Abstract
The transcription factor Pax6 is an important developmental regulator. Spatiotemporal control of Pax6 expression during embryogenesis is crucial for regulating distinct aspects of cortical development. Here, we report that Trim11, a member of the TRIM/RBCC protein family of E3 ubiquitin ligases, interacts with Pax6 and mediates Pax6 degradation via the ubiquitin–proteasome system. Trim11 overexpression decreases endogenous Pax6 protein levels and represses Pax6 functions, including Pax6-dependent transactivation and neurogenesis. Abrogation of endogenous Trim11 expression in the developing cortex increases the level of insoluble forms of Pax6 and enhances apoptosis. We provide evidence that the B30.2 domain of Trim11 is essential for the clearance of insoluble cell proteins. Furthermore, we show that the expression of Trim11 is directly regulated by Pax6 in developing cortex in vivo. Our findings indicate that an autoregulatory feedback loop between Trim11 and Pax6 maintains a balance between the levels of Pax6 and Trim11 proteins in cortical progenitors, having an essential role for the Pax6-dependent neurogenesis.
Keywords: Cortex, neurogenesis, Pax6, ubiquitin–proteosome system
Pax6 is a member of the Pax family of transcription factors that contains a DNA-binding paired-box and homeobox domain. In animals, including humans, Pax6 plays a key role in development, regulating organogenesis of the eye, pancreas, and brain (Callaerts et al. 1997; Simpson and Price 2002). In each of these organs, Pax6 acts as a potent cell fate determinant. Functional studies have highlighted the fact that the level of Pax6 protein is important in determining developmental outcome. Thus, in the absence of functional Pax6, as in the Small eye mutant mouse, homozygous (Sey/Sey) embryos have no eyes and exhibit severe forebrain and cortical malformations, whereas heterozygous (Sey/+) embryos have smaller eyes with various ocular defects and a milder cortical phenotype (Hill et al. 1991; Schmahl et al. 1993; Stoykova et al. 1996). Transgenic mice carrying multiple copies of the human Pax6 gene have ocular abnormalities similar to those in Sey/Sey mice (Schedl et al. 1996). Thus, either a reduction or an increase in Pax6 gene dosage causes profound developmental defects in the eye, suggesting that Pax6 function is critically dependent on a defined Pax6 threshold (Hill et al. 1991; Schedl et al. 1996).
In the developing cortex, Pax6 expression in pluripotent radial glial progenitors (RGs) imparts neurogenic potential (Gotz et al. 1998; Heins et al. 2002). In the absence of Pax6, RGs generate fewer than 50% of the normal number of neuronal cells; the presence of a single wild-type Pax6 allele is capable of moderating this phenotype (Schmahl et al. 1993). Recent evidence suggests that distinct spatiotemporal variations in the levels of Pax6 expression in cortical progenitors regulate specific processes during mammalian corticogenesis (for review, see Simpson and Price 2002; Guillemot et al. 2006; Mallamaci and Stoykova 2006). During early development in the mouse (embryonic days 10–13 [E10–E13]), RGs generate neuronal subtypes that migrate out and settle in lower layers of the cortex (layers 5 and 6), while later-born neurons (E13–E18) populate upper cortical layers (layers 2–4). Pax6 is expressed at much higher levels in early RGs, and appears to act exclusively in these early progenitors to control mitotic cell cycle parameters (Quinn et al. 2006). In the developing telencephalon, the highest level of Pax6 expression is confined to ventral and lateral pallium progenitors, which specify the morphogenesis of several amygdalar nuclei (Stoykova et al. 2000; Tole et al. 2005; Remedios et al. 2007). In the ventricular zone (VZ), Pax6 exhibits a strong rostrolateral (high) to caudomedial (low) expression gradient (Stoykova et al. 1996). When Pax6 function is abolished, caudal domains are enlarged at the expense of rostral cortical areas, which are severely diminished in size, suggesting the involvement of Pax6 in cortical arealization (Bishop et al. 2002; Muzio et al. 2002). In addition, depending on their endogenous Pax6 expression level, RG cells are distinctly sensitive to transgenic elevation of Pax6, which causes either apoptosis or enhanced neurogenesis in vivo (Berger et al. 2007). Collectively, the available evidence suggests that the maintenance of correct Pax6 levels in progenitor cells in distinct cortical domains and during different developmental time windows has a pivotal role in corticogenesis and forebrain patterning. However, the molecular mechanisms that control Pax6 cellular content have not been described.
Here, we show that the Ring finger E3 ubiquitin ligase, Trim11, physically interacts with Pax6 and mediates Pax6 ubiquitination, promoting Pax6 degradation by the ubiquitin proteasome system (UPS). Trim11-mediated Pax6 degradation inhibits Pax6 transcriptional activity and impairs Pax6-dependent neurogenesis. We show that silencing endogenous Trim11 in the mouse cortex promotes an accumulation of Pax6 inclusion bodies and increases apoptosis. We also present the first evidence that the B30.2 domain of Trim11 is responsible for the clearance of cellular insoluble proteins, including insoluble Pax6, thus defining a novel and important role for Trim11 in protein quality control (PQC). Furthermore, we demonstrate that overexpression of Pax6 induces Trim11 expression in vivo. Our novel findings indicate that the autoregulatory loop between Pax6 and Trim11 balances the expression of both proteins and plays an essential role in cortical neurogenesis.
Results
Trim11 interacts with Pax6
To identify proteins that interact with Pax6 and possibly modulate the activity of Pax6 during corticogenesis, we used a yeast two-hybrid assay to screen a mouse embryonic (E15.5) cortical cDNA library using Pax6ΔPD (PD domain-deleted Pax6) as a bait (Fig. 1A). Out of a library containing 2.5 × 106 transformants, four sequences obtained from a total of 116 independent clones were found to code for Trim11 (Fig. 1B). Sequences isolated from two positive colonies encoded full-length Trim11 cDNA (Trim11/FL or Trim11), which contains a Ring domain, a coiled-coil domain (CC), a B-box (BB), and a B30.2 domain. The shortest isolated sequence corresponded to a complete CC, flanked by the BB and a partial B30.2 domain. Both full-length clones contained a point mutation in the C terminus.
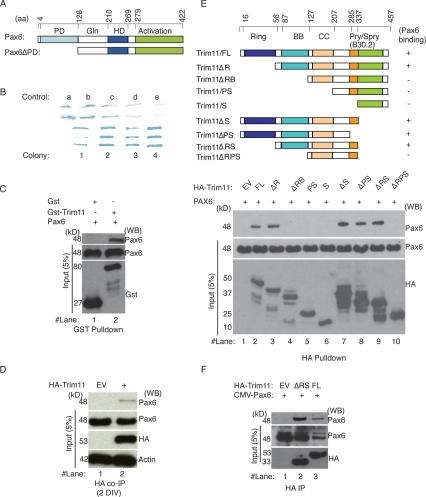
Figure 1.
Pax6 interacts physically with Trim11. (A) The schematics depict the functional domains of the Pax6 protein. Pax6 contains a glycine-rich region (Gln), an activation C-terminal domain, and two DNA-binding domains: the paired-box domain (PD) and the paired-type homeodomain (HD). A Pax6 construct lacking the paired-box domain (Pax6ΔPD) was used as a bait in yeast two-hybrid screens. (B) Five different controls (a–e) were used in the preformed two-hybrid yeast screen, as recommended by the manufacturer. Note that intensity of the blue signals in controls a–e represent very strong, strong, moderate, weak, and none of the interaction strength, respectively. Four colonies representing independent clones strongly positive for Pax6 binding revealed potential interactions between Trim11 and Pax6. (C) The interaction of Trim11 and Pax6 was assessed by a GST pull-down assay using purified GST-Trim11 and lysates of HeLa cells transfected with CMV-Pax6 expression construct. The top blot shows eluated proteins after the pull-down experiments, the bottom blots show the input proteins. (D) HA-Trim11 plasmids were electroporated into primary cortical cells prepared from E12.5 embryos. After 2 DIV, the interaction of Trim11 and endogenous Pax6 was proven by HA Coimmunoprecipitation. (E) The top schematic illustrates the structure of full-length (FL) Trim11, which consists of Ring (R), B-box1/B-box2 (BB), Coiled-coil (CC), and B30.2 domains. To map Trim11 domains that bind Pax6, a series of HA-tagged Trim11 deletion constructs was generated and used in HA pull-down assays employing lysates of HeLa cells transfected with a CMV-Pax6 expression construct. (F) The interaction of Trim11/FL and Trim11ΔRS with Pax6 were confirmed by coimmunoprecipitation assays performed in HeLa cells after transfection with HA-tagged full-length Trim11 (FL), the deletion Trim11ΔRS construct or EV in presence of proteasome inhibitor, MG132 (20 μM). The top blot shows eluted proteins after immunoprecipitation with Pax6 antibody; the bottom blots show the input proteins.
To confirm that Trim11 binds to Pax6, we performed GST pull-down assays using purified GST-fused full-length Trim11 (GST-Trim11) and lysates of Pax6-transfected HeLa cells. As shown in Figure 1C (lane 2), Pax6 physically interacted with GST-Trim11; no binding was detected between GST and Pax6 (Fig. 1C, lane 1). Because the Trim11 antibodies generated by us (as well as the two tested commercially available Trim11 antibodies) were not working well, in order to prove the interaction of Trim11 and Pax6 in vivo we performed experiments in which HA-Trim11 plasmid was electroprated into primary cortical cultures prepared from E12.5 embryo brains and cultured for 2 d in vitro (DIV). After HA-driven coimmunoprecipitation, we found that indeed Trim11 immunoprecipitates the endogenous cellular Pax6 protein (Fig. 1D).
