agar. The indicated MCF10A stable cells were plated in soft agar and allowed to grow for 3 wk, after which colonies were stained with crystal violet and counted. Pictures of the stained colonies were presented in higher magnification to show the colony size reduction by CTGF shRNAs.
The function of endogenous YAP and TEAD in CTGF expression was examined by YAP or TEAD1/3/4 knockdown in ACHN cells, which have elevated YAP activity due to a mutation of Sav, a key component of the Hippo pathway (Tapon et al. 2002). RNAi specificity and efficiency were confirmed by quantitative RT–PCR (Supplemental Fig. S3A) and Western blot (Fig. 3G). We found that knockdown of either YAP or TEAD1/3/4 caused a dramatic reduction of both CTGF mRNA (Fig. 3F) and protein (Fig. 3G). We next examined the function of CTGF in mediating the cellular function of YAP. Similar to the knockdown of YAP and TEAD1/3/4, knockdown of CTGF significantly inhibited ACHN cell growth (Fig. 3H). These data further demonstrate the functional significance of TEAD1/3/4 and CTGF as important downstream targets of YAP in the Hippo pathway in cell growth regulation. Furthermore, knockdown of CTGF in the YAP-5SA-expressing MCF10A cells decreased the acini growth and reversed the complex-shaped and rough surface morphology in 3D culture (Fig. 3I). However, CTGF knockdown did not reverse the EMT-like morphology in monolayer culture. These results indicate that CTGF plays an important role in the growth-promoting function but may not be required for the EMT-inducing activity of YAP.
We also tested the effect of CTGF knockdown in the anchorage-independent growth potential of YAP-5SA-overexpressing MCF10A cells. Although CTGF knockdown did not completely block the anchorage-independent growth of YAP-5SA-overexpressing MCF10A cells, it significantly decreased the number of colonies formed (Fig. 3J; Supplemental Fig. S3B) and dramatically reduced the colony size (Fig. 3J). However, expression of CTGF alone did not phenocopy the effects of YAP overexpression in MCF10A cells (data not shown). Therefore, we speculate that CTGF works with other YAP target genes to mediate the oncogenic transformation potential of YAP.
YAP/Yki and TEAD/Sd genetically interact to promote tissue growth in Drosophila
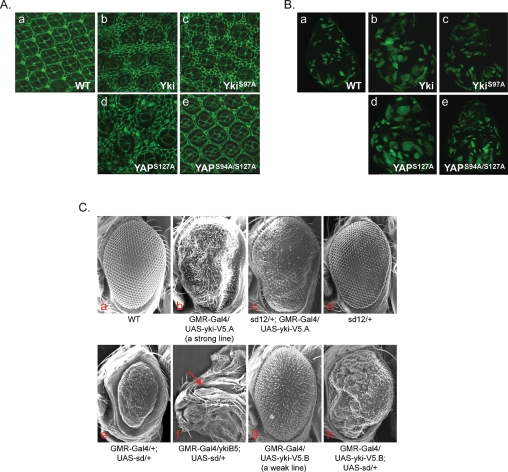
To investigate the function of TEAD in YAP-induced growth control, we generated transgenic flies that express human YAP-S127A (an active form) or YAP-S94A/S127A in developing eyes. YAP-S127A overexpression significantly increased eye size (Supplemental Fig. S4A, panels a,d) and the number of interommatidial cells (Fig. 4A, panels a,d). Mutation of S94A dramatically decreased the activity of YAP-S127A in promoting tissue growth (Fig. 4A, panel e; Supplemental Fig. S4A, panel e). Scalloped (Sd) is the only TEAD homolog in Drosophila. We found that Yki directly interacted with Sd in an in vitro binding assay (Supplemental Fig. S4B). Furthermore, Yki S97A mutation (equivalent to YAP-S94A) diminished its interaction with Sd. Moreover, this Sd-binding-defective Yki-S97A mutant was less potent in stimulating growth in vivo compared with wild-type Yki (Fig. 4A, panels a–c; Supplemental Fig. S4A, panels a–c). The functional defect of the TEAD-binding-deficient YAP/Yki was further confirmed by generating overexpression flip-out clones in the Drosophila larval wing discs as labeled by positive GFP expression (Fig. 4B). Both YAP-S127A and Yki are potent in stimulating tissue growth as individual clones, and the whole discs were generally larger than wild-type clones or discs (Fig. 4B, panels a,b,d). However, neither YAP-S94A/S127A nor Yki-S97A showed a similar level of growth-promoting effect (Fig. 4B, panels c,e). These data indicate that TEAD/Sd binding is important for the physiological function of YAP/Yki.
