Abstract
Histone acetylation and deacetylation are among the principal mechanisms by which chromatin is regulated during transcription, DNA silencing, and DNA repair. We analyzed patterns of genetic interactions uncovered during comprehensive genome-wide analyses in yeast to probe how histone acetyltransferase (HAT) and histone deacetylase (HDAC) protein complexes interact. The genetic interaction data unveil an underappreciated role of HDACs in maintaining cellular viability, and led us to show that deacetylation of the histone variant Htz1p at Lys 14 is mediated by Hda1p. Studies of the essential nucleosome acetyltransferase of H4 (NuA4) revealed acetylation-dependent protein stabilization of Yng2p, a potential nonhistone substrate of NuA4 and Rpd3C, and led to a new functional organization model for this critical complex. We also found that DNA double-stranded breaks (DSBs) result in local recruitment of the NuA4 complex, followed by an elaborate NuA4 remodeling process concomitant with Rpd3p recruitment and histone deacetylation. These new characterizations of the HDA and NuA4 complexes demonstrate how systematic analyses of genetic interactions may help illuminate the mechanisms of intricate cellular processes.
Keywords: Systems biology, histone, NuA4, acetylation, DNA repair
Post-translational modifications of histones control many DNA-related processes (Kouzarides 2007). Dynamic histone (de)acetylation regulates gene transcription and silencing, chromosome condensation, DNA replication, and preservation of DNA integrity via DNA damage repair (Millar and Grunstein 2006). There are over 20 known histone acetyltransferases (HATs) and histone deacetylases (HDACs) in Saccharomyces cerevisiae; virtually all function as protein complexes (Lee and Workman 2007; Shahbazian and Grunstein 2007). These activities are coordinated in the cell, and comprise a system that dynamically regulates chromatin state. Systems with this many components are difficult to analyze using conventional genetics and biochemical methods, although some large-scale attempts have been made (Collins et al. 2007; Mitchell et al. 2008). Comprehensive assessment of this system is further complicated by the inclusion of essential genes (e.g., the essential acetyltransferase ESA1), requiring suitable conditional or hypomorphic query alleles. Moreover, recent studies in higher organisms have shown that HATs and HDACs have many substrates apart from histones (Glozak and Seto 2007; Xu et al. 2007), hinting that such substrates may exist in yeast as well.
Several recent studies have demonstrated that comprehensive genetic interaction profiling can effectively resolve complex pathways into conceptually and experimentally tractable modules (Tong et al. 2004; Schuldiner et al. 2005; Pan et al. 2006; Collins et al. 2007). Intergenic interactions can be either aggravating (negative), such as synthetic fitness or lethality defects (SFL), or alleviating (positive) such as synthetic rescue (SR). The genes involved can function either in a common essential pathway or in distinct but compensatory pathways converging on the same essential function (Hartman et al. 2001). Genetic interaction networks can be further organized into interacting functional modules based on a statistical analysis of the number of genetic interactions observed between sets of genes (Hartman et al. 2001; Tong et al. 2004; Schuldiner et al. 2005; Segre et al. 2005; Pan et al. 2006; Collins et al. 2007).
Here we present a comprehensive genetic interaction network of HAT and HDAC protein complexes in yeast, generated using “diploid-based Synthetic Lethality Analysis on Microarray” (dSLAM) (Pan et al. 2006). The high degree of connectivity in this network enabled classification of interactions across protein complexes and identification of gene sets that function together as modules. Analysis of these modules revealed that histone hyperacetylation is as deleterious as hypoacetylation, consistent with previous studies showing that balanced acetylation status is crucial for cell viability (Vogelauer et al. 2000). Genetic interactions between HDAC complex (Carmen et al. 1996) and complexes involved in acetylation or deposition of the histone H2A variant Htz1p led us to demonstrate that HDA is the previously unidentified deacetylase for Htz1p. Other genetic interactions revealed that Esa1p acetylates and stabilizes the nucleosome acetyltransferase of H4 (NuA4) subunit Yng2p, insights that helped formulate a new model for how the NuA4 complex undergoes rapid remodeling during the repair of DNA double-stranded breaks (DSBs). These new functions of the HDA and NuA4 complexes exemplify the value of comprehensive surveys of genetic interactions for exploring the roles of key protein complexes in controlling the dynamic balance of acetylation and deacetylation histones and other proteins.
Results
Features of the interaction network
We used dSLAM to determine genome-wide genetic interaction profiles of 38 query genes involved in histone (de)acetylation. Query genes included the HAT and HDAC catalytic subunits (Lee and Workman 2007) and associated protein complex subunits. Mutations used as queries included knockout deletions (KO) of 32 nonessential genes plus seven temperature-sensitive (Ts) or hypomorphic alleles of six essential genes (Fig. 1A; also see Supplemental Table S1 for a list of point mutations of the Ts and hypomorphic alleles). None of the essential genes had been used previously as query genes in genome wide studies. We validated 2823 unique pairwise genetic interactions involving 763 genes (∼12.5% of yeast genes) by tetrad dissection and/or random spore analysis. There were comprise 105 (∼4.2%) SR interactions, and 2718 SFL defects (Fig. 1A; see also Supplemental Fig. S1 for a high-resolution image of the entire clustogram; see Supplemental Table S2 for a complete list of genetic interaction pairs). Only 14% of the interactions identified here were reported previously. Essential query genes had 188 genetic interaction partners on average, whereas nonessential counterparts had 42, comparable with previous studies (Tong et al. 2004; Davierwala et al. 2005).
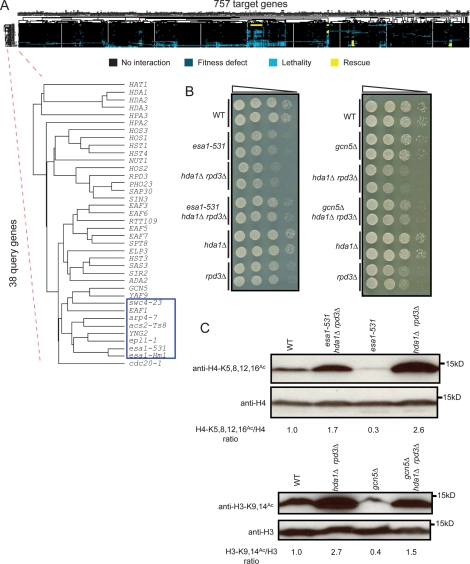
Figure 1.
