Abstract
The transcription factor PU.1 is an important regulator of hematopoiesis; precise expression levels are critical for normal hematopoietic development and suppression of leukemia. We show here that noncoding antisense RNAs are important modulators of proper dosages of PU.1. Antisense and sense RNAs are regulated by shared evolutionarily conserved cis-regulatory elements, and we can show that antisense RNAs inhibit PU.1 expression by modulating mRNA translation. We propose that such antisense RNAs will likely be important in the regulation of many genes and may be the reason for the large number of overlapping complementary transcripts with so far unknown function.
Keywords: Noncoding antisense RNA, upstream and intronic regulatory elements, coordinated expression of the target and regulator, translation stalling
Recent genomic studies are uncovering an abundance of bidirectional transcription of eukaryotic genes across metazoan genomes (Werner and Berdal 2005) and have revealed a large number of overlapping transcripts in ∼50% of all investigated cases (Cheng et al. 2005). These and other recent reports are changing the notion of natural antisense transcripts as transcriptional noise and rather suggest an important role as regulatory tools in the fine regulation of genes (Lee and Lu 1999; Sleutels et al. 2002; Katayama et al. 2005). However, very little is known about the precise function of natural antisense transcripts as well as the molecular mechanisms of their biogenesis.
The transcription factor PU.1 is a hematopoietic lineage-specific ETS family member (Moreau-Gachelin et al. 1988) that is absolutely required for normal hematopoiesis (Tenen 2003). The level of PU.1 expression is critical for specifying cell fate, and, if perturbed, even modest decreases in PU.1 can lead to leukemias and lymphomas (Moreau-Gachelin et al. 1988; DeKoter and Singh 2000; Anderson et al. 2002; Rothenberg and Anderson 2002; Dahl et al. 2003; Cook et al. 2004; Rosenbauer et al. 2004, 2006; Huang et al. 2008). Previous reports demonstrated regulation of the PU.1 gene through the proximal promoter (Chen et al. 1995) and an upstream regulatory element (URE) located −14 kb and −17 kb upstream of the transcription start sites (TSS) in mice and humans, respectively (Li et al. 2001; Rosenbauer et al. 2004; Okuno et al. 2005). Removal of the URE by homologous recombination resulted in 80% reduction of PU.1 expression in bone marrow compared with wild-type mice and development of leukemias and lymphomas (Rosenbauer et al. 2004, 2006).
In this study, we demonstrate that the PU.1 locus gives rise to both mRNA and natural noncoding antisense transcripts that are originating from an intronic promoter. We show that these antisense transcripts have a regulatory function and control PU.1 gene expression via the regulation of translation. We demonstrate that the URE is required to drive both sense and antisense transcription, and that the URE physically interacts with both sense and antisense promoters. Our results demonstrate a novel mechanism of control of gene regulation through coordinated expression of the target gene and its regulatory natural antisense transcripts.
Results
Identification of naturally occurring antisense transcripts overlapping the PU.1 coding region
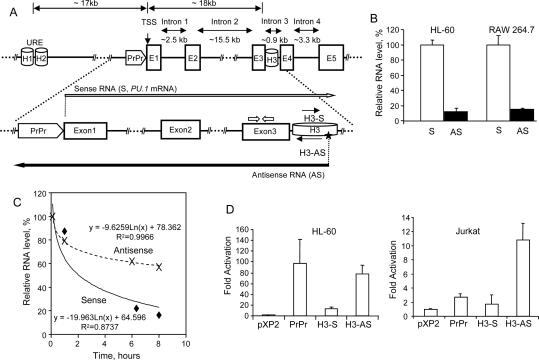
We discovered the presence of antisense transcripts in the intronic regions (introns 1, 2, and 3) and in the region upstream of the PU.1 mRNA TSS (Fig. 1A). These transcripts were found in a variety of PU.1 expressing and nonexpressing human and murine tissue culture and primary hematopoietic cells (Supplemental Fig. S1). 5′-RACE experiments revealed the antisense TSS (ATSS) in intron 3 (Supplemental Fig. S2). Interestingly, the ATSS was located within an evolutionarily conserved cis-element within intron 3 (H3) (Fig. 1A; Supplemental Fig. S3). Like the URE and the proximal promoter, H3 coincided with a previously reported DNase I-hypersensitive site (Li et al. 2001), which indicates regions bound by active transcription factor complexes. We could not detect antisense 3′ ends by 3′-RACE, probably due to the fact that these transcripts are heterogeneous in length and are very large in size, >9 kb. Northern blotting analysis revealed an almost equal amount of the antisense transcripts within the nuclear and cytoplasmic compartments and partial polyadenylation of the antisense RNAs; the majority of the detected antisense transcripts were not polyadenylated (data not shown). We quantified relative levels of the antisense RNAs by real-time PCR (Fig. 1B). The levels of antisense RNAs were ∼12%–15% of PU.1 mRNA level (sense transcript) in high-PU.1-expressing human and murine cells (HL-60 and RAW 264.7). Finally, we assessed the stability of sense and antisense transcripts. Noteworthy, the antisense RNA turned out to be stable, and its half-life was markedly higher than that of PU.1 mRNA (Fig. 1C).