Figure 4.
yki and scalloped genetically interact to control tissue growth and organ size. (A) The TEAD/Sd-binding-defective YAP and Yki are compromised in inducing extra interommatidial cells. Mid-pupal eye discs were stained with Discs large (Dlg) antibody to outline cells. The genotypes of the fly tissues are Wild-type (Canton S) (panel a), GMR-Gal4/UAS-yki-V5 (panel b), GMR-Gal4/UAS- ykiS97A-V5 (panel c), GMR-Gal4/UAS-Flag-YAPS127A (panel d), and GMR-Gal4/UAS-Flag-YAPS94A/S127A (panel e). (B) The TEAD/Sd-binding-defective YAP and Yki are compromised in inducing clone expansion. Wing imaginal discs containing 72-h-old control (panel a) or various YAP/Yki-overexpressing clones (panels b–e) were generated by flip-out and positively marked by GFP. The genotypes of the fly tissues are hsFLP/+; act>y+>Gal4, UAS-GFPS65T/+ (panel a), hsFLP/+; act>y+>Gal4, UAS-GFPS65T/UAS-yki-V5 (panel b), hsFLP/+; act>y+>Gal4, UAS-GFPS65T/UAS-ykiS97A-V5 (panel c), hsFLP/+; act>y+>Gal4, UAS-GFPS65T/UAS-Flag-YAPS127A (panel d), and hsFLP/+; act>y+>Gal4, UAS-GFPS65T/UAS-Flag-YAPS94A/S127A (panel e). (C) yki and scalloped genetically interact to control tissue growth and organ size. Genotypes of the fly tissues are indicated. SEM (scanning electron microscopy) images of adult eyes are shown in panels a–e, g, and h. A late pupal head is shown in panel f. The arrow in panel f indicates where a retina is normally expected to grow.
We next tested the genetic interaction between Yki and Sd. A strong loss-of-function allele of sd dominantly suppressed the enlarged and rough eye phenotypes caused by Yki overexpression (Fig. 4C, panels a–d). Thus, the level of Sd is critical for Yki to promote tissue growth. Overexpression of Sd caused small eyes (Fig. 4C, panel e), presumably due to a dominant-negative effect (Simmonds et al. 1998), but it did not result in lethality. This phenotype was strongly enhanced by reduction of yki levels, such that all of these flies died at the late pupal stage and had no eyes (Fig. 4C, panel f). Furthermore, coexpression of Yki with Sd suppressed the reduced eye phenotype caused by Sd overexpression (Fig. 4C, panels e,g,h). In fact, the eyes of animals overexpressing both Yki and Sd were enlarged more than those of animals that only expressed Yki. Therefore, Sd overexpression enhanced the Yki overexpression phenotypes. Together, these results indicate that Sd is a critical functional partner of Yki, a conclusion consistent with TEAD as a critical downstream target transcription factor of YAP.
Discussion
The Hippo pathway plays an important role in the regulation of cell and tissue growth (Saucedo and Edgar 2007). Dysregulation of this pathway, such as mutations in NF2, leads to human cancer (McClatchey et al. 1998). Acting at the end of the Hippo pathway is the YAP transcription coactivator, which is an oncogene capable of promoting cell growth, oncogenic transformation, and EMT in cultured cells. YAP overexpression increases organ size and causes cancer in transgenic mice (Dong et al. 2007). An important open question in the field is the transcription factor(s) that mediate the biological function of YAP. In this study, we demonstrated that the TEAD family transcription factors play an essential role in YAP-dependent gene expression and cell growth stimulation. The functional relationship between YAP and TEAD is conserved in Drosophila, in which Yki acts through Sd to regulate cell growth and organ size. During the preparation of this manuscript, it was reported that Sd mediates Hippo signaling downstream from Yki (Goulev et al. 2008; Wu et al. 2008; Zhang et al. 2008). These Drosophila studies are completely consistent with our Drosophila data and further support our conclusion that TEAD is a key transcription factor mediating YAP function in mammals.
Although both Yki and YAP promote cell and tissue growth in Drosophila and mammals, respectively, the genes induced by these two transcription coactivators are not identical. For example, cyclin E is induced by Yki overexpression in Drosophila but not by YAP overexpression in mammalian cells (Dong et al. 2007). We identified CTGF as a direct target gene of YAP-TEAD in mammalian cells. Interestingly, elevated CTGF levels have been detected in human cancers (Xie et al. 2001), and anti-CTGF antibody inhibited tumor growth and metastasis (Dornhofer et al. 2006). This supports a possible role of CTGF in mediating the growth-stimulating and oncogenic function of YAP-TEAD. Although CTGF appears to play an important role in YAP-induced cell growth, it may not be required for YAP-induced EMT. This indicates that other genes may be involved in the biological function of YAP. Consistently, the TEAD-binding-defective YAP-S94A mutant can still induce expression of a fraction of the YAP-regulated genes. Furthermore, overexpression of the Sd-binding-defective Yki-S97A elicits a significantly reduced but still obvious overgrowth in Drosophila eyes and wings. These observations indicate that additional transcription factors may be used by YAP/Yki to regulate cell and tissue growth.