Global features of genetic interaction patterns. (A) Full hierarchical clustering of genetic interaction patterns. Blue, yellow, and black boxes represent aggravating, alleviating, and no interaction, respectively. Only validated data were used in hierarchical clustering and all subsequent computational analysis. The dendrogram of query genes was expanded for visualization, which indicated the similarities of their interaction patterns. The cluster of essential query genes is highlighted inside the blue box. (B) esa1-531 and gcn5Δ partially rescue the growth defect of hda1Δ rpd3Δ double mutants. Growth of each strain was assessed by plating four 10-fold serial dilution on SC–Ura medium (for esa1-531 rescue experiment) or on YPD medium (for gcn5Δ rescue experiment) at 30°C, a semipermissive temperature for esa1-531. (C) esa1-531 and gcn5Δ partially reversed hyperacetylation of histone H4 and H3, respectively, in hda1Δ rpd3Δ double mutant at 30°C, a semipermissive temperature for esa1-531. The acetylation levels of H4 K5, H4 K8, H4 K12, and H4 K16, and H3 K9 and H3 K14 were analyzed by immunoblot.
We arranged query genes by hierarchical clustering based on genetic interaction pattern similarities (Eisen et al. 1998). The essential query genes involved in histone acetylation formed a compact cluster, indicating that their patterns of genetic interaction were correlated (Fig. 1A). By contrast, essential genes involved in different biological processes shared little correlation with each other: Two examples are shown in Supplemental Figure S2 (CDC20 is the essential coactivator of anaphase-promoting complex, while CDC45 is an essential DNA replication initiation factor). The interaction profiles identified using Ts alleles did not appear to be dominated by temperature effects, since the genetic interaction profile of esa1-Hm1, a hypomorphic allele of ESA1, with a lower growth rate than wild-type allele at 30°C, most closely resembled that of the Ts allele esa1-531.
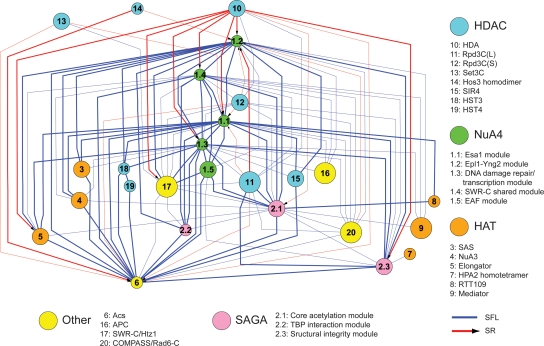
To discern relationships among genes at a higher level of organization, we examined interactions among sets of genes whose products might function together as a complex. We inferred membership in a complex from the subunit composition of known protein complexes or from highly interconnected patterns of genetic interactions indicating functional similarity. These complexes were regarded as nodes in a network in which the edges connected those pairs of nodes exhibiting more genetic interactions than expected by chance. Figure 2 shows a subnetwork highlighting HATs and HDACs (Supplemental Fig. S3 shows a comprehensive network; Supplemental Table S3 shows the complete data set). The resulting complex-to-complex network was highly connected. The close interactions among HATs and HDACs suggest that cells must maintain global acetylation levels within a certain range for viability. There were many aggravating interactions between different functional modules collectively comprising the same protein complex. Prominent SFL (blue) network hubs (see Supplemental Fig. S4 for degree distribution of network and hub definition) included the NuA4 and SAGA (Spt–Ada–Gcn5–acetyltransferase) complexes and ACS2 (encoding an essential acetyl-CoA synthetase supplying the nucleocytosolic acetyl-CoA pool) (Takahashi et al. 2006).
Figure 2.
The derived complex-to-complex genetic interaction network for HATs and HDACs. Nodes represent protein complexes and functional modules. Pairs of nodes were linked by an edge if they had significantly more genetic interactions than expected by chance. Node sizes proportional to the number of subunits in the corresponding complex or module; edge thicknesses signifies the statistical significance of the enrichment in genetic interactions (thick and thin edges indicate P < 10−16 and 10−16 ≤ P < 10−5, respectively).
The HDA complex was the most prominent alleviating (SR, red) hub, showing strong alleviating interactions with the NuA4, SAGA, and Elongator complexes. Not all deacetylase complexes had alleviating interactions with these complexes; for example, large and small forms of the Rpd3 complex [Rpd3C(L) and Rpd3C(S), respectively] had alleviating interactions only with NuA4 core acetylation machinery and aggravating interactions with most other HAT complexes, despite a broader histone substrate spectrum than the HDA complex. This finding suggests an unexpectedly detrimental effect of the HDA complex when the balance of chromatin acetylation and deacetylation is tilted. Moreover, the strong aggravating interaction between the HDA complex and Rpd3C mutants, and the fact that the growth defect and histone hyperacetylation of a hda1Δ rpd3Δ double mutant can be partially rescued by esa1-531 or gcn5Δ (Fig. 1B,C), supports the idea that these two complexes are collectively (and redundantly) responsible for bulk cellular histone deacetylation, and suggests an overlooked role of deacetylation in maintaining cell viability.
Finally, our genetic interaction maps were enriched with genes annotated by Gene Ontology (GO) as having vacuolar/endosomal (Supplemental Fig. S5; see also Supplemental Table S4 for a full list of enriched GO annotations), which is also revealed in the comprehensive complex-to-complex network (Supplemental Fig. S3). Similar enrichments have been observed in other studies (Takahashi et al. 2006; Mitchell et al. 2008). Previous expression microarray experiments of major HATs and HDACs (Choy and Kron 2002; Robyr et al. 2002; Le Masson et al. 2003; Huisinga and Pugh 2004; Zhang et al. 2004; Durant and Pugh 2006) identified no significant change of transcription of the key genes required for vacuolar/endosomal function. The genetic interaction and transcriptional profiles suggest possible roles of these HATs and HDACs in regulating extranuclear functions through mechanisms other than regulation of transcription, recalling an apparent role of Elongator in regulating polarized exocytosis (Rahl et al. 2005), and the importance of the nuclear pore complex in controlling transcription (Akhtar and Gasser 2007) and the targeting of DNA DSBs to the nuclear periphery for efficient repair (N.J. Krogan, pers. comm.).