To map the region of Trim11 that interacts with Pax6, we prepared a series of HA-tagged Trim11 deletion constructs and expressed them in HeLa cells (Fig. 1E, scheme). In HA pull-down assays of Pax6-transfected HeLa cell lysates (Fig. 1E, middle image), Pax6 was found to interact with Trim11 fragments that included both BB and CC domains (Fig. 1E, lanes 2,3,7–9), but not with those that lacked the BB domain (Fig. 1E; lanes 4–6,10). Notably, we observed that a fragment including only the CC domain (Trim11ΔRB) is not able to bind to Pax6 (see Fig. 1E, lane 4). Evidence has been presented that a fragment containing the BB and the N-terminal of the CC domain is responsible for interactions of many other members of the Trim protein family with their interacting partners (Meroni and Diez-Roux 2005). Together, these findings suggest that the interaction between Trim11 and Pax6 possibly requires both the BB and at least the N-terminal of the CC domain.
To confirm that the Trim11 fragment, containing BB and CC domains, interacts with Pax6 in mammalian cells, we cotransfected HeLa cells with HA-Trim11/FL, HA-Trim11ΔRS, or empty vector (EV) and an expression plasmid for mouse Pax6 cDNA (CMV-Pax6). Consistent with the results from GST pull-down assays, both HA-Trim11/FL and HA-Trim11ΔRS were specifically coimmunoprecipitated with Pax6 (Fig. 1F, lanes 2,3), while no binding was seen in EV-transfected cells (Fig. 1F, lane 1).
Trim11 transcripts are ubiquitously distributed in developing mouse tissue at E12.5, but similar to Pax6, they are most abundantly present in the proliferative neuroepithelium throughout the central nervous system (Reymond et al. 2001). Accordingly, HA-tagged Trim11 was diffusely distributed in the nucleus and cytoplasm of HeLa cells (Supplemental Fig. S1, G). Notably, after cotransfection of CMV-Pax6 and HA-Trim11 constructs in HeLa, we found that these proteins were colocalized to the nucleus (Supplemental Fig. S1J–L), suggesting that Pax6 and Trim11 interact in vivo.
Trim11 induces ubiquitination and modulates the steady-state levels of Pax6 protein
Recent evidence has shown that Trim11 acts as an E3 ubiquitin protein ligase for Humanin and ARC105, mediating their degradation by the UPS (Niikura et al. 2003; Ishikawa et al. 2006). Given that Trim11 interacts with Pax6, we next tested whether Trim11 also has a role in controlling Pax6 degradation. We first analyzed the half-life of Pax6 in HeLa cells transfected with either Trim11/FL or EV, using the cycloheximide-chase approach. Quantifications of the relative protein contents indicated that the half-life of Pax6 was decreased much faster in cells overexpressing Trim11 than in EV-transfected cells (as shown in the diagram and the table in Fig. 2A). To confirm that Trim11 is involved in controlling Pax6 stability, we cotransfected HeLa cells with expression vectors for CMV-Pax6 with either HA-Trim11 or EV. Western blot analysis revealed that Pax6 protein was indeed substantially degraded in the presence of Trim11, depending on the length of the interaction time between both proteins, from severely diminished protein level (Fig. 3A, lanes 1,2; transfected cells were cultured for 24 h) to a complete Pax6 degradation (Fig. 2C, lanes 1,2; transfected cells were cultured for 48 h).
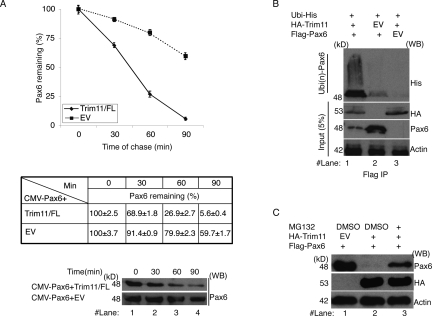
Figure 2.
Trim11 induces ubiquitination of Pax6 and modulates steady-state levels of Pax6 protein. (A) Cycloheximide chase assays were used to assess the effect of Trim11 on the steady-state level of Pax6. Western blot shows the level of Pax6 in HeLa cells after cotransfection with the CMV-Pax6 expression construct and either Trim11/FL or EV (dashed line), respectively. Note the progressive and substantial decrease in the level of Pax6 in the presence of Trim11. The graphic and the table show the quantified relative values of Pax6 protein level in Western blot analysis at different time points upon a Cycloheximide block of protein synthesis. Amounts of Pax6 protein before the Cycloheximide treatment are designated as 100% in the graphic. The relative amount of Pax6 protein was measured in three independent experiments. The values are expressed as mean ± SD (error bars). (B) Ubiquitination assays were performed in HeLa cells cotransfected with plasmids, as indicated. Western blot analysis of proteins eluted with an anti-His antibody after precipitation with Flag-tagged Pax6 revealed a much stronger smeared band of ubiquitinated Pax6 (lane 1, top) in the presence of Trim11, compared with controls without Trim11 or Pax6 (lanes 2,3 top, respectively). In the bottom blot (against anti-Pax6 antibody) note the substantial reduction in the level of Pax6 protein upon transfection of HA-Trim11. (C) Confirmation of UPS-mediated degradation of Pax6 using proteasome inhibition assays. HeLa cells were transfected with plasmids and treated with proteasome inhibitor (MG132) as indicated. In the presence of MG132, Pax6 levels recovered significantly (cf. levels of Pax6 protein in lanes 2 and 3 of the top blot). In B and C, blotting with an anti-HA antibody shows that an equal amount of Trim11 protein was used in each experiment; blotting with an anti-actin antibody demonstrates equal loading of total protein on SDS-PAGE.
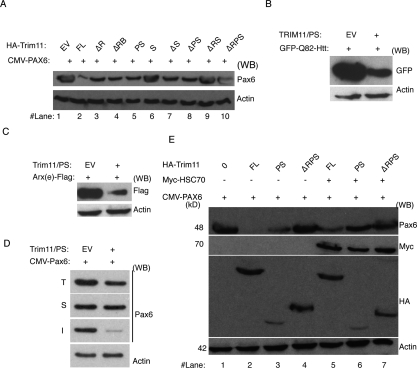
Figure 3.
Trim11 domain B30.2 is responsible for clearance of insoluble cellular proteins. (A) HeLa cells were cotransfected with deletion constructs of Trim11 (shown in Fig. 1D) and the CMV-Pax6 expression plasmid and the level of Pax6 protein was determined by quantifying band densities on Western blots. Note that the B30.2 domain of Trim11 (Trim11/PS, lane 5) efficiently degraded Pax6. (B,C) Trim11/PS also efficiently degraded both GFP-Q82-Htt (B) and Arx(e)-Flag (C). (D) Trim11/PS promoted the degradation of insoluble Pax6 (image I), but did not affect soluble Pax6 (image S). (E) B30.2-containing constructs were cotransfected with Pax6 in the presence or absence of Hsc70 expression plasmids. The presence of Hsc70 inhibited Trim11/B30.2-mediated degradation of Pax6. (A–E) The blot at bottom (anti-actin antibody) demonstrates equal loading of total protein on SDS-PAGE.
Next, we determined whether Pax6 could serve as a substrate for the ubiquitin ligase activity of Trim11 by assaying for ubiquitinated Pax6 in HeLa cells. To accomplish this, we overexpressed HA-Trim11 together with Flag-Pax6 and His-tagged ubiquitin (His-Ub) in the presence of the proteasome inhibitor, MG132. Pax6 was then immunoprecipitated from cell lysates with an anti-Flag antibody and immunoprecipitates were analyzed for the appearance of Ub-conjugated forms of Pax6 (Fig. 2B). In Western blots of immunoprecipitates, a large amount of ubiquitinated Pax6 protein was detected only in the presence of Trim11 (Fig. 2B, smear in lane 1), suggesting that Trim11 mediates ubiquitination of intracellular Pax6. Finally, to determine whether the degradation of ubiquitinated Pax6 is truly mediated by Trim11 through UPS, we compared Pax6 expression levels in HeLa cells after cotransfection of Pax6 and Trim11 in the presence or absence of the proteasome inhibitor MG132. In the absence of MG132, Pax6 protein expression was largely abolished in parallel with an elevation in Trim11 levels (Fig. 2C, lanes 1,2). Conversely, in the presence of MG132, Pax6 levels were substantially restored (Fig. 2C, lane 3). Collectively, these results indicate that Trim11 mediates degradation of Pax6 via UPS.
B30.2 domain of Trim11 accelerates the degradation of insoluble proteins
RING finger-containing E3 ligases interact both with E2 enzymes (through RING domains) and substrates (via substrate-binding domains) to bring the two proteins into close proximity and allow transfer of an ubiquitin moiety from E2 to substrate (Hershko and Ciechanover 1998). To examine the contribution of specific Trim11 domains to Trim11-mediated degradation of Pax6, we cotransfected HeLa cells with CMV-Pax6 and HA-tagged Trim11 deletion constructs (Fig. 1E) or EV control plasmid. Western blot analysis indicated that the full length of Trim11 (containing RING domain) most effectively mediated Pax6 degradation (Fig. 3A, lane 2). Surprisingly, however, we found that all Trim11 constructs containing the B30.2 domain (Trim11/FL, Trim11ΔR, Trim11ΔRB, Trim11/PS) (Fig. 3A, lanes 2–5) were capable of promoting Pax6 degradation. This was true even for constructs that lacked both the E3 ligase activity-containing RING domain and the BB/CC domains (the Pax6-binding domains, Trim11/PS). The B30.2 domain has been reported to interact with both proteasome subunits and ubiquitin (Suzumori et al. 2003), raising the possibility that Trim11 might have a more general role in regulating cell protein content. To test this hypothesis, we determined whether the Trim11/PS construct, which contains only the B30.2 domain, could also promote the degradation of GFP-Q82-Htt, a pathological polyglutamine-expanded form of Huntingtin and Arx(e)-Flag, which has polyalanine tract expansions (Nasrallah et al. 2004; Miller et al. 2005). Both of those proteins are ubiquitinated and form aggregates of insoluble proteins in the cell nucleus (Nasrallah et al. 2004; Miller et al. 2005). Remarkably, the B30.2 domain of Trim11 could efficiently degrade not only Pax6, but also both GFP-Q82-Htt and Arx(e)-Flag proteins (Fig. 3B,C). These results suggest that the Trim11/B30.2 domain is involved globally in the clearance of insoluble and ubiquitinated proteins, indicating a likely role in PQC.