Materials and methods
Cell culture, transfection, and retroviral infection
HEK293 cells, HEK293-T cells, NIH-3T3 cells, and ACHN cells were cultured in DMEM (Invitrogen) containing 10% FBS (Invitrogen) and 50 μg/mL penicillin/streptomycin (P/S). MCF10A cells were cultured in DMEM/F12 (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 10 μg/mL insulin, 100 ng/mL cholera toxin, and 50 μg/mL P/S. Transfection with lipofectamine was performed according to the manufacturer’s instructions.
To generate wild-type or mutant YAP-expressing stable cells, retrovirus infection was performed by transfecting 293 Phoenix retrovirus packaging cells with empty vector or pQCXIH-YAP constructs. Forty-eight hours after transfection, retroviral supernatant was supplemented with 5 μg/mL polybrene, filtered through a 0.45-μm filter, and used to infect MCF10A or NIH-3T3 cells. Thirty-six hours after infection, cells were selected with 200 μg/mL hygromycin (Roche) in culture medium.
Lentiviral shRNA cloning, production, and infection
To generate YAP, TEAD1/3/4, or CTGF knockdown cells, oligonucleotides were cloned into pLKO.1 with the AgeI/EcoRI sites (Moffat et al. 2006). TEAD1/3/4 shRNAs were designed in a region identical in TEAD1, 3, and 4. The sequences of the oligonucleotides are as follows: YAP #1-sense, 5′-CCGGCTG GTCAGAGATACTTCTTAACTCGAGTTAAGAAGTATCTCT GACCAGTTTTTC-3′; YAP #1-antisense, 5′-AATTGAAA AACTGGTCAGAGATACTTCTTAACTCGAGTTAAGAAGT ATCTCTGACCAG-3′; YAP #2-sense, 5′-CCGGAAGCTTT GAGTTCTGACATCCCTCGAGGGATGTCAGAACTCAAA GCTTTTTTTC-3′; YAP #2-antisense, 5′-AATTGAAAAAAA GCTTTGAGTTCTGACATCCCTCGAGGGATGTCAGAACT CAAAGCTT-3′; TEAD1/3/4 #1-sense, 5′-CCGGATGATCA ACTTCATCCACAAGCTCGAGCTTGTGGATGAAGTTGATC ATTTTTTC-3′; TEAD1/3/4 #1-antisense, 5′-AATTGAAAA AATGATCAACTTCATCCACAAGCTCGAGCTTGTGGATG AAGTTGATCAT-3′; TEAD1/3/4 #2-sense, 5′-CCGGGAT CAACTTCATCCACAAGCTCTCGAGAGCTTGTGGATGAA GTTGATCTTTTTC-3′; TEAD1/3/4 #2-antisense, 5′-AATTG AAAAAGATCAACTTCATCCACAAGCTCTCGAGAGCTTGT GGATGAAGTTGATC-3′; CTGF #1-sense, 5′-CCGGAAAT CTCCAAGCCTATCAAGTCTCGAGACTTGATAGGCTTGG AGATTTTTTTTC-3′; CTGF #1-antisense, 5′-AATTGAAAA AAAATCTCCAAGCCTATCAAGTCTCGAGACTTGATAGGC TTGGAGATTT-3′; CTGF #2-sense, 5′-CCGGCTGCACCAG CATGAAGACATACTCGAGTATGTCTTCATGCTGGTGCA GTTTTTC-3′; CTGF #2-antisense, 5′-AATTGAAAAACTG CACCAGCATGAAGACATACTCGAGTATGTCTTCATGCT GGTGCAG-3′.
Plasmids were propagated in and purified from Stbl2 competent cells (Invitrogen). The infection process was similar to that of retroviral infection except that the lentiviral packaging plasmids psPAX2 and pMD2.G were cotransfected into HEK293-T cells for virus production. Cells were selected in 5 μg/mL puromycin in culture medium.
ChIP and ChIP-on-chip
ChIP-on-chip and genome-wide location analysis were performed as previously described (Yu et al. 2007). Briefly, cells were cross-linked, lysed, and sonicated to generate DNA fragments with an average size of 0.5 kb. ChIP was performed using 5 μg of antibodies against YAP, TEAD1, AR, or control IgG. ChIP-enriched DNA, along with input whole lysate DNA, were subjected to a ligation-mediated PCR step to generate enough DNA materials, which were then labeled with fluorescent dyes and hybridized to a promoter microarray according to the manufacturer’s protocols (Agilent Technologies). The hybridization intensity was extracted using the Agilent Feature Extraction Software. The bound probes were determined at a cut-off P-value of XDEV, which is a scaled log-ratio value generated from single-gene error model, <0.001.