HDA deacetylates Htz1-K14Ac
Htz1p is an H2A variant isoform in yeast with important roles in transcription, DNA replication, chromosome segregation, and the delineation of heterochromatin boundaries (Santisteban et al. 2000; Meneghini et al. 2003; Krogan et al. 2004a; Dhillon et al. 2006). Htz1p is exchanged for H2A by the SWR-C chromatin remodeling complex (Krogan et al. 2003; Kobor et al. 2004; Mizuguchi et al. 2004), and acetylation of Htz1p is required for this process (Millar et al. 2006). Htz1p and SWR-C share similar genetic interaction patterns, and thus have been assigned to the same functional module (Krogan et al. 2003; Collins et al. 2007). It is known that NuA4 and SAGA acetylate Htz1p at all four lysine residues in its N-terminal tail (K3, K8, K10, K14), with K14 being the most prominent acetylation site, but no Htz1p HDAC has been identified previously (Keogh et al. 2006; Millar et al. 2006). Htz1p-dependent genes tend to reside in small clusters called Htz1-activated domains (HZADs), and several of them overlap with Hda1-affected subtelomeric domains (HASTs) in subtelomeric chromatin (Meneghini et al. 2003). The two domains share the common feature of Hda1p-directed hypoacetylation of histone H3, which dampens the expression of their constituent genes.
Previous studies revealed synthetic lethality between HTZ1 and three genes of NuA4 (EAF1, EAF5, and EAF7), as well as components of Rpd3C(L) (Krogan et al. 2003; Kobor et al. 2004). Our dSLAM data demonstrated strong aggravating genetic interactions between the SWR-C/Htz1p module and all the functional modules of NuA4 and SAGA (Fig. 3A), as expected. We also observed a significant alleviating genetic interaction between the SWR-C/Htz1p module and the HDA complex (Fig. 3A). Deletion of HDA1 rescued the slow growth phenotype of the htz1Δ mutant and also reversed its sensitivity to the genotoxic agents hydroxyurea (HU) and methyl methanesulfonate (MMS) and to the microtubule-interfering drug benomyl (Fig. 3B). Transcription of many Htz1p-dependent genes was also restored in the hda1Δ htz1Δ double mutant, including subtelomeric genes both in and out of HASTs and genes far away from telomeres (Supplemental Table S5). The ability of Htz1p to form a boundary adjacent to the silent mating-type cassette HMR was preserved in the hda1Δ strain (Supplemental Fig. S6). In contrast to the other htz1Δ phenotypes, deletion of HDA1 could not rescue the boundary function defect of htz1Δ that limits spread of silent chromatin from HMR locus (Supplemental Fig. S7; Meneghini et al. 2003).
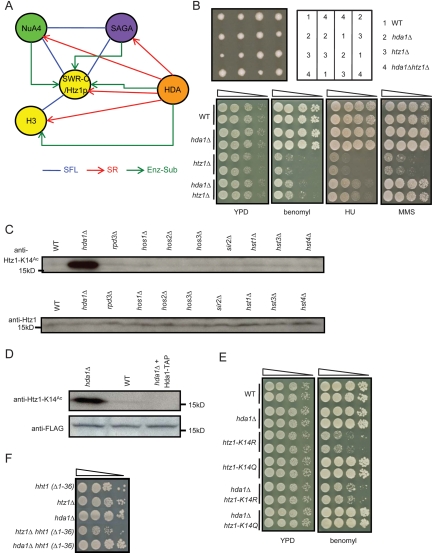
Figure 3.
HDA complex deacetylates Htz1-K14Ac in vitro and in vivo. (A) Overlap of the enzyme–substrate relationship with genetic interactions. A schematic model depicts genetic and enzyme–substrate interactions between HATs (NuA4 and SAGA), HDACs (HDA complex), and their substrates (Htz1p and histone H3). (B) Deletion of HDA1 rescues the slow-growth and drug-sensitivity phenotype of htz1Δ. Four tetrads dissected from an hda1Δ/HDA1 htz1Δ/HTZ1 diploid strain on YPD medium are shown. Drug sensitivities were assessed by plating 10-fold serial dilution on YPD medium containing HU (200 mM), MMS (0.03%), or benomyl (15 μg/mL). (C,D) Hda1p deacetylates Htz1-K14Ac in vivo and in vitro. The K14 acetylation level in wild-type and HDAC mutant strains was analyzed by immunoblot. (D) In vitro deacetylation activity of Hda1-TAP purified by tandem affinity purification was assessed by incubating it with Flag-tagged Htz1p purified from hda1Δ strain and then monitoring the K14 acetylation level by immunoblot. No protein was added to the control samples. (E) Deletion of HDA1 rescues the sensitivity of htz1-K14R to benomyl. The experimental conditions are described in B. (F) Genetic interactions of an N-tail deletion of histone H3 [hht1(Δ1–36)] with hda1Δ (alleviating) and htz1Δ (aggravating) mutants, suggesting that histone H3 reside in the same functional module as Htz1p.
Based on these observations, we investigated whether the HDA complex deacetylates Htz1p biochemically. Acetylation level of Htz1p at Lys 14 dramatically increased in vivo in the hda1Δ mutant, but not in any other HDAC deletion strain (Fig. 3C). Addition of affinity-purified Hda1p to Flag-tagged Htz1p purified from the hda1Δ strain sharply diminished the increased level of K14 acetylation (Fig. 3D). Deletion of HDA1 also reversed the sensitivity of htz1-K14R to benomyl (Fig. 3E; Keogh et al. 2006), which suggests Hda1p may also deacetylate other lysine residues. These results showed that the HDA complex, previously regarded as an H3 and H2B-specific HDAC (Wu et al. 2001), also counteracts the effects of NuA4 and SAGA by removing a critical acetyl group from Htz1p.