Figure 1.
Both strands of the PU.1 gene are transcribed, and homology region H3 confers promoter activity in the antisense orientation. (A, top panel) Linear diagram showing the positions of the PU.1 gene locus homology regions URE (H1 + H2), proximal promoter (PrPr), and H3, as well as exons 1–5 (E1–E5). (Bottom panel) Diagram showing the position of the sense- and antisense-specific primers in exon 3 (block arrows), and the ATSS (star), corresponding to antisense RNA (AS; long black arrow). The long white arrow indicates the orientation of the sense RNA (S; PU.1 mRNA). Two short black arrows indicate orientation of the homology region H3 fragment–luciferase constructs (H3-S and H3-AS, sense and antisense, respectively). (B) Quantification of the PU.1 gene antisense transcripts by real-time strand-specific RT–PCR in HL-60 and RAW 264.7 cells. Strand-specific RT was performed for sense RNA (PU.1 mRNA) and for antisense RNA (AS) using sense- and antisense-specific primers (as shown in A), followed by PCR with the respective primer pair (see the Supplemental Material for oligonucleotide sequences and the Materials and Methods and legend to Supplemental Fig. S1 for a detailed protocol for strand-specific RT–PCR). Expression values are shown as the percent of sense transcript expression (100%). Mean values and standard deviation (error bars) based on three real-time PCR runs are displayed. (C) Antisense RNAs (AS) are more stable than sense RNAs (S). Shown is a time course of sense and antisense RNA levels isolated from bone marrow mononuclear cells derived from wild-type mice. Cells were treated with Actinomycin D (Alexis Biochemicals) at 10 μg/mL. An equal amount of viable cells (0.5 million), counted after Ficoll gradient purification, were collected at 0, 1, 6.5, and 8 h of treatment. Total RNA was isolated as described in the Materials and Methods and subjected to sense- and antisense-specific quantitative RT–PCR. The primers used for quantitative RT–PCR are described in the Supplemental Material, “Primers Used for Real-Time RT–PCR,” and shown in Supplemental Figure S1A. Correlation coefficients (R2) and equations of the exponential regression curves are indicated. (D) Homology region H3 confers promoter activity in the antisense orientation. The 215-bp fragment of homology region H3 was inserted into the pXP2 luciferase reporter vector in both sense (H3-S) and antisense (H3-AS) orientation. The PU.1 proximal promoter (PrPr) construct was described previously (Li et al. 2001). The pXP2 luciferase vector alone, as well as vectors including the PU.1 proximal promoter or H3 constructs were transiently transfected into HL-60 (left panel) and Jurkat (right panel) cells. Shown is the fold change in luciferase activity compared with pXP2 alone. Data are presented as the mean ± standard deviation based on three to six data points for each construct.
Expression of both sense and antisense PU.1 gene RNAs is dependent on the URE
Little is known about the molecular details of what regulates the biosynthesis of natural antisense RNA transcripts. To test for a possible cis-regulatory function of the PU.1 intron 3 fragment, we analyzed luciferase constructs fused to the entire conserved H3 region (Fig. 1A). The results demonstrate that H3 has promoter activity and can activate the luciferase gene in both high-PU.1-expressing (HL-60) and in nonexpressing (Jurkat) cells. Importantly, elevated activity was observed only when H3 was oriented in the antisense direction (Fig. 1D).