Three-dimensional culture of MCF10A cells
The 3D culture of MCF10A cells was done as described (Debnath et al. 2003). Briefly, Growth Factor Reduced Matrigel was layered onto eight-well glass chamber slides to make a reconstituted basement membrane. MCF10A cells were seeded on top of that at a concentration of 5000 cells/well in assay medium containing 2% Matrigel and 5 ng/mL EGF. Cells were cultured in a 5% CO2 humidified incubator at 37°C. The medium was replaced every 4 d.
Acknowledgments
We thank Drs. Marius Sudol for pCMV-Flag-YAP2 and pM-ErbB4-CTFΔK constructs, David M. Sabatini for the pLKO.1 vector, Sean Carroll for pGST-sd, Duojia Pan for pGal4-yki, Hongjiao Ouyang for RUNX2 and 6× OSE2-luc reporter, Jing Yang for EMT marker antibodies, and Taocong Jin for assistance on gene expression microarray. We thank Dr. Georg Halder for hsFLP; act>y+>GFP-S65T, the Bloomington Drosophila Stock Center for sd12 and UAS-sd fly strains, and the Developmental Studies Hybridoma Bank at the University of Iowa for Dlg antibody. This work is supported by grants from NIH (to K.L.G.), NIH (to C.Y.W.), NSF (to Z.C.L.), and Rackham Graduate School, University of Michigan (to B.Z.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1664408.
References
- Anbanandam A., Albarado D.C., Nguyen C.T., Halder G., Gao X., Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc. Natl. Acad. Sci. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Totty N.F., Irwin M.S., Sudol M., Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14–3–3 and attenuation of p73-mediated apoptosis. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., Brummelkamp T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., Brugge J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornhofer N., Spong S., Bennewith K., Salim A., Klaus S., Kambham N., Wong C., Kaper F., Sutphin P., Nacamuli R., et al. Connective tissue growth factor-specific monoclonal antibody therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 2006;66:5816–5827. doi: 10.1158/0008-5472.CAN-06-0081. [DOI] [PubMed] [Google Scholar]
- Edgar B.A. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fossdal R., Jonasson F., Kristjansdottir G.T., Kong A., Stefansson H., Gosh S., Gulcher J.R., Stefansson K. A novel TEAD1 mutation is the causative allele in Sveinsson’s chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum. Mol. Genet. 2004;13:975–981. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- Goulev Y., Fauny J.D., Gonzalez-Marti B., Flagiello D., Silber J., Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the Hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Hao Y., Chun A., Cheung K., Rashidi B., Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2007;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Hariharan I.K., Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu. Rev. Genet. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- Harvey K., Tapon N. The Salvador-Warts-Hippo pathway—An emerging tumour-suppressor network. Nat. Rev. Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Harvey K.F., Pfleger C.M., Hariharan I.K. The Drosophila Mst ortholog, Hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K., Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Kitagawa M. A Sveinsson’s chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem. Biophys. Res. Commun. 2007;361:1022–1026. doi: 10.1016/j.bbrc.2007.07.129. [DOI] [PubMed] [Google Scholar]
- Komuro A., Nagai M., Navin N.E., Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Lai Z.C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- McClatchey A.I., Giovannini M. Membrane organization and tumorigenesis–the NF2 tumor suppressor, Merlin. Genes & Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- McClatchey A.I., Saotome I., Mercer K., Crowley D., Gusella J.F., Bronson R.T., Jacks T. Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes & Dev. 1998;12:1121–1133. doi: 10.1101/gad.12.8.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K., et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Overholtzer M., Zhang J., Smolen G.A., Muir B., Li W., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo L.J., Edgar B.A. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- Simmonds A.J., Liu X., Soanes K.H., Krause H.M., Irvine K.D., Bell J.B. Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes & Dev. 1998;12:3815–3820. doi: 10.1101/gad.12.24.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Harvey K.F., Bell D.W., Wahrer D.C., Schiripo T.A., Haber D.A., Hariharan I.K. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Udan R.S., Kango-Singh M., Nolo R., Tao C., Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Vassilev A., Kaneko K.J., Shu H., Zhao Y., DePamphilis M.L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes & Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J., Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J., Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xie D., Nakachi K., Wang H., Elashoff R., Koeffler H.P. Elevated levels of connective tissue growth factor, WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res. 2001;61:8917–8923. [PubMed] [Google Scholar]
- Yagi R., Chen L.F., Shigesada K., Murakami Y., Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Rhodes D.R., Tomlins S.A., Cao X., Chen G., Mehra R., Wang X., Ghosh D., Shah R.B., Varambally S., et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B., Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]