The alleviating genetic interactions between hda1 and htz1 suggested the existence of parallel substrates for Hda1p, which would be predicted to display SFL interactions with the htz1Δ mutant and SR interactions with the hda1Δ mutant. Histone H3 is a well-known substrate of HDA complex (Wu et al. 2001), and an N-tail deletion strain [hht1 (Δ1-36)] shows the expected genetic interaction pattern (Fig. 3F). Moreover, hht1 (Δ1-36) has genetic interaction with 88 out of 129 known genetic interaction partner genes of htz1 [see Supplemental Table S6 for a complete list of genetic interactions of hht1 (Δ1-36) and htz1]. These observations suggest that Htz1p, a histone H2A variant, and histone H3 share redundant functions.
Functional organization of NuA4—a new look
NuA4, the only essential HAT in yeast, acetylates the N-terminal tails of H2A, H4 and Htz1p. The processes are critical for regulating gene transcription, limiting the spread of silent heterochromatin and repairing DNA DSBs (Doyon and Cote 2004). Yeast cells lacking normal NuA4 arrest at the G2/M phase in a RAD9-dependent manner (Choy and Kron 2002). Esa1p is the essential catalytic subunit of the NuA4 complex. It forms the core acetylation machinery with Epl1p and Yng2p, termed Piccolo NuA4, to maintain global histone H4 and H2A acetylation in vivo (Boudreault et al. 2003; Selleck et al. 2005). Impaired function of any of the three component genes drastically decreases catalytic activity of the complex (Smith et al. 1998; Allard et al. 1999; Clarke et al. 1999; Choy and Kron 2002; Boudreault et al. 2003).
We found that the lethality of the esa1-531 and epl1-1 Ts alleles and the growth defect of yng2Δ could each be suppressed by deletion of either HDA1 or RPD3 (Fig. 2). In other words, growth defects resulting from impaired NuA4 acetylation were rescued by compensatory loss of either of the corresponding HDACs. We expected Ts mutants esa1-531 and epl1-1 to cluster most tightly but surprisingly, epl1-1 clustered with yng2Δ (Fig. 4A). Also, to our surprise, esa1-531 had a distinct response to the HDAC inhibitor trichostatin A (TSA) to that of epl1-1 and yng2Δ. TSA at 100 μM, which inhibits most HDAC activity of Hda1p and Rpd3p in vitro (Carmen et al. 1999), restored growth of epl1-1 and yng2Δ to that of wild type, whereas esa1-531 lethality was only partially rescued (Fig. 4B). This suggested a broader substrate spectrum for Esa1p than for NuA4 per se; thus, Esa1p might act independently of other NuA4 components. Conversely, nicotinamide (a chemical that inhibits another major type of HDAC named sirtuin) (Denu 2005) had little effect on the three mutants (Supplemental Fig. S8), and there were few SR interactions between sirtuin and NuA4 genes (Fig. 2).
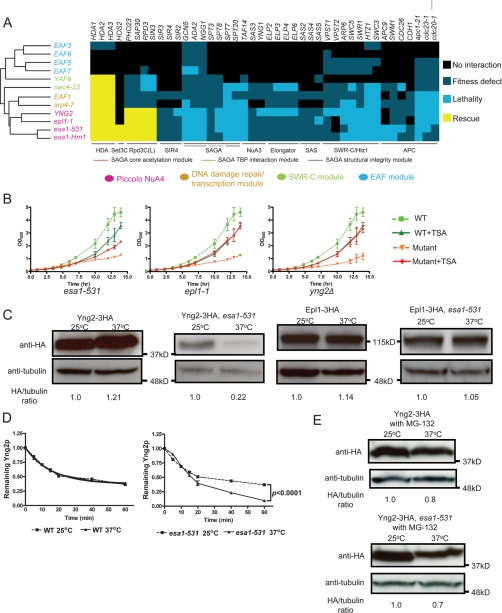
Figure 4.
Functional dissection of the NuA4 complex. (A) Hierarchical clustering of NuA4 component genes based on genome-wide genetic interaction patterns shows functional association. Font colors indicate distinct functional modules (see legend). Subsets of genetic interactions between NuA4 and other complexes were organized for visualization. (B) Core acetylation machinery mutants are differentially sensitive to TSA. Growth curves of esa1-531, epl1-1, and yng2Δ mutants in SC–Ura medium at 37°C (restrictive temperature) with or without TSA (100 μM) are shown. Error bars indicate ±1 SEM from three biological replicates. (C) ESA1 stabilizes and controls the protein abundance of Yng2p. Wild-type (WT) and esa1-531 strains stably expressing tagged Yng2p or Epl1p were grown at 25°C (permissive temperature) to OD600 ∼0.3, then shifted to 37°C or kept for 4 h at 25°C; whole-cell extracts (WCEs) were then collected and probed with anti-HA, and band intensities were quantified. (D) Yng2-3HA turnover kinetics in wild-type and esa1-531 strains investigated by cycloheximide turnover experiments. Cells were grown at 25°C to OD600 ∼0.3, then either shifted to 37°C or kept at 25°C, and treated with 0.1 μg/mL cycloheximide simultaneously. Equal amount of cells were collected at indicated time points and analyzed by immunoblot as shown in Supplemental Figure S9A. The fraction of Yng2-3HA remaining after cycloheximide addition was plotted. Error bars indicate ±1 SEM. (E) Yng2p is stabilized by MG-132 in esa1-531 cells. Wild-type and esa1-531 cells stably expressing tagged Yng2p were incubated with MG-132 (75 μM) for 4 h at 25°C (permissive temperature) or 37°C (restrictive temperature), after which WCEs were analyzed by immunoblot.
Based on the distinct behavior of ESA1 relative to other genes encoding core NuA4 acetylation machinery, we hypothesized that Esa1p performs functions beyond its well-established global chromatin acetylation activity in the context of Piccolo NuA4. We discovered that the abundance of Yng2-3HA (but not Epl1-3HA) depended on normal ESA1 function (Fig. 4C). We also showed that this effect involved the prevention of proteasome-mediated protein degradation (Fig. 4D; Supplemental Fig. S9A) and not transcriptional activation (Supplemental Fig. S9B), since Yng2p is stabilized by the proteasome inhibitor MG-132 (Fig. 4E).