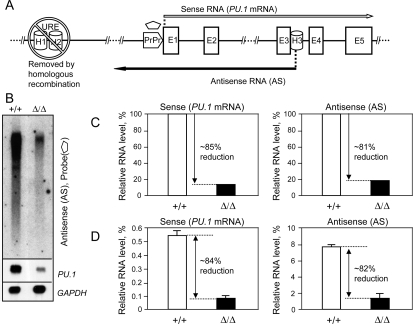
To test whether the distant PU.1 gene regulatory element URE was also involved in regulation of PU.1 antisense RNAs, we measured antisense transcript levels in cells derived from the UREΔ/Δ mouse model, in which the URE had been genetically deleted in vivo (Fig. 2A; Rosenbauer et al. 2004), and compared these levels with that in wild-type animals. We found an almost identical reduction of PU.1 sense and antisense transcripts in mutant animals (∼85% and ∼81%, respectively, [Fig. 2B–D] and ∼78% and ∼74%, respectively [Supplemental Fig. S4]), demonstrating that the URE is involved in regulation of both RNAs. The reduction in antisense RNA was not due to destabilization, because the pattern of polyadenylated antisense transcripts in the UREΔ/Δ mouse model (Rosenbauer et al. 2004) was largely identical to that observed in wild-type animals (Supplemental Fig. S5B,C). Furthermore, the direct assessment of stability of total antisense RNAs in wild type and UREΔ/Δ again revealed no significant difference (Supplemental Fig. S5D). In summary, our data demonstrate that the biogenesis of PU.1 gene mRNA as well as antisense RNAs rely on the URE.
Figure 2.
The URE is instrumental in PU.1 antisense RNA synthesis. (A) The linear diagram shows the position of the PU.1 gene locus homology regions: the URE (H1 + H2), removed by homologous recombination (Rosenbauer et al. 2004), proximal promoter (PrPr), and H3; initiation sites and orientation of the sense (long white arrow) and antisense (long black arrow) transcripts; and the position of the antisense-specific probe (pentagon). (B) Northern blot analysis illustrates down-regulation of the antisense RNAs in the UREΔ/Δ mouse model (Rosenbauer et al. 2004). RNAs were isolated from bone marrow of wild-type (+/+) and hypomorphic (Δ/Δ) animals. Two micrograms of polyA(+) were run side by side and hybridized with the probe specific to antisense RNAs (pentagon, A). The top panel demonstrates down-regulation of antisense RNAs. The middle panel demonstrates down-regulation of the PU.1 sense mRNA in the UREΔ/Δ mouse model, using a double-stranded murine PU.1 cDNA probe. The bottom panel depicts GAPDH mRNA levels as loading controls (see the Supplemental Material for the probe information). (C,D) The URE regulates expression of both sense and antisense RNAs. Relative sense and antisense RNA levels in bone marrow of wild type and animals with an URE deletion were quantitated by Northern blot phosphorescence imaging (normalized to GAPDH mRNA) (C) and, as corroboration, by strand-specific RT–PCR (graph values calculated as mean ± standard deviation of three real-time RT–PCR runs, normalized to GAPDH mRNA, measured by TaqMan real-time analysis) (D). See the Supplemental Material for the TaqMan probes and primers used for strand-specific RT–PCR.
The URE interacts with both the sense and the antisense promoters
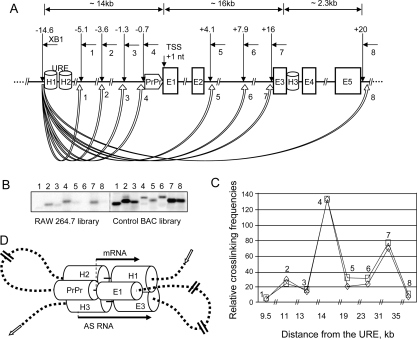
We tested whether the URE exerts its functional role through a direct interaction with H3 by using chromosome conformation capture (3C), which is based on cross-linking between distant chromosomal elements (Dekker et al. 2002; Tolhuis et al. 2002; Spilianakis et al. 2005). We applied a modified quantitative 3C protocol to the murine macrophage RAW 264.7 cell line. In these experiments, we quantified the cross-linking efficiencies between the distant regulatory element URE and several regions in the PU.1 locus extending from 5 kb upstream of to 20 kb downstream from the TSS (Fig. 3A). To avoid problems with the nonlinearity of PCR, we developed a simple and accurate approach exploiting Southern blot hybridization combined with further phosphorescence imaging (described in detail in the Supplemental Material). 3C experiments were also performed with primary human granulocytes and HL-60 promyelocytic cells (Supplemental Fig. S6; data not shown) with similar results. The different experiments demonstrated that specific signals were only observed with primers located at the URE, the proximal promoter, and the conserved H3 intronic element, but not at intervening genomic sequences (Fig. 3C).