Proteins of cell extracts could be separated to insoluble and soluble fraction, the last one representing the pool of physiological active proteins (Alves-Rodrigues et al. 1998; Muchowski and Wacker 2005). To determine whether the portion of Pax6 degraded by the Trim11/B30.2 corresponds to insoluble Pax6, we cotransfected HeLa cells with Pax6 and Trim11/PS plasmids (or EV) and analyzed Pax6 levels in Triton X-100-soluble and Triton X-100-insoluble lysate fractions corresponding to native proteins and inclusion bodies, respectively. Surprisingly, B30.2 did not affect the level of soluble Pax6 protein (Fig. 3D, image S); however, it almost entirely abolished the insoluble Pax6 fraction (Fig. 3D, image I). Thus, the B30.2 domain of Trim11 alone is apparently able to specifically promote the degradation of insoluble proteins.
Up-regulation of molecular chaperones decreases the level of insoluble cellular proteins by partitioning them into a productive folding pathway (Alves-Rodrigues et al. 1998; McClellan et al. 2005; Muchowski and Wacker 2005; Rubinsztein 2006). To investigate whether chaperone-dependent protein folding could inhibit B30.2 domain-mediated degradation of insoluble Pax6, we cotransfected Pax6 and various truncation mutant constructs of Trim11 together with a mammalian Myc-tagged Hsc70 (Hsc70-Myc) expression plasmid (or EV) in HeLa cells. All constructs that included the Trim11/B30.2 domain promoted a substantial decrease in Pax6 levels compared with EV controls or to constructs lacking Ring and B30.2 domains (Trim11ΔRPS) (Fig. 3E, cf. lanes 2,3 and 1,4). In the presence of Hsc70, Pax6 levels were significantly recovered (Fig. 3E, cf. lanes 2,3 and 5,6), indicating that the molecular chaperone largely prevented Trim11/B30.2 domain-mediated degradation of insoluble Pax6.
Collectively, our findings indicate that, in addition to its previously documented E3 ligase-dependent functions, Trim11, through its B30.2 domain, plays an essential role in promoting the degradation of insoluble ubiquitinated proteins, thus representing an important component of the cellular PQC.
The proteasome mediates degradation of Pax6 during corticogenesis
As noted above, Pax6 is an important neurogenic factor for cortical RG progenitors that has phenotypic consequences exhibiting strong dosage effects. We first asked whether the proteasome system contributes to the previously reported dynamic changes in Pax6 expression in the developing cortex. We measured Pax6 transcript and protein levels during cortical neurogenesis (E11–E18) using qPCR and Western blotting analyses, respectively. In accord with previous data, the levels of both Pax6 transcript and Pax6 protein were high in the early RG progenitors (E11–E14) and gradually decreased thereafter (Fig. 4A). Interestingly, however, the level of Pax6 protein appeared to decline faster, leading to a marked discrepancy between the levels of Pax6 mRNA and Pax6 protein during the later developmental stages.
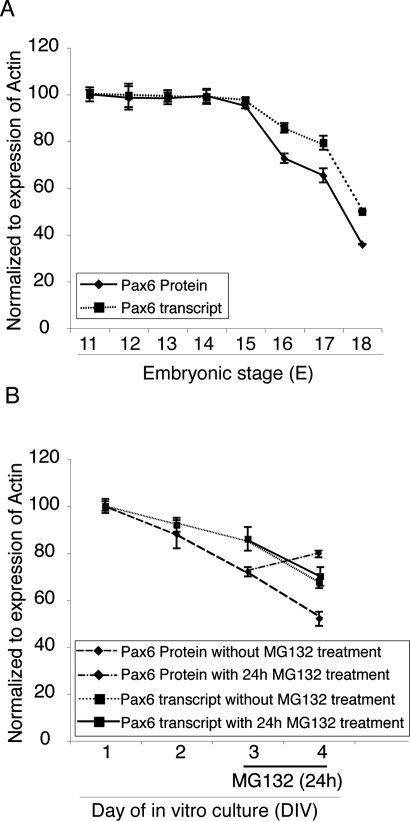
Figure 4.
Pax6 protein level in cortical progenitors is regulated by the proteasome system. (A) The expression of Pax6 in isolated embryo cortices during progression of corticogenesis (E11–E18) was quantified at both RNA and protein levels. Note that after stage E14.5, there was a discrepancy between Pax6 mRNA and protein levels. (B) A more intensive decline in Pax6 protein levels relative to mRNA levels was detected during the progressive neuronal differentiation of primary cortical cultures in vitro. Note that treatment with the proteasome inhibitor, MG132, resulted in a significant elevation in Pax6 protein level at the 4 DIV stage. The results of Western blot and RNA quantification are presented as means ± SD (error bars) from three independent experiments (with each RNA quantification carried out in triplicate).
To determine whether this decline in Pax6 protein levels involved protein degradation by the proteasome system, we first quantified Pax6 mRNA and Pax6 protein in primary cortical cells isolated from the mouse E12.5 cortex and cultured for 1–4 DIV. Similar to the results obtained in vivo, during the progression of neuronal differentiation in vitro, Pax6 protein levels declined more rapidly than did the level of Pax6 transcript (Fig. 4B). Treatment of cortical progenitor cells with the proteasome inhibitor, MG132, led to a significant increase in the level of Pax6 protein at 4 DIV, but had no effect on Pax6 transcript levels (Fig. 4B), indicating that endogenous Pax6 protein content is indeed modulated by the UPS during corticogenesis.
Trim11 influences Pax6-dependent transcriptional activity
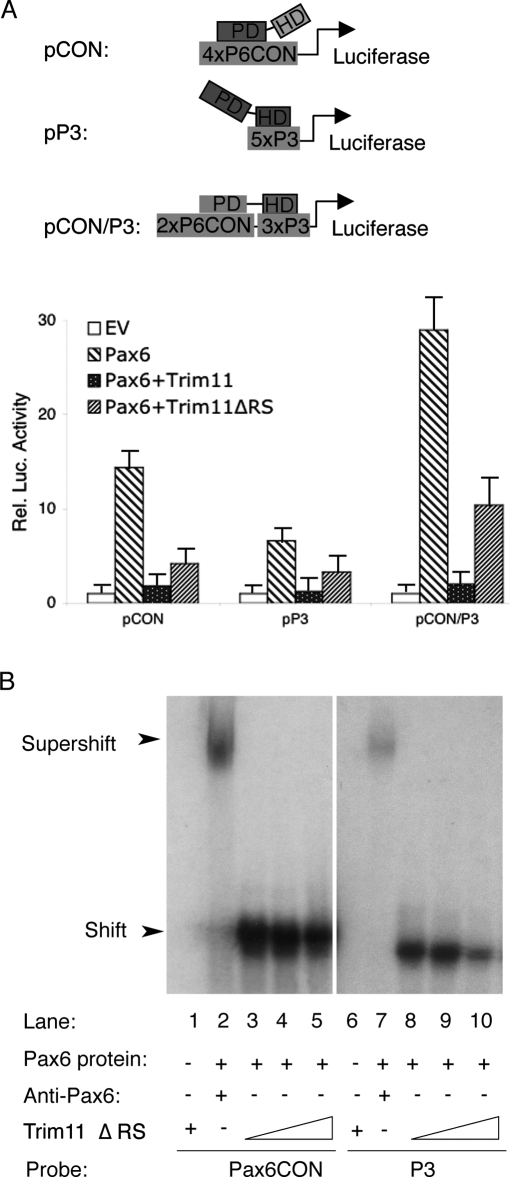
Because protein–protein interactions influence the transactivation function of transcription factors, we next examined the effect of Trim11/Pax6 interactions on the transcriptional activation of Pax6-dependent reporter constructs (Epstein et al. 1994). For this purpose, HeLa cells were cotransfected with the luciferase reporter constructs pCON or pP3, which contain the P6CON and P3 consensus binding sequences for Pax6 PD and HD domains, respectively, or the pCON/P3 chimeric construct, which contains fragments of both P6CON and P3 (Fig. 5A). Consistent with previous reports (Epstein et al. 1994), Pax6 alone strongly transactivated pCON, pP3, and pCON/P3 reporter constructs. Remarkably, the presence of Trim11 almost completely repressed transactivation of all tested reporter constructs (Fig. 5A). Most surprisingly, the Trim11ΔRS fragment (without Ring and B30.2 domains), although interacting with Pax6, does not mediate Pax6 degradation (Figs. 1E [lane 9], 3A [lane 9]) and profoundly inhibited Pax6-dependent reporter transactivation (diagram in Fig. 5A). This finding suggest that a Trim11 fragment that includes the BB and CC domains may exert its inhibitory effects by preventing access of Pax6 to its consensus binding sequences. To investigate this possibility, we performed gel shift assays using Pax6 protein with P6CON and P3 probes in the presence or absence of the Trim11ΔRS fragment. In the absence of Trim11ΔRS peptides, Pax6 bound strongly to both P6CON and P3 probes (Fig. 5B, lanes 3,8). Remarkably, Pax6 binding was progressively inhibited by increasing amounts of Trim11ΔRS (Fig. 5B, lanes 3–5,8,9). Pax6-specific binding by P6CON or P3 was confirmed by incubating with an anti-Pax6 antibody, which resulted in a super shift of the probe–Pax6 protein–Pax6 antibody complex (Fig. 5B, lanes 2,7). These findings suggest that interaction between Pax6 and Trim11 may cause conformational changes that prevent the accessibility of Pax6 protein to its consensus target sequences. However, it should be also noted that we observed a strong reduction of Pax6 transcriptional activity in the reporter assay in HeLa cells (Fig. 5A), implicating that Trim11/Pax6 interactions in HeLa might affect, in addition, other processes; e.g., binding of Pax6 to its activators.