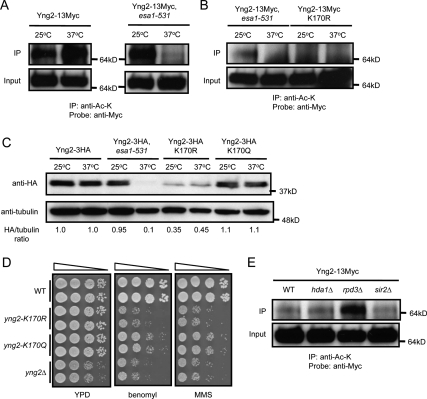
To further investigate the mechanism of Esa1p on regulating the protein stability of Yng2p, we examined the acetylation status of Yng2-Myc by immunoprecipitation with mouse monoclonal anti-acetylated lysine. To our surprise, a large proportion of Yng2p was acetylated in vivo (Fig. 5A). The signal of Yng2p acetylation could be efficiently competed away with acetylated BSA (Supplemental Fig. S10A), and mouse monoclonal anti-HA could not pull down acetylated Yng2p to any detectable level (Supplemental Fig. S10B), indicating the specificity of the detected acetylation signals. The acetylation level of Yng2p diminished dramatically in an esa1-531 mutant at restrictive temperature (Fig. 5A), suggesting its acetylation depends on normal ESA1 function. A candidate lysine residue (K170) of Yng2p for acetylation was identified by tandem mass spectrometry (Supplemental Figure S11), which was confirmed by the loss of acetylation of Yng2p when substituting K170 with arginine (K170R), a mutation blocking acetylation (Fig. 5B). A K170R mutation constitutively destabilized Yng2p, whereas substituting K170 with glutamine (K170Q, a mutation mimicking constitutive acetylation) did not cause detectable change of protein abundance relative to the wild type, but rendered it insensitive to the effects of an esa1-531 Ts mutation (Fig. 5C). The K170R and K170Q mutants were each hypersensitive to benomyl and MMS (Fig. 5D), consistent with dynamic acetylation and deacetylation of Yng2p affecting its normal function. A screen of several known HDAC mutants identified increased acetylation of Yng2p in rpd3Δ mutant (Fig. 5E), suggesting that deacetylation of Yng2p in vivo depends on the balance of normal ESA1 and RPD3 activities.
Figure 5.
Acetylation and deacetylation of Yng2p controls its protein stability and function. (A) Yng2p is acetylated in vivo through an Esa1p-dependent mechanism. Wild-type (WT) and esa1-531 strains stably expressing Myc-tagged Yng2p were grown at 25°C (permissive temperature) to OD600 ∼0.3, then shifted to 37°C or kept for 4 h at 25°C; WCEs were then collected, immunoprecipitated with anti-Ac-K and probed with anti-Myc. (B) K170 is the major acetylated lysine residue of Yng2p in vivo. K170 was identified by tandem mass spectrometry. Substitution of K170 with arginine (K170R) diminishes acetylation of Yng2p. (C) Effects of K170 substitutions on Yng2p stability. Cells were grown in conditions describes in Figure 3A. WCEs were collected and probed with anti-HA, and band intensities were quantified. K170R mutation causes decreased protein abundance of Yng2p, while K170Q causes no detectable change. (D) Effects of K170 substitutions on Yng2p function. Drug sensitivities were assessed by plating 10-fold serial dilution on YPD medium containing MMS (0.03%) or benomyl (15 μg/mL). (E) Yng2p is deacetylated through an Rpd3p-dependent mechanism in vivo. Cells were grown at 30°C to OD600 ∼0.6, then WCEs were collected, immunoprecipitated with anti-Ac-K and probed with anti-Myc.
The genetic interaction profile of other NuA4 components provided additional information on the functional organization of the NuA4 complex (Fig. 4A). Arp4p is an actin-related gene shared among the NuA4, Ino80, and SWR-C chromatin remodeling complexes, and Arp4p is important for recruiting NuA4 to DSBs and specifically regulating the transcription of ESA1-dependent genes (Galarneau et al. 2000; Downs et al. 2004). ARP4 was previously assigned to the same module with three other genes (ACT1, SWC4, and YAF9) also shared between NuA4 and SWR-C (Doyon and Cote 2004; Auger et al. 2008). However, our genetic interaction data indicated that ARP4 has a closer functional relationship to EAF1 than to SWC4 and YAF9 (Fig. 4A). This finding agrees with previous chemical genomics surveys implicating EAF1 in DNA damage repair (Parsons et al. 2004). Based on this, we assign ARP4 and EAF1 to the DNA damage repair and transcription regulation module, keeping SWC4 and YAF9 in the SWR-C module.
Molecular choreography at DSBs
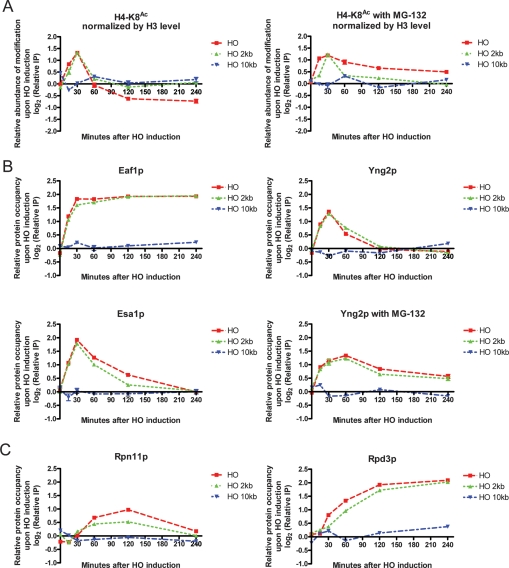
Given that Esa1p-dependent acetylation and Rpd3p-dependent deacetylation controls the protein stability of Yng2p, which is essential for the enzymatic activity of Piccolo NuA4 on acetylating nucleosomal histones (Boudreault et al. 2003; Selleck et al. 2005), we investigated the influence of these post-translational modifications on DSB chromatin dynamics. We used a galactose-inducible DSB induction system to monitor protein species and chromatin state at the DSB. The DSB, introduced by HO endonuclease at the mating type locus (MAT), can only be repaired by nonhomologous end joining in this strain (Lee et al. 1998). In agreement with a previous study (Downs et al. 2004), we found transient hyperacetylation of histone H4 followed by a distinct hypoacetylation phase near the DSB (Fig. 6A). This change and others reported below were observed near the break and 2 kb distal to it but not 10 kb away. The recruitment of NuA4 to DNA DSBs is important for local acetylation of histone H4 (Bird et al. 2002; Downs et al. 2004; Tamburini and Tyler 2005), but the recruitment kinetics of Esa1p, Epl1p, and Yng2p were distinct from that of bulk NuA4 (Fig. 6B; Supplemental Fig. S12). Whereas Eaf1p, the only NuA4-specific subunit protein not shared with any other protein complex (Auger et al. 2008), remained steadily enriched near the DSB, Esa1p, Epl1p, and Yng2p were initially recruited with kinetics similar to Eaf1p but were evicted from the broken chromatin region at distinct rates thereafter. Addition of MG-132, a proteasome inhibitor that prevents Yng2p degradation (Fig. 4E), greatly delayed eviction of Yng2p from the DSB, suggesting an ubiquitin based displacement mechanism (Fig. 6B). Consistent with altered Yng2p recruitment kinetics, MG-132 also inhibited Ac-H4 depletion (Fig. 6A). In contrast, MG-132 did not affect histone H3 eviction kinetics (Supplemental Fig. S13). Rpn11p, a metalloprotease subunit of the 19S regulatory particle of the 26S proteasome lid (Verma et al. 2002), is concomitantly recruited to the DSB, suggesting that the proteasome is responsible for degrading Yng2p locally at the DSB (Fig. 6C).