Figure 3.
Quantitative 3C assays demonstrate the close physical proximity of the URE to the proximal promoter and H3 antisense promoter. (A) Diagram showing the genomic position of the homology regions (H1–H3 and proximal promoter [PrPr]), position and orientation of the primers used in the final 3C PCRs (common primer from the URE region: XB1; variable downstream primers: short horizontal arrows marked as 1–8), and locations of XbaI restriction enzyme sites (vertical arrows); block arrows marked as 1–8 represent calculated interaction frequencies. (B,C) Southern blot analysis of the linear amplification of the control and test libraries. Interaction frequencies were calculated by dividing the test 3C library (in the murine macrophage RAW 264.7 line) signals by control library (BAC) signals. The graphs represent results of two independent PCR/Southern blot experiments. (D) Schematic model of spatial organization of the regulatory elements of the PU.1 gene. The model depicts (1) the close physical proximity of the PU.1 gene locus homology regions (the URE [H1 and H2], proximal promoter [PrPr], and H3) and (2) the initiation and orientation of sense and antisense transcripts (long arrows). Short block arrows indicate genomic orientation of the PU.1 gene locus.
As depicted in Figure 3D, the results of the 3C experiments demonstrate that the URE controls the levels of sense and antisense transcription by physically interacting with the proximal promoter and the intronic conserved element, H3.
PU.1-encoded antisense transcripts negatively regulate PU.1 expression
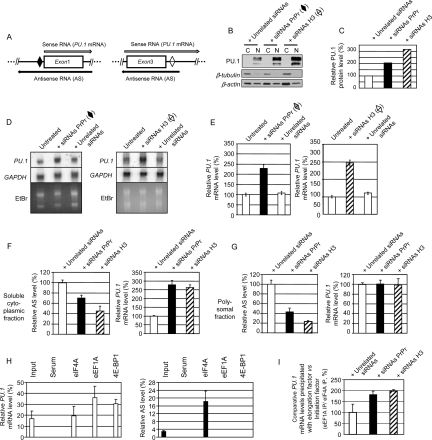
To test for a potential regulatory role of the antisense RNAs in PU.1 expression, we performed RNAi experiments with siRNAs targeting human PU.1 antisense RNAs using chemically modified strand-specific dsRNAs, thus avoiding recognition by complexes initiating cellular stress responses (Stealth RNAi; Invitrogen). Two target regions were chosen (Fig. 4, indicated as rhomboids). The first region was located 5′ upstream of the sense TSS, within the proximal promoter, and the second within homology region H3. Strikingly, the proximal promoter-specific and H3-specific siRNAs induced increases in PU.1 protein levels of ∼105% and ∼210%, respectively (Fig. 4B,C) and increases in PU.1 mRNA levels of 125 ± 11% (P < 0.001) and ∼145 ± 12% (P < 0.001), respectively (Fig. 4D,E). Similar results were obtained with siRNAs specifically targeting PU.1 antisense RNAs in U937, RAJI (human) and RAW 264.7, 416B (murine) cell lines (Supplemental Fig. S7). These results demonstrate that the PU.1 antisense transcripts act as negative regulators of PU.1 protein and PU.1 gene mRNA levels in both species. This observation raises the possibility that targeting these antisense transcripts could be used as a novel method to increase PU.1 levels and might provide a novel therapeutic tool to reverse the reduction in PU.1 expression levels that were reported in leukemias and lymphomas (Moreau-Gachelin et al. 1988; DeKoter and Singh 2000; Dahl et al. 2003; Cook et al. 2004; Rosenbauer et al. 2004, 2006; Mueller et al. 2006; Steidl et al. 2006, 2007).