Figure 5.
Trim11 inhibits the transcriptional activity of Pax6. (A) Transcriptional activity of Pax6 protein was measured in HeLa cells transfected with luciferase-reporter plasmids containing the consensus Pax6-binding sequences, P6CON (pCON), P3 (pP3), or a chimera of the two (pCON/P3). Cells were cotransfected with a Pax6 expression vector (or EV), in various combinations with Trim11 and Trim11ΔRS, as indicated. Note that both Trim11 and Trim11ΔRS profoundly repressed Pax6-dependent activation of all reporter constructs. Luciferase activity was measured in triplicate and expressed as mean ± SD (error bars). (B) EMSA assay was performed using in vitro translated Pax6 and Trim11ΔRS proteins and 32P-labeled P6CON and P3 probes. Pax6 binding to P6CON and P3 was progressively inhibited by increasing amounts of Trim11ΔRS.
Together, our findings suggest that in addition to its role for post-transcriptional degradation of Pax6 protein, Trim11 appears to inhibit the transcriptional activity of Pax6, possibly in part by preventing the binding of Pax6 to its consensus sequences. The more pronounced inhibition of Pax6 transcription activity by Trim11/BC might involve in addition other mechanism(s).
Trim11 controls endogenous Pax6 level
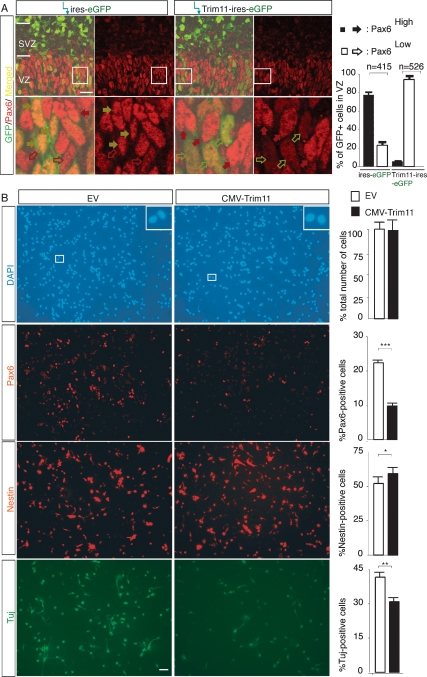
To test whether Trim11 regulates the level of endogenous Pax6 protein in vivo, we electroporated either Trim11 expression plasmid (Trim11-ires-eGFP) or EV (-ires-eGFP) into E12.5 embryo brains and, after 3 d, cortical sections from treated and untreated hemispheres were examined immunohistochemically for Pax6 expression and neuronal differentiation. We found that electroporation with EV (-ires-eGFP) did not affect the level of the endogenous Pax6 (Supplemental Fig. S3A) or the number of proliferating cells in cultures (Supplemental Fig. S4C,D), which allowed the use of untreated hemispheres as internal controls in some of the assays. After immunostaining for GFP and Pax6 antibodies, the intensity of the fluorescent signals were examined by a confocal microscopy and quantified (using ImageJ software; NIH freeware) (Supplemental Fig. S3). This approach allowed us to subdivide the GFP+/Pax6+ cells in VZ into two subpopulations, highly expressing Pax6 (Pax6high, pointed with red filled arrowheads, intensity of fluorescent signals >30 pixels), and with a low/or undetectable Pax6 (Pax6low, pointed with red empty arrowheads, intensity of fluorescent signals <30 pixels; Supplemental Fig. S3). After Pax6/GFP double immunostaining, we found that in the cells treated with EV (-ires-eGFP) 78.6 ± 3.5% were expressing Pax6 at a high level and 22.4 ± 4.7% showed a low/neglectible Pax6 expression level (415 GFP+/Pax6+ cells, four sections). The cortex, treated with Trim11-ires-eGFP vector, however, contained only 5.6 ± 0.3% Pax6high-positive cells and 94.4 6 ± 4 0.3% Pax6low cells (526 GFP+/Pax6+ cells, four sections). Noteworthy, after electroporation with EV, GFP+/Pax6+ cells (yellow filled arrowheads) showed similar Pax6 expression as the neighboring GFP-negative cells (red filled arrows). However, after electroporation of Trim11-ires-eGFP plasmid, the Pax6 expression level was substantially diminished in the majority of GFP+/Pax6+ cells (yellow empty arrowheads) as compared with GFP-negative neighbors. These findings indicate that Trim11 is acting cell-autonomously in regulation of the Pax6 level. It should be noted that the diminished endogenous Pax6 level was not due to apoptosis, since TUNEL assays showed that there was no significant difference in DNA fragmentation between cells of the Trim11-treated and untreated hemispheres (Supplemental Fig. S7A). Together, these findings suggest that Trim11 negatively regulates in vivo the endogenous Pax6 level in cortical progenitors through a cell-autonomous mechanism.
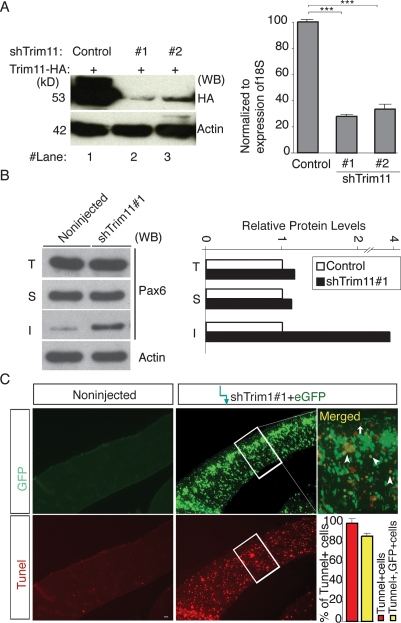
To provide additional support for the inferences drawn based on our in vivo Trim11 overexpression studies, we studied the effects of knocking down endogenous Trim11 levels using RNA silencing. In control experiments, transfection of a short hairpin interfering RNA constructs, shTrim11#1 and shTrim11#2, efficiently decreased the levels of Trim11 exogenously expressed from a cotransfected HA-Trim11 plasmid in NIH-3T3 cells (Fig. 7A, below) and Neuro2A cells (data not shown). The shTrim11 construct was similarly effective in primary cortical cell cultures, where electroporation of shTrim11#1 and shTrim#2 decreased Trim11 mRNA to 27% ± 1.2 or 28.5 ± 2.3% of controls, respectively (Fig. 7A, diagram [below]). Using the in vivo electroporation experimental paradigm described above, we electroporated shTrim11#1 plasmid and examined proteins extracted from each hemisphere (pooled from three individual embryos) by Western blot. We found that Pax6 levels in the shTrim11#1-injected hemispheres were 16.92% higher than those in the contralateral, noninjected control side (Fig. 7B, below). These results are consistent with the data from the Trim11 overexpression assays and support the conclusion that Trim11 modulates endogenous Pax6 protein level in vivo.
Figure 7.
Silencing of Trim11 leads to an increase of Pax6 inclusion bodies and apoptosis. (A) NIH-3T3 cells were cotransfected with Trim11-HA in the absence (lane 1) or presence (lanes 2,3) of a two distinct short hairpin RNAi constructs against Trim11 (shTrim11#1, lane 2, shTRim11#2, lane 3). Immunoblotting with HA-antibody (to the left) demonstrates highly efficient Trim11 silencing at the protein level with both shTrim11 plasmids. The diagram to the right represents a qPCR analysis of the level of Trim11 mRNA after electroporation of the two shTrim11 plasmids in primary cortical cell cultures (at 2 DIV) expressed relative to 18S levels. (***) P < 0.005; n = 6. (B) Western blot analysis of protein extracts (total, T; soluble, S; and insoluble, I fraction) from embryonic hemispheres treated in vivo with shTrim11#1 and immunostained with Pax6 antibody. Compared with the control, the experimental hemispheres contain 1.2- and fourfold more Pax6 soluble and insoluble protein, respectively. Relative levels of Pax6 in total, soluble and insoluble fractions are presented in the diagram to the right. (C) TUNEL assay on brain sections of the animals from the same experiment shows enhanced apoptosis in shTrim11#1-treated hemispheres. Bar, 50 μm.
To exclude the possibility that the changes in endogenous Pax6 protein levels associated with Trim11 gain-of-function (GOF) or Trim11 silencing conditions were due to alteration of the Pax6 transcription, we used qPCRs to quantify mRNA levels of Pax6- and Pax6(5a), which encode the two active vertebrate isoforms of Pax6. Despite clear effects on the level of Pax6 protein in both Trim11 GOF and Trim11 silencing experiments, the levels of Pax6 and Pax6(5a) mRNA were unchanged in samples from electroporated and contralateral hemispheres in the two experimental designs (Supplemental Fig. S2A,B). Thus, expression of Trim11 accelerates the degradation of Pax6 protein, but does not affect the transcription of the Pax6 gene.
Activation of Trim11 represses Pax6-dependent neuronal differentiation
As noticed above, overexpression of Trim11 in cortical progenitors in vivo decreased the number and the level of Pax6+ cells. Immunostaining with the proliferation marker, Ki67, indicated that the number of Ki67+ cells was significantly increased in the Trim11-transfected hemispheres (24.8 ± 0.7% and 16.5 ± 0.9% Ki67-positive cells in Trim11-treated and Trim11-untreated hemispheres, respectively; Supplemental Fig. S4). Because nestin+ cells and newly produced neurons are attached to RG fibers, it is difficult to obtain an accurate estimate of the number of nestin and Tuj1-positive cells in vivo. Accordingly, we used a CMV-Trim11 construct to overexpress Trim11 in E12.5 primary cortical cultures in vitro and analyzed cell differentiation after 3 DIV (Fig. 6B). Trim11 activation consistently led to a substantial decrease in the number of Pax6-positive cells (9.8 ± 0.4% in Trim11-transfected cells vs. 22.2 ± 0.5% in EV-transfected controls). Immunostaining with the progenitor marker, nestin, indicated a slight enhancement of the number of nestin-positive cells in CMV-Trim11-transfected cells (57.6 ± 1.7%) compared with controls (52.7 ± 2.1%) (Fig. 5B). In contrast, labeling with neuronal marker TuJ-1 indicated that CMV-Trim11-transfected cultures contained fewer neurons (30.9 ± 1.0% TuJ-1 positive cells) compared with controls (41.7% ± 1.2%) (Fig. 5B). In addition, overexpression of Trim11 did not affect the number of GFAP-positive cells under the same experimental conditions (data not shown). These results are in full agreement with the enhanced number of proliferating progenitors and abrogation of neuronal differentiation reported to occur in the cortex of the Pax6/Small eye mutant (Stoykova et al. 1997; Gotz et al. 1998; Warren et al. 1999; Heins et al. 2002; Quinn et al. 2006)
Figure 6.