Figure 6.
Post-translational protein modification of the NuA4 core acetylation machinery choreographs DSB repair. (A) MG-132 alters the kinetics of histone H4 acetylation near an HO-induced DNA DSB. Chromatin immunoprecipitation (ChIP) assays with anti-H4-K8Ac were used to assess kinetics of histone H4 acetylation. Enrichment of histone H4-K8Ac was quantified by RT–PCR and normalized to the GEA2 internal control and also the local abundance of histone H3 (Supplemental Fig. S11). (B) NuA4 core acetylation machinery components show distinctive kinetics of recruitment to an HO-induced DNA DSB. The enrichment of tagged Esa1p, Yng2p, and Eaf1p after HO induction was assessed by ChIP assays with anti-HA. GAL-HO pdr5Δ strains were used in MG-132 experiments. (C) Proteasome and Rpd3C are recruited to an HO-induced DSB. Enrichment of tagged Rpn11p and Rpd3p after HO induction was assessed by ChIP with anti-HA.
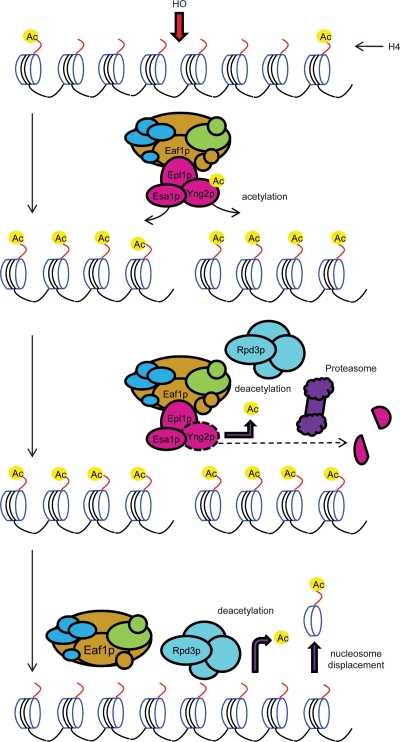
A dynamic system with H4 acetylation followed by deacetylation might be crucial for efficient DSB repair is suggested by the hypersensitivity of GAL-HO strains to MG-132 upon DSB induction by galactose (Supplemental Fig. S14). Rpd3p was recruited to DSBs preceding deacetylation of histone H4 and eviction of Yng2p (Fig. 6C), and addition of MG-132 did not change the kinetics of Rpd3p recruitment (Supplemental Fig. S15). By contrast, Hda1p and Hos2p were not enriched at the DSB (Supplemental Fig. S16). These data suggest that in addition to conducting global nucleosomal acetylation, Piccolo NuA4 is elaborately remodeled independently of the rest of NuA4 locally at DSBs and that this molecular choreography is governed by dynamic post-translational modification occurring specifically at DSBs (Fig. 7).
Figure 7.
Schematic model for the dynamic regulation of acetylation status near DSBs. Upon introduction of a DSB, such as that caused by HO endonuclease at the HO recognition site, NuA4 is actively recruited focally and rapidly hyperacetylates nearby histone H4. The function of the core acetylation machinery of NuA4 is disrupted when Yng2p is deacetylated through an Rpd3C-depedent mechanism followed by degradation by proteasome, followed by eviction of Esa1p and Epl1p. Three distinct processes appear to be at play to facilitate transition to the hypoacetylation phase crucial for proper DSB repair: (1) The breakdown of Piccolo NuA4 stops ongoing acetylation, (2) Rpd3C is recruited and could actively remove acetyl groups, and (3) ATP-dependent chromatin remodeling complexes, which conduct active nucleosome displacement.
Discussion
In this study we systematically surveyed the functional associations among genes that dynamically regulate histone acetylation and deacetylation in yeast, generating a comprehensive network of genetic interactions. The inclusion of six essential query genes significantly enhanced our ability to identify functionally important target genes.
Our analysis focused on the abstracted network of functional modules and protein complexes rather than those of individual genes. The network was highly connected, and revealed a close functional relationship between HAT and HDAC complexes, indicating that these complexes share common essential cellular functions despite the fact that most of them modify distinct spectra of histone lysine residues. The HDA complex was the most distinct SR hub, pointing to major counterbalancing effects on the NuA4, SAGA, and Elongator HAT complex, which indicates that the HDA complex removed the largest amount of acetyl groups. By contrast, the Rpd3C had aggravating interactions with most of the HAT complexes, except Piccolo NuA4; these interactions are consistent with the cooperation between Rpd3C(S) and various HATs in regulating transcriptional elongation (Carrozza et al. 2005; Keogh et al. 2005; Li et al. 2007), and also the role of Rpd3C in governing histone H4 deacetylation following an acetylation conducted by NuA4 in the vicinity of DNA DSBs.