Figure 4.
RNAi targeting antisense transcripts leads to an increase in PU.1 protein and PU.1 mRNA levels associated with decreased levels of polysomal antisense transcripts. (A) Diagrams showing position of sites in the proximal promoter and homology region H3 used to design complementary siRNA (black and white rhomboids, respectively). Long arrows indicate orientation of the sense (PU.1 mRNA) and antisense (AS) transcripts. (B,C) Knockdown of antisense transcripts using siRNAs complementary to RNAs covering the proximal promoter and homology region H3 in HL-60 cells leads to ∼105% and ∼210% increases in PU.1 protein, respectively. Cytoplasmic and nuclear extracts (marked C and N, respectively) (15 μg each) isolated from cells transfected with antisense-specific and unrelated siRNAs were used for Western blot analysis. (B, top panel) Result of probing the membrane with PU.1 antibody. (Bottom panels) Controls used were β-tubulin and β-actin. (C) Relative PU.1 protein levels in nuclear fractions of cells transfected with antisense-specific siRNAs and unrelated siRNAs. The vertical bars indicate the relative amount of PU.1 protein in nuclear fractions normalized to β-actin and shown as the percent of PU.1 protein levels in cells transfected with unrelated siRNAs (100%) (NIH ImageJ version 1.37k). (D,E) Knockdown of antisense transcripts using siRNAs complementary to the proximal promoter or homology region H3 in HL-60 cells leads to 125% ± 11% (P < 0.001) and 145% ± 12% (P < 0.001) increase in PU.1 mRNA, respectively. (D) Northern blot analysis of the PU.1 mRNA levels in total RNA samples using a double-stranded human PU.1 cDNA probe. GAPDH mRNA levels and ethidium bromide staining were used as loading controls (see the Supplemental Material for probe information). (E) Histograms showing relative PU.1 gene mRNA levels in cells either untreated or transfected with antisense-specific siRNAs or unrelated siRNAs (Invitrogen, Cat. No. 12935-400) as a control, normalized to 18S. Expression values are shown as the percent of PU.1 mRNA levels in untreated cells (100%). Mean values and standard deviations (error bars) based on three real-time PCR runs are displayed. (F) Knockdown of antisense transcripts using siRNAs complementary to the proximal promoter (PrPr) and homology region H3 in HL-60 cells leads to ∼30% and ∼55% decreases in PU.1 antisense RNAs, respectively (left panel) in the cytoplasmic subfractions, and to ∼175% and ∼160% increases in PU.1 sense mRNA, respectively (right panel). Expression values of PU.1 mRNA and antisense RNAs in the soluble cytoplasmic subfractions isolated from the cells transfected with antisense-specific siRNAs are shown as the percent of those transfected with unrelated siRNAs (100%). Mean values and standard deviation (error bars) based on four experiments are displayed. (G) Knockdown of antisense transcripts using siRNAs complementary to the proximal promoter and homology region H3 in HL-60 cells leads to ∼58% and ∼78% decreases in PU.1 antisense RNAs, respectively (left panel), and to no changes in PU.1 gene mRNA levels (right panel) in the polysomal fractions. Expression values of PU.1 mRNA and antisense RNAs in the polysomal fractions isolated from the cells transfected with antisense-specific siRNAs are shown as the percent of those transfected with unrelated siRNAs (100%). Mean values and standard deviations (error bars) based on four experiments are displayed. (H) Immunoprecipitation of sense/antisense PU.1 RNAs-translation factor complexes from total polysomal fraction RNA–translation factor complexes (pRIP) using antibodies against eIF4A (translation initiation factor), eEF1A (translation elongation factor), and 4E-BP1 (inhibitor of translation initiation). HL-60 cells were HCHO-cross-linked, and total polysomal fraction was isolated as shown in Supplemental Figure 9A. pRIP was performed as described in the Supplemental Material. Histograms show relative PU.1 gene mRNA and antisense RNA levels in the immunoprecipitates, normalized to GAPDH (left and right panels, respectively). Mean values and standard deviations (error bars) based on three real-time PCR runs are displayed. (I) Antisense PU.1 gene RNAs function by stalling the translation between the initiation and elongation steps. HL-60 cells were treated with antisense-specific and unrelated siRNAs, cross-linked (1% HCHO, 10 min), and the polysomal fractions was analyzed by pRIP with antibodies against eIF4A and eEF1A followed by real-time RT–PCR.