Activation of Trim11 in cortical progenitors leads to a decrease in endogenous Pax6 protein and suppression of neurogenesis. (A) A Trim11 expression construct (Trim11-ires-EGFP) or EV (-ires-eGFP) were electroporated into the cortex of E12.5 embryos in utero and analyzed 3 d later. The strong GFP signal indicates that transfection efficiency with electroporation was high. After double-immunostaining for GFP and Pax6 antibody, the strength of the Pax6-fluorescent signal of confocal microscopy images was quantified by ImageJ software (NIH). Based on the detected strength of the fluorescent signal, two subpopulations of progenitors in VZ were easily distinguishable; namely, subpopulation with highly expressed Pax6 (Pax6high; pixel values >30; pointed with filled arrows) and subpopulation with a low or undetectable Pax6 level (Pax6low; pixel values <30; pointed with empty arrows, see also Supplemental Fig. S3). The EV (ires-GFP)-injected hemispheres had 78% ± 3.5% cells with Pax6high and 22.4% ± 4.7% cells with Pax6low expression level (out of 526 GFP+ cells per four sections). The electroporated with Trim11-ires-GFP plasmid hemispheres contained only 5.6 ± 0.3% cells with Pax6high and 94.4 ± 4.3% Pax6low-positive progenitors (out of 415GFP+ cells per four sections). The high-magnification images illustrate that the level of Pax6 is specifically down-regulated only in GFP+ cells of the Trim11-ires-GFP-transfected cells (yellow empty arrows), while nontransfected cells (GFP−) (red filled arrows) or GFP+ cells in hemispheres transfected with the EV (-ires-GFP) (yellow filled arrows) show an unaltered Pax6 expression. Bar, 20 μm. (B) Primary cell cultures derived from E12.5 cortical tissue were electroporated with a CMV-Trim11 expression or EV plasmids. After 3 DIV, immunohistochemical analyses were performed using antibodies that label cortical progenitors (anti-nestin) or neurons (anti-TuJ-1). Counterstaining with DAPI indicated that the cells were healthy under both experimental conditions, and the cultures contained an almost equal number of cells (428 ± 27 and 417 ± 35 cells per each experiment of CMV-Trim11 and EV-transfected cells, respectively). The activation of Trim11 in primary cortical cells caused a decrease in the number of Pax6-positive cells, increase in the number of nestin-positive progenitors, and diminishing in TuJ-1-positive neurons (from 41.7 ± 1.2% to 30.9 ± 1.0%). The chart to the right of each panel shows the respective calculated percentage of immunopositive cells. Results are presented as marker-specific cells expressed as a percentage (mean ± SD) of the total number of DAPI-stained cells. Asterisks denote statistical differences between the indicated groups. (*) P < 0.05; (**) P < 0.01; (***) P < 0.005; n = 4 for each group. Bar, 50 μm.
To determine whether the observed impairment of neurogenesis induced by overexpression of Trim11 is specifically related to the neurogenic function of Pax6, we performed similar in vitro experiments as described above in primary cortical cultures obtained from the cortex of homozygous Sey/Sey mice (Hill et al. 1991). Remarkably, in the absence of Pax6, there was no significant change in the number of TuJ-1 or nestin-positive cells in CMV-Trim11-transfected cultures from the mutant cortex (Supplemental Fig. S5). Together, these findings indicate that by inhibiting the transcriptional activity of neurogenic factor Pax6 and facilitating the degradation of Pax6 protein, Trim11 indirectly modulates cortical neurogenesis.
Silencing of Trim11 results in an elevation of Pax6 inclusion bodies and apoptosis
The role of Trim11 in controlling Pax6 levels identified here, particularly Trim11 participation in the clearance of Pax6 inclusion bodies in vitro (see Fig. 3), led us to examine whether silencing Trim11 in vivo might also promote accumulation of Pax6 inclusion bodies. Following electroporation of the shTrim11#1 and shTrim11#2 constructs, total protein in pooled samples from three treated or untreated hemispheres was separated into soluble and insoluble fractions and probed for Pax6 by Western blot. Interestingly, the levels of insoluble Pax6 in inclusion bodies were approximately fourfold and threefold higher in shTrim11#1 and shTrim11#2-treated hemispheres than in untreated hemispheres, respectively (Fig. 7B; Supplemental Fig. S6, image I), while the levels of soluble Pax6 were increased by only ∼1.2–fold, with both Trim11 knockdown constructs (Fig. 7B; Supplemental Fig. S6, image S).
By degrading misfolded proteins, the UPS plays a crucial role in PQC. Inhibition of the PQC system normally tends to promote the accumulation of inclusion bodies and, ultimately, apoptosis (Alves-Rodrigues et al. 1998; McClellan et al. 2005; Muchowski and Wacker 2005; Rubinsztein 2006). Therefore, we performed TUNEL assays to determine whether Trim11 silencing in cortical cells in vivo also causes apoptotic cell death, and found that apoptosis was extensive in hemispheres treated with shTrim11(s) (Fig. 7C; Supplemental Fig. S6B). Remarkably, coelectroporation of shTrim11 plasmids together with the overexpression construct (Trim11-ires-eGFP) could completely rescue Trim11 knockdown-mediated apoptosis, supporting further the idea that the cell death in shTrim11-treated cortices is specifically due to silencing of Trim11 level (Supplemental Fig. S6B). We showed previously that upon conditional transgenic elevation of Pax6 (JoP6;Emx1Cre mice), cortical progenitors with distinct endogenous Pax6 levels respond with either p53-independent or premature differentiation (Berger et al. 2007). To determine whether the conditional Pax6 overexpression in vivo, like Trim11 silencing, leads to an increase in inclusion bodies, we examined soluble and insoluble fractions in cortical extracts from JoP6;Emx1Cre mice by Western blot. We found that Pax6-inclusion bodies accumulated in vivo in Pax6-overexpressing cortical progenitors, which exhibited a 4.87-fold enhancement in insoluble Pax6 fraction compared with controls (Supplemental Fig. S8). Together, these findings indicate that either diminishing Trim11 levels or a strong elevation of transgenic Pax6 in cortical progenitors could result in the accumulation of insoluble Pax6 protein.
Trim11 is a downstream target gene of Pax6
Interestingly, we furthermore found that Trim11 mRNA was substantially increased (1.9-fold) in the E12.5 cortex of these double transgenic mice relative to controls (Fig. 8A). In contrast, levels of Trim11 transcripts in cortical tissues, isolated from Sey/+ and Sey/Sey embryos at stages E12.5, were decreased to 64% and 43%, respectively, as compared with that of the wild-type control (Fig. 8A). These data indicate that the expression of Trim11 in vivo depends on the Pax6 dosage. Examination of a region of the mouse genomic sequence for the Trim11 gene that included 5 kb upstream of the starting codon of Trim11 revealed the presence of two putative Pax6-binding sites, BS(a) and BS(b), located at positions +5076–5096 and +11832–11852, respectively, within intron 3 (Fig. 8B). EMSA assays indicate that Pax6 did indeed bind to probes containing both Pax6-binding site sequences (Fig. 8C, lanes 2,8). Probe binding was completely abolished after mutation of these two Pax6-binding site sequences (Fig. 8C, lanes 4–6,10–12). Specificity of probe binding to Pax6 was confirmed by incubation with an anti-Pax6 antibody, which resulted in a super shift of the DNA–Pax6 protein–Pax6 antibody complex (Fig. 8C, lanes 3,9). To determine whether BS(a) and BS(b) are occupied by Pax6 protein in vivo, we performed ChIP assays using an extract from the mouse E15.5 cortex. In chromatin precipitated with the Pax6 antibody, two fragments (253 and 276 bp) were found at locations 4899–5151 and 11741–12016, respectively, which include the regions containing the Pax6-binding sites (Fig. 8D, lane 4). In contrast, a region (929–1208) located outside of intron 3 used as an external control was not amplified (Fig. 8D, lane 2). Together, these data indicate that the Pax6 protein binds to intron 3 of Trim11 both in vitro and in vivo.
Figure 8.
Pax6 regulates Trim11 transcription. (A) Results from qPCR analysis of Trim11 mRNA level in cortical extracts from Pax6/Small mutant homozygous (Sey/Sey), heterozygous (Sey/+), and wild-type embryos at E12.5 indicated a tight correlation between the cell dosage of Pax6 and the Trim11 expression level. In addition, the level of Trim11mRNA is elevated in the cortex of E12.5 embryos of transgenic mice conditionally overexpressing Pax6 in developng cortex (JoP6;EmxCre;) (Berger et al. 2007). The qPCR results are expressed relatively to GAPDH levels. (**) P < 0.001; n = 6. (B) The scheme depicts a region of the Trim11 gene, showing the relative position of two (a,b) potential Pax6-binding sites (BS) within intron 3 and the location of an external control (Ext) used in the chromatin immunoprecipitation (ChIP) assay (see below). A comparison of Pax6-binding site sequences (BS-wt) and the corresponding mutated sequence (BS-mut, underlined letters) is also presented. Mismatches relative to the Pax6 consensus sequence (P6CON) are presented in lowercase letters. (C) EMSA assay demonstrating that Pax6 bound strongly and specifically to both consensus binding sites identified in the Trim11 genomic sequence. (D) ChIP assay indicating that the Pax6 antibody specifically precipitated chromatin regions of the Trim11 genomic sequence that contain the Pax6-binding sites (lane 4), but did not precipitate chromatin outside of the Pax6-binding site (lane 2). (Lane 3) No binding was detected in the absence of the Pax6 antibody. (E) Histogram representing the ability of Pax6 to transactivate Trim11 minimal promoter-controlled reporters in HeLa cells cotransfected with CMV-Pax6. Mutation of the Pax6-binding site completely abolished Pax6-dependent reporter activity of the Trim11 minimal promoter. Luciferase activity was measured in triplicate and expressed as mean ± SD (error bars). (F) Proposed model of the autoregulatory feedback loop established by the genetic interplay between Trim11 and Pax6 (see Discussion).