Our results also revealed the general finding that the HDA and Rpd3 complexes together define the major HDAC activities that counteract the HAT activity of the NuA4 and SAGA complexes, and that these complexes provide the bulk control of the dynamic balance of global histone acetylation and deacetylation essential to cell viability. Requirement of histone acetylation by various HATs for maintaining cell viability have been well studied (Smith et al. 1998; Zhang et al. 1998; Allard et al. 1999; Clarke et al. 1999; Howe et al. 2001). Our data suggest that hyperacetylation of histone H3 and H4 in the hda1Δ rpd3Δ double mutant is as detrimental to cell viability as hypoacetylation, and can be rescued when the responsible acetylase activity is repressed.
In addition to its effect on many aspects of chromosome biology, a relationship between global histone acetylation/deacetylation and vacuolar function is also revealed by these studies. This relationship raises the possible existence of nonhistone substrates of HATs and HDACs in yeast.
The genetic interaction profile of htz1Δ and follow-up experiments led us to conclude that the HDA complex, previously known to acetylate histones H3 and H2B, removes the acetyl group from K14 and possibly also other lysine residues of the N-tail of Htz1p. However, lack of reliable antibodies limited our ability to test the acetylation level of lysine residues other than K14. We also propose that histone H3 and Htz1p reside in the same functional module based on their similar genetic interaction profiles. However, although being less enriched in the promoter region and defective in blocking telomeric heterochromatin spreading, an unacetylatable Htz1p mutant (htz1-K3,8,10,14R) is insensitive to genotoxic agents lethal to htz1Δ, suggesting that acetylation is important in some but not all aspects of Htz1p function (Babiarz et al. 2006; Millar et al. 2006). The genetic interactions between HTZ1 and its corresponding HATs (ESA1 and GCN5) and HDAC (HDA1) suggest that these complexes not only affect Htz1p modification but also modulate the essential pathway through additional unknown mechanisms.
In addition to examining functional relationships among protein complexes, synthetic genetic interaction profiles can be used for dissecting more elaborate protein complexes like NuA4 into separate functional modules. A recent genetic interaction survey of a subset of nonessential NuA4 subunits revealed that Eaf1p is important for maintaining the integrity of NuA4 complex (Mitchell et al. 2008). Here, comprehensive incorporation of most essential and nonessential NuA4 subunit genes led to many new findings. The genetic interaction profile of esa1-531 revealed new functions of Esa1p beyond its well-known nucleosomal acetylation activity. For example, we found that the enzymatic activity of Piccolo NuA4, the core acetylation machinery of NuA4, was maintained by control of Esa1p on the protein turnover of Yng2p through an acetylation-dependent mechanism. Deacetylation of Yng2p was dependent on Rpd3p, which potentially precedes the degradation of Yng2p by proteasome. Tandem mass spectrometry and further biochemical experiments confirmed that K170 is the major acetylated lysine residue of Yng2p. The hypersensitivity of both yng2-K170R and yng2-K170Q mutants (mutations mimicking constitutive deacetylation and acetylation, respectively) to benomyl and MMS suggests that dynamic acetylation and deacetylation is important to its normal function in DSB repair. Moreover, the recruitment kinetics of the Piccolo NuA4 subunits to an HO-induced DSB is distinct from the rest of NuA4. NuA4 is actively recruited focally and rapidly hyperacetylates nearby histone H4 at a DSB. We propose that the dynamic protein turnover of Yng2p mediated by an acetylation–deacetylation cycle with cooperative recruitment of Rpd3C and proteasome at DSBs disrupts the enzymatic activity of Piccolo NuA4, which stops ongoing acetylation. This finding is consistent with previous findings showing that proteasome is involved in the repair of DSBs (Krogan et al. 2004b). The recruited Rpd3C further removes acetyl groups from histone H4, and ATP-dependent chromatin remodeling complexes actively conduct nucleosome displacement (Tsukuda et al. 2005). These three mechanisms allow for dynamic hyperacetylation and subsequent hypoacetylation of histone H4 nearby facilitating DSB repair (Fig. 7).
In this study, we applied genetic interaction analysis on a large scale as a general approach for analyzing the complexities of histone (de)acetylation in yeast. New functions of the NuA4 and HDA complexes were identified, and a potential nonhistone substrate of Esa1p and Rpd3p was found. Extensions of this powerful strategy to mammalian systems will certainly be of interest in light of recent advances in high-throughput methodologies based on RNA interference (Silva et al. 2008).
Materials and methods
Full experimental details and data analysis methods are provided in the Supplemental Material.
Ts alleles creation
The method to generate Ts alleles was performed as described (Huang et al. 2008). To construct a cloning vector, the 5′ promoter and 3′ terminator regions of an essential gene were cloned into a CEN plasmid with URA3 marker. A pool of mutations was generated in a cassette containing the promoter, the open reading frame, and the terminator regions by manganese-driven errorprone PCR reaction. The mutagenized PCR products and the digested cloning vector (to create a gap between the promoter and the terminator) were then cotransformed into a haploid-convertible heterozygous YKO strain of the corresponding essential gene. Vectors harboring mutated cassettes were generated by recombinational gap repair. Cells containing a library of mutations were sporulated and then subjected to selection for Ura+-G418R phenotype in magic medium without uracil (MM–Ura; SC–LeuHis–Arg–Ura + canavanine + G418). These spores were then incubated at 25°C or 37°C to screen for a Ts phenotype.
dSLAM and genetic interaction target gene validation
The synthetic lethality screen was performed as described previously (Pan et al. 2006). A pool of haploid-convertible heterozygous diploid YKO library was transformed either once with a URA3 knockout cassette for nonessential query genes, or sequentially with a natMX knockout cassette followed by YCplac33 harboring a Ts allele for essential query genes. The resulting heterozygous double-mutant pool was then subjected to selection for a mixed population of single and double mutants as the control pool and a pure population of double mutants as the experimental pool in magic medium with the appropriate combination of selecting drugs. Three different semipermissive temperatures were tested for optimizing the screen conditions for essential query genes. Strains with a control/experiment ratio ≥2 in either UPTAG or DNTAG were selected for validation either with random spore analysis or tetrad dissection.