Mechanism of action of PU.1 gene antisense RNAs
There are several pathways through which PU.1 gene antisense RNAs might regulate PU.1 gene expression in hematopoietic cells. One potential pathway is RNAi (Matzke and Birchler 2005), in which RNA silencing can be achieved by induction of DNA methylation (RNA-directed DNA methylation) (Morris et al. 2004) brought about by siRNAs. We and others have observed an inverse correlation between levels of PU.1 promoter methylation and PU.1 mRNA expression in both human and murine cells (Amaravadi and Klemsz 1999; A.K. Ebralidze and D.G. Tenen, unpubl.). However, we did not detect naturally occurring siRNAs homologous to the exonic and intronic segments of the PU.1 gene by high-resolution Northern blot analyses (Supplemental Fig. S8).
We therefore examined whether antisense RNAs were regulating PU.1 translation. To directly test this hypothesis, we determined changes in PU.1 mRNA and antisense RNAs in different cellular compartments before and after treatment with siRNAs against antisense RNAs (Supplemental Fig. S9A). The purity of the fractions was confirmed by TaqMan and Northern blot analyses of U1 and U2 small nuclear RNAs (snRNAs) (Supplemental Fig. S9B,C). We again found increases in PU.1 protein and PU.1 mRNA levels upon siRNA treatment in total cellular extracts (Supplemental Fig. S10A,B and C, left bar graph). No down-regulation of the antisense RNAs in the nuclear fractions of the samples treated with the proximal promoter-specific and H3-specific siRNAs (Supplemental Fig. S10D, right bar graph) was observed, suggesting that the antisense RNAi-mediated effect occurred mainly in the nonnuclear compartment, which is in agreement with a previous report (Zeng and Cullen 2002). However, our results show a clear down-regulation of antisense RNAs in the soluble cytoplasmic subfractions (∼30% and ∼55% in samples treated with proximal promoter- and H3-specific siRNAs, respectively) (Fig. 4F, left bar graph), and, most importantly, in the polysome-enriched fractions (∼69% and ∼77% in samples treated with proximal promoter- and H3-specific siRNAs, respectively) (Fig. 4G, left bar graph). This indicates that this fraction contains both sense as well as antisense RNA. Most importantly, unlike in total, nuclear, and soluble cytoplasmic fractions (Supplemental Fig. S10C,D, left bar graphs; Fig. 4F, right bar graph, respectively), we did not observe increases in PU.1 sense transcripts in fractions enriched in polysome-bound mRNAs (Fig. 4G, right bar graph), strongly suggesting that the increased levels of PU.1 protein in the siRNA-treated cells are due to an increased ratio of sense versus antisense transcripts in the polysomes and not a down-regulation of mRNA levels, leading to a net increase in translation of PU.1 mRNA. Our results therefore support a model in which the antisense transcript negatively regulates the PU.1 gene in myeloid cells through translational interference. To elucidate the mechanism of translational interference exerted by antisense transcripts, we performed RNA immunoprecipitation on the purified cross-linked polysomal fraction (pRIP) with antibodies against three translation factors: eIF4A (regulates translation initiation), eEF1A (important for translation elongation), and 4E-BP1 (inhibitor of translation initiation) as described (Gilbert et al. 2004; Shamovsky et al. 2006). We detected in vivo complex formation between PU.1 mRNA and all three factors (Fig. 4H, left panel). In contrast, antisense RNAs formed complexes only with the translation initiation factor eIF4A (Fig. 4H, right panel). To test whether antisense RNA interferes with the formation of elongating eEF1A–mRNA complexes, we combined RNAi experiments with pRIP (Fig. 4I). This experiment showed that this was indeed true, since knockdown of the antisense transcripts led to an ∼89%–97% increase in PU.1 mRNA association with the elongation factor eEF1A in vivo. These experiments suggest that antisense RNAs exert their regulatory function through stalling translation between the initiation and elongation steps. Further molecular studies will be required to unveil the precise mechanism of how the binding of eEF1A is inhibited.