To determine whether Pax6 binding to these specific sites in intron 3 of Trim11 is sufficient to drive transcription, we generated two minimal reporter constructs, pBS(a)-wt and pBS(b)-wt, incorporating the genomic regions containing the corresponding sites upstream of a luciferase reporter. To confirm the functional significance of each sequence, we additionally prepared mutant constructs, pBS(a)-mut and pBS(b)-mut, in which the pBS(a) (5′-TGGAGG-3′) and pBS(b) (5′-TTAAGC-3′) sites were both mutated to 5′-CTCGAG-3′ (Fig. 8B,E). Cotransfection of HeLa cells, which do not express Pax6, with a CMV-Pax6 expression plasmid and either of the wild-type reporter constructs strongly enhanced luciferase activity, whereas control cells transfected with the empty minimal reporter construct displayed a very low basal level of luciferase activity (Fig. 8E). The mutation of the Pax6-binding sites in these minimal promoters completely abolished Pax6-dependent activity in the reporter assay (Fig. 8E). These data indicate that the DNA sequences identified within intron 3 of the mouse Trim11 gene do indeed constitute Pax6-binding sites. A comparison of the Trim11 locus from mouse, rat, chimpanzee, and human revealed that the evolutionary conserved BS(b) fragment is located in a large block of highly conserved sequences (data not shown), suggesting that a genomic sequence including BS(b) is possibly a regulatory element of Trim11. Although further experiments will be required to fully characterize the enhancer activity of intron 3 of Trim11, the available evidence strongly suggests that Pax6 directly regulates the expression of Trim11 in vivo.
Discussion
While considerable effort has been devoted to characterizing Pax6 expression patterns and genetic regulatory elements (Callaerts et al. 1997; Simpson and Price 2002), mechanisms that act post-transcriptionally to regulate spatiotemporal variations in Pax6 protein levels in the developing cortex have not yet been defined. In this study, we demonstrated an important role for Trim11 in regulating the ubiquitination and proteasome-mediated degradation of Pax6 protein. To be recognized by the proteasome, protein substrates are covalently modified with ubiquitin in a process that involves the consecutive activities of enzymes designated E1, E2, and E3, the last of which determines substrate specificity (Hershko and Ciechanover 1998). Trim11 belongs to this latter class of proteins, possessing E3 ligase activity that allows it to transfer ubiquitin from E2 enzymes to specific substrates, including Humanin and ARC105 (Niikura et al. 2003; Ishikawa et al. 2006). Our present findings indicate that Pax6 is a novel substrate for the Trim11 E3 ligase activity, which is important in regulating Pax6 degradation. Several lines of evidence support this conclusion. First, Trim11 catalyzes the generation of Ub-conjugated Pax6. Second, Trim11-mediated degradation of Pax6 is significantly blunted by treatment with a proteasome inhibitor. Third, expression of Trim11 controls the endogenous level of Pax6 in the embryonic cortex. Finally, both in vivo Trim11 GOF and Trim11 silencing experimental paradigms demonstrated that Trim11 controlled Pax6 protein stability without affecting the transcription of either major Pax6 isoform.
Available data indicate that members of the Trim/RBCC protein family exert multiple functions in different cell types (Meroni and Diez-Roux 2005). Interestingly, subclasses of this protein are involved in epigenetic control, gene silencing, and control of transcription, rather than the canonically assumed E3 ubiquitin ligase function (Meroni and Diez-Roux 2005). Our analysis of the function of Trim11 subdomains indicated that the Trim11 fragment containing the BB and CC domains played a crucial role in Pax6 binding, and also mediated UPS-dependent Pax6 proteolysis and inhibited the transcriptional activity of Pax6 (Supplemental Fig. S9). Recent data provides evidence that several E3 ligases (e.g., CHIP, Parkin, and Dorfin) may participate in the clearance of misfolded disease-related proteins (Kitada et al. 1998; Cyr et al. 2002; Niwa et al. 2002; Elsasser and Finley 2005; McClellan et al. 2005). The proposed mechanism involves recognition of the specific misfolded protein, ubiquitination, and transport to the proteasome for degradation. Interestingly, the Trim11/B30.2 domain has been reported previously to interact with both proteasome subunits and ubiquitin (Suzumori et al. 2003). Surprisingly, we found that the B30.2 domain does not bind to native Pax6, but nevertheless mediates proteasome-dependent degradation of Pax6 inclusion bodies. We also demonstrated that the B30.2 domain of Trim11 specifically recognizes misfolded forms of Huntingtin and Arx proteins, mediating their degradation. Collectively, these findings suggest that Trim11 has dual functions: (1) As a RING E3 ligase, Trim11 degrades specific substrates, as shown for Humanin, ARC105 (Niikura et al. 2003; Ishikawa et al. 2006), and Pax6 (this study); and (2) as a “ubiquitin receptor” (Alves-Rodrigues et al. 1998; Elsasser and Finley 2005; Hicke et al. 2005), Trim11 appears to play a global role in PQC, transporting ubiquitinated, misfolded proteins, including Huntingtin, Arx, and as demonstrated here, Pax6, to the proteasome for proteolysis (Supplemental Fig. S9). Thus, mechanisms that regulate intracellular Trim11 content could offer potential targets for therapeutic intervention in protein misfolding-related diseases.
Protein misfolding is a result of conformational changes caused, for example, by biosynthetic errors, overtranslation, genetic mutation, or inadequate post-translational modifications (Frydman and Hartl 1996; Schubert et al. 2000). Molecular chaperones are specialized in the repair of such misfolded proteins (AlvesRodrigues et al. 1998; McClellan et al. 2005; Muchowski and Wacker 2005; Rubinsztein 2006). Achieving the correct balance between protein folding by chaperones and degradation of misfolded proteins by the proteasome is critical for cell viability. The detected significant amount of insoluble Pax6 form in the Trim11 loss-of-function (LOF) experiments and after Pax6 overexpression in cortical progenitors in vivo, subsequently causing apoptosis, possibly resulted from the above reasons. We demonstrated here that the Hsc70 chaperone inhibits B30.2-mediated degradation of insoluble Pax6 protein, suggesting that the interplay between Hsc70 and Trim11 may determine whether nascent or misfolded Pax6 proteins are salvaged by chaperone-mediated repair or targeted for elimination by the UPS. An interesting question for future studies is whether such a direct association exists between Trim11 and other molecular chaperons and, if so, what role do such interactions play.
Increasing evidence suggests that the UPS is involved in cortical neurogenesis and cell fate specification. For instance, factors having important roles in progenitor differentiation, such as P35, Dab1, and Notch, are substrates for degradation by the proteasome system (Patrick et al. 1998; Qiu et al. 2000; Arnaud et al. 2003; Bock et al. 2004). Also, targeted mutation of UBR1 and UBR2, encoding two E3 ubiquitin ligases in the N-end rule pathway, causes severe impairment of neurogenesis (An et al. 2006). Furthermore, ubiquitin C-terminal hydrolase L1 influences the morphology of neural progenitor cells and modulates their differentiation (Sakurai et al. 2006). In this study, we present evidence that Trim11 could repress Pax6-dependent neurogenesis in primary cortical cultures as well as in vivo in the developing cortex by mediating Pax6 degradation via the UPS. By interacting with Pax6, Trim11 represses Pax6-dependent transactivation of promoters containing Pax6 consensus binding sequences, possibly including the Ngn2 enhancer and the Er81 promoter, two important Pax6 downstream targets in neurogenesis (Scardigli et al. 2003; Tuoc and Stoykova 2008). Thus, by facilitating Pax6 degradation or inhibiting the transcriptional activity of Pax6 on downstream target genes, Trim11 negatively regulates the neurogenic function of Pax6 in developing cortex as demonstrated in this study (Fig. 8F, see model; Supplemental Fig. S9).
Transcription factors and the UPS have been shown to display intracellular “cross-talk”, forming autoregulatory feedback loops that balance their respective activities (Wu et al. 1993; Leng et al. 2003). Previous results from Pax6 GOF experiments in transgenic mice showed that Pax6 protein levels do not increase in proportion to gene copy number or Pax6 mRNA levels (Berger et al. 2007; Manuel et al. 2007). Instead, Pax6 protein levels are maintained close to physiological levels (Supplemental Fig. S8), suggesting the existence of a negative feedback mechanism that serves to stabilize Pax6 protein levels (Manuel et al. 2007). Based on the presented results, we propose that the autoregulatory loop between Trim11 and Pax6 assures a maintenance of the Pax6 protein level at a physiological level in developing cortex, thereby preventing a condition in which the Pax6 level would be dramatically elevated, leading to misfolding of part of the protein (see Fig. 8F). The proposed model suggests that if native Pax6 is elevated above the physiological limit, the Pax6 protein activates the transcription of Trim11 and Trim11 lowers down the cellular Pax6 protein via the proteosome system, thus reassuring the physiological condition. As long as the established basal Pax6–Trim11 regulatory loop is fully closed, the balance tends to favor Pax6 levels that maintain a healthy biological state and normal neurogenesis. In situations when the cellular level of Trim11 is severely dysregulated (by still unknown mechanisms), the fate of the cortical progenitors could be affected as demonstrated here in Trim11GOF and Trim11 knockdown experiments; namely, (1) extreme elevation of Trim11 in the progenitors leads to impaired neurogenesis via a drastic diminishing of the Pax6 level in cortical progenitors and down-regulation of the expression of Pax6 downstream targets, important for neurogenesis (e.g., possibly Tfs Ngn2, Er81), and (2) pronounced down-regulation of Trim11 level in the progenitors leads to extensive progenitor apoptosis partially due to a drastic elevation of the endogeneous Pax6 level far above its physiological level, which forms insoluble bodies and causes progenitor apoptosis. Establishment of autoregulatory feedback loops between genes that act as developmental regulators is an important regulatory mechanism in many biological systems (Hasty et al. 2002). Our findings demonstrate that Trim11 and the neurogenic transcriptional factor, Pax6, participate in such a genetic interplay, which appears to be crucially important in cortical neurogenesis.