Data access
Microarray data were submitted to GEO with accession code GSE9771.
URLs
BioGRID database: http://www.thebiogrid.org; MIPS database: http://mips.gsf.de; Saccharomyces genome database: http://www.yeastgenome.org.
Acknowledgments
We thank W. Zachariae for providing the cdc20-1 Ts allele; E.M. Cooper and J. Dai for gifts of reagents; members of the Boeke laboratory for valuable discussions throughout the course of the work; and P.B. Meluh, A. Norris, K.A. O’Donnell, and O.J. Rando for their critical comments on the manuscript. Y.Q. is an IBM predoctoral fellow. X.P. was a fellow of the Leukemia and Lymphoma Society. This work was supported by NIH Roadmap grant “Technology Center for Networks and Pathways” (U54 RR 020839) and grant R01 HG 02432 to J.D.B.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1679508.
References
- Akhtar A., Gasser S.M. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Allard S., Utley R.T., Savard J., Clarke A., Grant P., Brandl C.J., Pillus L., Workman J.L., Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger A., Galarneau L., Altaf M., Nourani A., Doyon Y., Utley R.T., Cronier D., Allard S., Cote J. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell. Biol. 2008;28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz J.E., Halley J.E., Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes & Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.W., Yu D.Y., Pray-Grant M.G., Qiu Q., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Boudreault A.A., Cronier D., Selleck W., Lacoste N., Utley R.T., Allard S., Savard J., Lane W.S., Tan S., Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes & Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen A.A., Rundlett S.E., Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- Carmen A.A., Griffin P.R., Calaycay J.R., Rundlett S.E., Suka Y., Grunstein M. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Choy J.S., Kron S.J. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 2002;22:8215–8225. doi: 10.1128/MCB.22.23.8215-8225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A.S., Lowell J.E., Jacobson S.J., Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Davierwala A.P., Haynes J., Li Z., Brost R.L., Robinson M.D., Yu L., Mnaimneh S., Ding H., Zhu H., Chen Y., et al. The synthetic genetic interaction spectrum of essential genes. Nat. Genet. 2005;37:1147–1152. doi: 10.1038/ng1640. [DOI] [PubMed] [Google Scholar]
- Denu J.M. The Sir 2 family of protein deacetylases. Curr. Opin. Chem. Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Dhillon N., Oki M., Szyjka S.J., Aparicio O.M., Kamakaka R.T. H2A.Z functions to regulate progression through the cell cycle. Mol. Cell. Biol. 2006;26:489–501. doi: 10.1128/MCB.26.2.489-501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J.A., Allard S., Jobin-Robitaille O., Javaheri A., Auger A., Bouchard N., Kron S.J., Jackson S.P., Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Doyon Y., Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Durant M., Pugh B.F. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:2791–2802. doi: 10.1128/MCB.26.7.2791-2802.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau L., Nourani A., Boudreault A.A., Zhang Y., Heliot L., Allard S., Savard J., Lane W.S., Stillman D.J., Cote J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- Glozak M.A., Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- Hartman J.L.T., Garvik B., Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Howe L., Auston D., Grant P., John S., Cook R.G., Workman J.L., Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes & Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Sucgang R.S., Lin Y.Y., Shi X., Boeke J.D., Pan X. Plasmid-chromosome shuffling for non-deletion alleles in yeast. Nat. Methods. 2008;5:167–169. doi: 10.1038/nmeth.1173. [DOI] [PubMed] [Google Scholar]
- Huisinga K.L., Pugh B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Mennella T.A., Sawa C., Berthelet S., Krogan N.J., Wolek A., Podolny V., Carpenter L.R., Greenblatt J.F., Baetz K., et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes & Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J.2004A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin PLoS Biol. 2E131 . doi: 10.1371/journal. pbio.0020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. 2004a;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Lam M.H., Fillingham J., Keogh M.C., Gebbia M., Li J., Datta N., Cagney G., Buratowski S., Emili A., et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell. 2004b;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- Le Masson I., Yu D.Y., Jensen K., Chevalier A., Courbeyrette R., Boulard Y., Smith M.M., Mann C. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell. Biol. 2003;23:6086–6102. doi: 10.1128/MCB.23.17.6086-6102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.K., Workman J.L. Histone acetyltransferase complexes: One size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J.L. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Meneghini M.D., Wu M., Madhani H.D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Millar C.B., Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- Millar C.B., Xu F., Zhang K., Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes & Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell L., Lambert J.P., Gerdes M., Al-Madhoun A.S., Skerjanc I.S., Figeys D., Baetz K. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol. Cell. Biol. 2008;28:2244–2256. doi: 10.1128/MCB.01653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Parsons A.B., Brost R.L., Ding H., Li Z., Zhang C., Sheikh B., Brown G.W., Kane P.M., Hughes T.R., Boone C. Integration of chemical–genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- Rahl P.B., Chen C.Z., Collins R.N. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Robyr D., Suka Y., Xenarios I., Kurdistani S.K., Wang A., Suka N., Grunstein M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Santisteban M.S., Kalashnikova T., Smith M.M. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Segre D., Deluna A., Church G.M., Kishony R. Modular epistasis in yeast metabolism. Nat. Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- Selleck W., Fortin I., Sermwittayawong D., Cote J., Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 2005;25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Silva J.M., Marran K., Parker J.S., Silva J., Golding M., Schlabach M.R., Elledge S.J., Hannon G.J., Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Eisen A., Gu W., Sattah M., Pannuti A., Zhou J., Cook R.G., Lucchesi J.C., Allis C.D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., McCaffery J.M., Irizarry R.A., Boeke J.D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Tamburini B.A., Tyler J.K. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tsukuda T., Fleming A.B., Nickoloff J.A., Osley M.A. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., Koonin E.V., Deshaies R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Wu J., Suka N., Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- Wu J., Suka N., Carlson M., Grunstein M. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- Xu W.S., Parmigiani R.B., Marks P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- Zhang W., Bone J.R., Edmondson D.G., Turner B.M., Roth S.Y. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Richardson D.O., Roberts D.N., Utley R., Erdjument-Bromage H., Tempst P., Cote J., Cairns B.R. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell. Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]