Discussion
Functional role of PU.1 noncoding antisense RNAs
In this study, we present evidence that RNA-mediated translational interference is not restricted to microRNAs, but can also be mediated by long, noncoding natural antisense RNAs. We hypothesize that such antisense transcripts serve to keep the levels of PU.1 from being too high in expressing cells. Antisense transcripts were also found in cells not expressing PU.1 mRNA, similar to what has been observed with the c-fms locus (Tagoh et al. 2004). This may serve as an additional mechanism to keep PU.1 completely shut off in inappropriate cells such as T cells in which any expression would lead to adverse affects (Anderson et al. 2002). Given the number of sense–antisense transcription units recently discovered (Cheng et al. 2005), it is likely that such a mechanism will be exploited for the regulation of other genes.
Materials and methods
Detailed descriptions of methods are in the Supplemental Material.
Primary cells, cell lines, and transient transfection
Primary human granulocytes were isolated by dextran sedimentation followed by double immunomagnetic selection with CD15. PU.1-expressing human bipotential myeloid lines HL-60 and U937 and murine macrophage RAW 264.7, and PU.1 nonexpressing human Jurkat T-cell and murine T lymphocytic BW5147 and murine macrophage RAW 264.7 cell lines were obtained from American Type Culture Collection. The PU.1-expressing murine myeloid progenitor line 416B was described previously (Dexter et al. 1979; Li et al. 2001). Transfections of HL-60 and Jurkat cells were performed by electroporation. Primary bone marrow cells were isolated from mice harboring deletions of the URE (Rosenbauer et al. 2004).
RNAi knockdown of antisense transcripts
siRNA sequences, synthesis, and conditions for transfection are described in detail in the Supplemental Material.
Preparation of nuclear, cytoplasmic, and polysomal RNAs
Biochemical fractionation of nuclei was performed by hypotonic lysis with detergent followed by Dounce homogenization followed by sucrose sedimentation. Polysomes were isolated from cytoplasmic extracts by sucrose sedimentation. We used methods derived from the protocols developed for nuclei isolation by Blobel and Potter (1966), for cytoplasmic RNA preparation by del Prete et al. (2007), and for polysome isolation by Fennoy et al. (1997) with minor modifications.
3C
Conventional 3C protocol. We used the 3C procedure developed by Dekker and coworkers for detection of genomic loci interactions (Dekker et al. 2002).
Modified quantitative 3C protocol. We developed a modified 3C protocol using Southern blotting of PCR products and specific probes that can only hybridize with the specifically amplified ligation products. Quantitation of hybridization signals was performed using a Storm PhosphorImager (GE Healthcare). The method is described in detail in the Supplemental Material.
The sequences of the oligonucleotides and DNA probes used in Northern blotting, 5′- and 3′−RACE, real-time and strand-specific PCR, and 3C (including quantitation of restriction enzyme efficiency) are found in the Supplemental Material.
Acknowledgments
We acknowledge Phil Sharp, Henry Koon, Christopher Carpenter, Anna Krichevsky, Bertie Gottgens, and John Rinn, as well as Steffen Klippel, David Gonzalez, Hanna Radomska, Dana Neel, and other members of the Tenen laboratory for helpful discussions; Arnaud Mailleux for help with graphics; fellowships from the Lymphoma Research Foundation and a Young Investigators Grant from the Helmholtz Association to F.R.; grants from the Leukaemia Research Fund UK to C.B.; grants from the NIH and Canada Research Chair to K.B.; and NIH grant CA41456 to D.G.T.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1654808.