Materials and methods
Animal
Small-eye (allele Sey), JoP6, Emx1-Cre mice were maintained in a C57BL6/J background. CD1 mice were used for in utero experiments and to prepare primary cortical cells. Animals were handled in accordance with the German Animal Protection Law and with the permission of the Bezirksregierung Braunschweig.
Plasmids and antibody
A list of plasmids and antibodies with detailed descriptions is provided in the Supplemental Material).
Yeast two-hybrid screen
The yeast two-hybrid screen was performed using the ProQuest two-hybrid system (Invitrogen), according to the manufacturer's instructions. Briefly, the truncated mouse Pax6 cDNA construct, Pax6ΔPD (Fig. 1A), corresponding to amino acid residues 129–422, was expressed as a GAL4 DNA-binding fusion protein and used as bait to probe an embryonic (E15.5) mouse cortex cDNA library (EvoQuest Custom cDNA library, Invitrogen). A total of 2.5 × 106 transformants were used in mating assays to identify Pax6ΔPD-interacting proteins. Clones expressing potential Pax6-interacting proteins were selected on the basis of their ability to grow on plates lacking leucine, tryptophan, and histidine, and supplemented with 65 mM 3-aminotriazole. All independent clones selected were assayed for β-galactosidase activity and confirmed by sequencing.
GST, HA pull-down, and HA coimmunoprecipitation assay
HeLa cells were transfected with CMV-Pax6 and maintained in culture for 48 h, and then proteins were extracted using the M-PER mammalian protein-extraction reagent (Pierce). Equal amounts of mammalian-expressed proteins were added to bacterially expressed, purified GST or GST-Trim11, and incubated for 2 h at 4°C in pull-down buffer containing 20 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA, 0.1% NP-40, and protease inhibitor cocktail (Roche).
For HA pull-down experiments, HeLa cells were transiently transfected with deletion constructs of HA-fused Trim11 and cultured for 48 h. Lysates containing these HA-fused proteins were cross-linked to anti-HA immobilized resin (Roche). Lysates of Pax6-transfected HeLa cells were added to protein-coupled resin and incubated for an additional 2 h.
For HA coimmunoprecipitation experiments, HeLa cells were transiently cotransfected with CMV-Pax6 and HA-Trim11, or HA-Trim11vector was electroporated into primary cortical cells prepared from E12.5 embryos. Forty-eight hours later, cells were harvested and proteins were immunoprecipitated using the ProFound HA Tag IP/Co-IP kit (Pierce), according to the manufacturer’s protocol. The experiments were carried out in the presence of the proteasome inhibitor, MG132 (20 μM). Input and eluted proteins from pull-down and coimmunoprecipitation assays were separated by SDS-PAGE and probed with the corresponding antibodies in Western blot analyses.
EMSA and ChIP assay
EMSA were described previously (Tuoc and Stoykova 2008). Immunoprecipitations were performed on chromatin extracted from embryonic (E15.5) mouse cortices, using a ChIP assay kit, according to the manufacturer's instructions (Upstate Biotechnology). Polyclonal Pax6 antibodies (BABCO) or preimmune serum (control) were used for immunoprecipitations (10 μg antibody per immunoprecipitation). Primer sequences are listed in Supplemental Table 1. A detailed description is provided in the Supplemental Material.
Cell cultures, transfection, luciferase assay
NIH-3T3, HeLa, and Neuro2A cells were maintained and cultured in DMEM medium plus 10% FCS. Cells were transfected using Lipofectamine 2000 according to the supplier’s guidelines. Luciferase activity in cell lysates was quantified using a luciferase assay system (Promega) according to the manufacturer’s instructions.
Measurement of the half-life of Pax6 protein
Twenty-four hours after transfecting HeLa cells with the indicated combination of plasmids, cycloheximide (20 μg/mL) was added. Protein levels were determined by collecting cells at different time points and performing immunoblotting, as described above. The relative amount of Pax6 protein was quantified densitometrically using ImageJ software.
In vivo ubiquitination assay
HeLa cells were transiently transfected with mammalian expression plasmids for His-Ub, HA-Trim11, and Flag-Pax6, as indicated. Twenty four hours after transfecting, MG132 (20 μM) was added and cells were incubated for an additional 8 h. After harvesting, cell lysates were immunoprecipitated using the Flag-tagged Protein Immunoprecipitation Kit (Sigma) according to the manufacturer's protocol. Input proteins and proteins eluted from immunoprecipitates were probed by Western blotting.
Analysis of steady-state protein levels
HeLa cells were cotransfected with CMV-Pax6 and either HA-Trim11 or EV. Twenty-four hours after transfecting, cells were treated with MG132 (20 μM) for an additional 24 h before harvesting. Lysates of harvested cells were separated by SDS-PAGE and probed for Pax6 protein by Western blotting using an anti-Pax6 antibody. As a control for equal loading, blots were subsequently reprobed with an anti-β-actin antibody.
Preparation of detergent-soluble and detergent-insoluble fractions
Mouse brains or primary cortical cells were collected and homogenized in five volumes of ice-cold 0.25 M sucrose buffer (50 mM Tris-HCl at pH 7.4, 1 mM EDTA) containing protease inhibitors. Homogenates were centrifuged at 500g for 10 min at 4°C, and the resulting supernatants were lysed with an equal volume of cold sucrose buffer containing 1% Triton X-100. Lysates were centrifuged at 13,000g for 15 min at 4°C to separate supernatants (fractions soluble in 0.5% Triton X-100) and pellets. Pellets were resuspended in 1% SDS in PBS (Triton X-100-insoluble fractions).
Generation and characterization of Trim11 RNAi vector
Oligonucleotide were selected, synthesized commercially, and cloned into the pSilencer-2.0 vector (Ambion), which places shRNA expression under the control of the U6 promoter. In this study, seven Trim11 shRNA plasmids (shTrim11) and a control sequence lacking homology with any known mRNA were used. All shRNA plasmids were cotransfected with HA-Trim11 into NIH-3T3 and Neuro2A cells or electroporated into primary cortical cells and cultured for 2–3 d. Western blotting and qPCR were used to analyze the efficiency of exogenous and endogenous Trim11 knockdown, respectively, by shTrim11(s). Two shTrim11 (shTrim11#1 and shTrim11#2) with the sequence 5′-GGTTTACTGCAGGACATTA-3′ and 5′-GTGTAAAGAAAC TGCCAAC-3′ were found to most efficiently knock down Trim11 expression and were used in all subsequent experiments.
Primary culture of cortex tissues and electroporation
Preparation of primary cortical cells and culture conditions have been described previously (Gotz et al. 1998; Tuoc and Stoykova 2008). The procedure is described in detail in the Supplemental Material.
In utero electroporation
In utero electroporation was performed using 2.5 μg/μL endotoxin-free DNA plasmids prepared using Endo Free Plasmid (Qiagen). In knockdown experiments shown in Figure 7 and Supplemental Figure S6A, an eGFP plasmid was coinjected with shTrim11 or EV at a concentration ratio of 1:4. In knockdown and rescue experiments as shown in Supplemental Figure S6B, eGFP plasmids were coinjected with shTrim11 at a concentration ratio of 1:1. E12.5–E13.5 CD1 mice were anesthetized by intraperitoneal injection of Ketamine/Xylazine (100/10 mixture, 0.1 mg per gram body weight). The uterine horns were carefully exposed through the incision using O-ringed forceps and humidified gauze pads. Plasmids were injected with 0.05% Fast Green (Sigma) into the telecephalic lateral ventricles of embryos using pulled glass capillaries. Five electrical pulses were applied at 30–35 V using platinum tweezer electrodes (CUY650P3, BEX Co., Ltd.) and an ECM-830 BTX square-wave electroporator (CUY21, BEX Co., Ltd). The uterine horns were then replaced in the abdominal cavity and skins were sutured using a surgical needle and thread. Three days after electroporation, pregnant mice were sacrificed and embryos were processed for histological analysis.
Immunohistochemistry, TUNEL assay
Protocols were described previously (Gotz et al. 1998; Berger et al. 2007). The procedures are described in detail in the SupplementalMaterial.
qPCR analysis
Total RNA isolated from the cortex was quantified spectrophotometrically and used in quantitative RT–PCR analyses employing the QuantiTect Rev.Transcription and the QuantiTect SYBR Green PCR Kits (Qiagen). The assays were performed in triplicate and normalized to internal 18S or GAPDH. Primer sequences are listed in Supplemental Table 1.
Statistical analysis
Results are presented as means ± standard deviation (SD); the number of experiments is indicated in the figure legends. Statistical differences were evaluated using Student’s t-tests.
Acknowledgments
We gratefully acknowledge S. Schlott and M. Daniel for outstanding technical assistance, L. Luo for his helpful advice, discussions, and B. Kasapoglu for her participation at the beginning of the study. We are very grateful to F. Cecconi for critical comments on the manuscript. We thank T. Niikura (Keio University School of Medicine), D. Bohmann (University of Rochester Medical Center), C. Krull (University of Missouri-Colombia), J. Golden (University of Pennsylvania), and J. Keller (University of Kentucky) for providing reagents. This work was supported by the Max Planck Gesellschaft and the DFG- CMPB.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.471708.