References
- Amaravadi L., Klemsz M.J. DNA methylation and chromatin structure regulate PU.1 expression. DNA Cell Biol. 1999;18:875–884. doi: 10.1089/104454999314737. [DOI] [PubMed] [Google Scholar]
- Anderson M.K., Weiss A.H., Hernandez-Hoyos G., Dionne C.J., Rothenberg E.V. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- Blobel G., Potter V.R. Nuclei from rat liver: Isolation method that combines purity with high yield. Science. 1966;154:1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Chen H., Ray-Gallet D., Zhang P., Hetherington C.J., Gonzalez D.A., Zhang D.E., Moreau-Gachelin F., Tenen D.G. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene. 1995;11:1549–1560. [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Cook W.D., McCaw B.J., Herring C., John D.L., Foote S.J., Nutt S.L., Adams J.M. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood. 2004;104:3437–3444. doi: 10.1182/blood-2004-06-2234. [DOI] [PubMed] [Google Scholar]
- Dahl R., Walsh J.C., Lancki D., Laslo P., Iyer S.R., Singh H., Simon M.C. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPα ratio and granulocyte colony-stimulating factor. Nat. Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- del Prete M.J., Vernal R., Dolznig H., Mullner E.W., Garcia-Sanz J.A. Isolation of polysome-bound mRNA from solid tissues amenable for RT–PCR and profiling experiments. RNA. 2007;13:414–421. doi: 10.1261/rna.79407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter T.M., Allen T.D., Scott D., Teich N.M. Isolation and characterisation of a bipotential haematopoietic cell line. Nature. 1979;277:471–474. doi: 10.1038/277471a0. [DOI] [PubMed] [Google Scholar]
- Fennoy S.L., Jayachandran S., Bailey-Serres J. RNase activities are reduced concomitantly with conservation of total cellular RNA and ribosomes in O2-deprived seedling roots of maize. Plant Physiol. 1997;115:1109–1117. doi: 10.1104/pp.115.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Kristjuhan A., Winkler G.S., Svejstrup J.Q. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- Huang G., Zhang P., Hirai H., Elf S., Yan X., Chen Z., Koschmieder S., Okuno Y., Dayaram T., Growney J.D., et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat. Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Lee J.T., Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Li Y., Okuno Y., Zhang P., Radomska H.S., Chen H., Iwasaki H., Akashi K., Klemsz M.J., McKercher S.R., Maki R.A., et al. Regulation of the PU.1 gene by distal elements. Blood. 2001;98:2958–2965. doi: 10.1182/blood.v98.10.2958. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Birchler J.A. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Moreau-Gachelin F., Tavitian A., Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- Morris K.V., Chan S.W., Jacobsen S.E., Looney D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Mueller B.U., Pabst T., Fos J., Petkovic V., Fey M.F., Asou N., Buergi U, Tenen D.G. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 2006;107:3330–3338. doi: 10.1182/blood-2005-07-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Huang G., Rosenbauer F., Evans E.K., Radomska H.S., Iwasaki H., Akashi K., Moreau-Gachelin F., Li Y., Zhang P., et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 2005;25:2832–2845. doi: 10.1128/MCB.25.7.2832-2845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F., Wagner K., Kutok J.L., Iwasaki H., Le Beau M.M., Okuno Y., Akashi K., Fiering S., Tenen D.G. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F., Owens B.M., Yu L., Tumang J.R., Steidl U., Kutok J.L., Clayton L.K., Wagner K., Scheller M., Iwasaki H., et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- Rothenberg E.V., Anderson M.K. Elements of transcription factor network design for T-lineage specification. Dev. Biol. 2002;246:29–44. doi: 10.1006/dbio.2002.0667. [DOI] [PubMed] [Google Scholar]
- Shamovsky I., Ivannikov M., Kandel E.S., Gershon D., Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- Sleutels F., Zwart R., Barlow D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., Flavell R.A. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Steidl U., Rosenbauer F., Verhaak R.G., Gu X., Ebralidze A., Otu H.H., Klippel S., Steidl C., Bruns I., Costa D.B., et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat. Genet. 2006;38:1269–1277. doi: 10.1038/ng1898. [DOI] [PubMed] [Google Scholar]
- Steidl U., Steidl C., Ebralidze A., Chapuy B., Han H.J., Will B., Rosenbauer F., Becker A., Wagner K., Koschmieder S., et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J. Clin. Invest. 2007;117:2611–2620. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagoh H., Schebesta A., Lefevre P., Wilson N., Hume D., Busslinger M., Bonifer C. Epigenetic silencing of the c-fms locus during B-lymphopoiesis occurs in discrete steps and is reversible. EMBO J. 2004;23:4275–4285. doi: 10.1038/sj.emboj.7600421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen D.G. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Werner A., Berdal A. Natural antisense transcripts: Sound or silence? Physiol. Genomics. 2005;23:125–131. doi: 10.1152/physiolgenomics.00124.2005. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Cullen B.R. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8:855–860. doi: 10.1017/s1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]