References
- Alves-Rodrigues A., Gregori L., Figueiredo-Pereira M.E. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci. 1998;21:516–520. doi: 10.1016/s0166-2236(98)01276-4. [DOI] [PubMed] [Google Scholar]
- An J.Y., Seo J.W., Tasaki T., Lee M.J., Varshavsky A., Kwon Y.T.2006Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway Proc. Natl. Acad. Sci. 1036212–6217. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud L., Ballif B.A., Cooper J.A. Regulation of protein tyrosine kinase signaling by substrate degradation during brain development. Mol. Cell. Biol. 2003;23:9293–9302. doi: 10.1128/MCB.23.24.9293-9302.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J., Berger S., Tuoc T.C., D’Amelio M., Cecconi F., Gorski J.A., Jones K.R., Gruss P., Stoykova A.2007Conditional activation of Pax6 in the developing cortex of transgenic mice causes progenitor apoptosis Development 134 13111322 [DOI] [PubMed] [Google Scholar]
- Bishop K.M., Rubenstein J.L., O'Leary D.D. Distinct actions of Emx1, Emx2, and Pax6 in regulating the specification of areas in the developing neocortex. J. Neurosci. 2002;22:7627–7638. doi: 10.1523/JNEUROSCI.22-17-07627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock H.H., Jossin Y., May P., Bergner O., Herz J. Apolipoprotein E receptors are required for reelin-induced proteasomal degradation of the neuronal adaptor protein Disabled-1. J. Biol. Chem. 2004;279:33471–33479. doi: 10.1074/jbc.M401770200. [DOI] [PubMed] [Google Scholar]
- Callaerts P., Halder G., Gehring W.J. PAX-6 in development and evolution. Annu. Rev. Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Cyr D.M., Hohfeld J., Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- Elsasser S., Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Epstein J., Cai J., Glaser T., Jepeal L., Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- Frydman J., Hartl F.U. Principles of chaperone-assisted protein folding: Differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- Gotz M., Stoykova A., Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Guillemot F., Molnar Z., Tarabykin V., Stoykova A. Molecular mechanisms of cortical differentiation. Eur. J. Neurosci. 2006;23:857–868. doi: 10.1111/j.1460-9568.2006.04626.x. [DOI] [PubMed] [Google Scholar]
- Hasty J., McMillen D., Collins J.J. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- Heins N., Malatesta P., Cecconi F., Nakafuku M., Tucker K.L., Hack M.A., Chapouton P., Barde Y.A., Gotz M. Glial cells generate neurons: The role of the transcription factor Pax6. Nat. Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicke L., Schubert H.L., Hill C.P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hill R.E., Favor J., Hogan B.L., Ton C.C., Saunders G.F., Hanson I.M., Prosser J., Jordan T., Hastie N.D., van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Tachikawa H., Miura Y., Takahashi N. TRIM11 binds to and destabilizes a key component of the activator-mediated cofactor complex (ARC105) through the ubiquitin–proteasome system. FEBS Lett. 2006;580:4784–4792. doi: 10.1016/j.febslet.2006.07.066. [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Leng R.P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Mallamaci A., Stoykova A. Gene networks controlling early cerebral cortex arealization. Eur. J. Neurosci. 2006;23:847–856. doi: 10.1111/j.1460-9568.2006.04634.x. [DOI] [PubMed] [Google Scholar]
- Manuel M., Georgala P.A., Carr C.B., Chanas S., Kleinjan D.A., Martynoga B., Mason J.O., Molinek M., Pinson J., Pratt T., et al. Controlled overexpression of Pax6 in vivo negatively autoregulates the Pax6 locus, causing cell-autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization. Development. 2007;134:545–555. doi: 10.1242/dev.02764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan A.J., Tam S., Kaganovich D., Frydman J. Protein quality control: Chaperones culling corrupt conformations. Nat. Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- Meroni G., Diez-Roux G. TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- Miller V.M., Nelson R.F., Gouvion C.M., Williams A., Rodriguez-Lebron E., Harper S.Q., Davidson B.L., Rebagliati M.R., Paulson H.L. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J. Neurosci. 2005;25:9152–9161. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski P.J., Wacker J.L. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Muzio L., DiBenedetto B., Stoykova A., Boncinelli E., Gruss P., Mallamaci A. Emx2 and Pax6 control regionalization of the pre-neuronogenic cortical primordium. Cereb. Cortex. 2002;12:129–139. doi: 10.1093/cercor/12.2.129. [DOI] [PubMed] [Google Scholar]
- Nasrallah I.M., Minarcik J.C., Golden J.A. A polyalanine tract expansion in Arx forms intranuclear inclusions and results in increased cell death. J. Cell Biol. 2004;167:411–416. doi: 10.1083/jcb.200408091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura T., Hashimoto Y., Tajima H., Ishizaka M., Yamagishi Y., Kawasumi M., Nawa M., Terashita K., Aiso S., Nishimoto I. A tripartite motif protein TRIM11 binds and destabilizes Humanin, a neuroprotective peptide against Alzheimer's disease-relevant insults. Eur. J. Neurosci. 2003;17:1150–1158. doi: 10.1046/j.1460-9568.2003.02553.x. [DOI] [PubMed] [Google Scholar]
- Niwa J., Ishigaki S., Hishikawa N., Yamamoto M., Doyu M., Murata S., Tanaka K., Taniguchi N., Sobue G. Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity. J. Biol. Chem. 2002;277:36793–36798. doi: 10.1074/jbc.M206559200. [DOI] [PubMed] [Google Scholar]
- Patrick G.N., Zhou P., Kwon Y.T., Howley P.M., Tsai L.H. p35, the neuronal-specific activator of cyclin-dependent kinase 5 (Cdk5) is degraded by the ubiquitin-proteasome pathway. J. Biol. Chem. 1998;273:24057–24064. doi: 10.1074/jbc.273.37.24057. [DOI] [PubMed] [Google Scholar]
- Qiu L., Joazeiro C., Fang N., Wang H.Y., Elly C., Altman Y., Fang D., Hunter T., Liu Y.C. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- Quinn J.C., Molinek M., Martynoga B.S., Zaki P.A., Faedo A., Bulfone A., Hevner R.F., West J.D., Price D.J. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev. Biol. 2006;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remedios R., Huilgol D., Saha B., Hari P., Bhatnagar L., Kowalczyk T., Hevner R.F., Suda Y., Aizawa S., Ohshima T., et al. A stream of cells migrating from the caudal telencephalon reveals a link between the amygdala and neocortex. Nat. Neurosci. 2007;10:1141–1150. doi: 10.1038/nn1955. [DOI] [PubMed] [Google Scholar]
- Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sakurai M., Ayukawa K., Setsuie R., Nishikawa K., Hara Y., Ohashi H., Nishimoto M., Abe T., Kudo Y., Sekiguchi M., et al. Ubiquitin C-terminal hydrolase L1 regulates the morphology of neural progenitor cells and modulates their differentiation. J. Cell Sci. 2006;119:162–171. doi: 10.1242/jcs.02716. [DOI] [PubMed] [Google Scholar]
- Scardigli R., Baumer N., Gruss P., Guillemot F., Le Roux I. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- Schedl A., Ross A., Lee M., Engelkamp D., Rashbass P., van Heyningen V., Hastie N.D. Influence of PAX6 gene dosage on development: Overexpression causes severe eye abnormalities. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- Schmahl W., Knoedlseder M., Favor J., Davidson D. Defects of neuronal migration and the pathogenesis of cortical malformations are associated with Small eye (Sey) in the mouse, a point mutation at the Pax-6-locus. Acta Neuropathol. (Berl.) 1993;86:126–135. doi: 10.1007/BF00334879. [DOI] [PubMed] [Google Scholar]
- Schubert U., Anton L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Simpson T.I., Price D.J. Pax6; a pleiotropic player in development. Bioessays. 2002;24:1041–1051. doi: 10.1002/bies.10174. [DOI] [PubMed] [Google Scholar]
- Stoykova A., Fritsch R., Walther C., Gruss P. Forebrain patterning defects in Small eye mutant mice. Development. 1996;122:3453–3465. doi: 10.1242/dev.122.11.3453. [DOI] [PubMed] [Google Scholar]
- Stoykova A., Gotz M., Gruss P., Price J. Pax6-dependent regulation of adhesive patterning, R-cadherin expression and boundary formation in developing forebrain. Development. 1997;124:3765–3777. doi: 10.1242/dev.124.19.3765. [DOI] [PubMed] [Google Scholar]
- Stoykova A., Treichel D., Hallonet M., Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzumori N., Burns K.H., Yan W., Matzuk M.M. RFPL4 interacts with oocyte proteins of the ubiquitin-proteasome degradation pathway. Proc. Natl. Acad. Sci. 2003;100:550–555. doi: 10.1073/pnas.0234474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tole S., Remedios R., Saha B., Stoykova A. Selective requirement of Pax6, but not Emx2, in the specification and development of several nuclei of the amygdaloid complex. J. Neurosci. 2005;25:2753–2760. doi: 10.1523/JNEUROSCI.3014-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc T.C., Stoykova A. Er81 is a downstream target of Pax6 in cortical progenitors. BMC Dev. Biol. 2008;8:23. doi: 10.1186/1471-213X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren N., Caric D., Pratt T., Clausen J.A., Asavaritikrai P., Mason J.O., Hill R.E., Price D.J. The transcription factor, Pax6, is required for cell proliferation and differentiation in the developing cerebral cortex. Cereb. Cortex. 1999;9:627–635. doi: 10.1093/cercor/9.6.627. [DOI] [PubMed] [Google Scholar]
- Wu X., Bayle J.H., Olson D., Levine A.J. The p53-mdm-2 autoregulatory feedback loop. Genes & Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]