Abstract
Influenza A and B viruses contain proton-selective ion channels, A/M2 and BM2, respectively, and the A/M2 channel activity is inhibited by the drugs amantadine and its methyl derivative rimantadine. The structure of the pore-transmembrane domain has been determined by both x-ray crystallography [Stouffer et al. (2008) Nature 451:596–599] and by NMR methods [Schnell and Chou (2008) Nature 451:591–595]. Whereas the crystal structure indicates a single amantadine molecule in the pore of the channel, the NMR data show four rimantadine molecules bound on the outside of the helices toward the cytoplasmic side of the membrane. Drug binding includes interactions with residues 40–45 with a polar hydrogen bond between rimantadine and aspartic acid residue 44 (D44) that appears to be important. These two distinct drug-binding sites led to two incompatible drug inhibition mechanisms. We mutagenized D44 and R45 to alanine as these mutations are likely to interfere with rimantadine binding and lead to a drug insensitive channel. However, the D44A channel was found to be sensitive to amantadine when measured by electrophysiological recordings in oocytes of Xenopus laevis and in mammalian cells, and when the D44 and R45 mutations were introduced into the influenza virus genome. Furthermore, transplanting A/M2 pore residues 24–36 into BM2, yielded a pH-activated chimeric ion channel that was partially inhibited by amantadine. Thus, taken together our functional data suggest that amantadine/rimantadine binding outside of the channel pore is not the primary site associated with the pharmacological inhibition of the A/M2 ion channel.
Keywords: amantadine inhibition of influenza virus, amantadine resistance, drug-binding site, influenza reverse genetics
Influenza A and B viruses are enveloped negative-strand segmented RNA viruses that cause epidemics, and for influenza A virus, pandemics. At the biochemical and cellular biological level the viruses have some common properties. These include virus particles using host cell surface expressed sialic acid as their receptor and entry into cells by endocytosis. Virus uncoating takes place in the lumen of the endosome and is mediated by the hemagglutinin glycoprotein which is triggered by the acidic environment to undergo a protein refolding event that causes the fusion of the viral envelope with the endosomal membrane (reviewed in ref. 1).
Influenza A and B viruses encode the A/M2 and BM2 integral membrane proteins (2, 3), respectively. A/M2 and BM2 are proton selective ion channels (4–6) that permit protons to enter virus particles during uncoating in endosomes. Acidification of the virus interior causes dissociation of the membrane (M1) protein from the ribonucleoprotein core (3, 7) a necessary step for import of the ribonucleoproteins into the nucleus (8). A/M2 and BM2 ion channel activity also modulates the pH of the trans-Golgi network in virus-infected cells equilibrating the acidic pH of the lumen of the Golgi with the cytoplasm (9, 10) (reviewed in ref. 11). The mature A/M2 protein consists of 96 residues with a 23-residue N-terminal extracellular domain, a single internal hydrophobic domain of 19 residues that acts as a TM domain and forms the pore of the channel, and a 54-residue cytoplasmic tail (2). The BM2 protein consists of a 7-residue ectodomain, a 19-residue TM domain, and an 82-residue cytoplasmic tail (3). It has been shown for both A/M2 and BM2 that the proteins form homotetramers (3, 12, 13). The ectodomain of A/M2 contains two cysteine residues that form disulfide bonds but neither of these disulfide bonds is needed for ion channel activity (14) nor for replication of the virus in tissue culture or mice (15). The functionally active A/M2 and BM2 channels are the homotetramers and each channel has a centrally located pore for proton conduction (16–21). The A/M2 and BM2 proton channel proteins of influenza A and B viruses are among the smallest bona fide ion channel proteins (with the properties of ion selectivity and activation), and they succeed in accomplishing the same function with only a meager similarity in primary amino acid sequence. The only homology between the amino acid sequences of these two proteins is found in the TM domain HXXXW motif of the inner membrane-spanning residues. Histidine 37 confers the high proton selectivity of the A/M2 channel (22–24) and tryptophan 41 acts as the channel gate, opening and closing the pore of the A/M2 channel (25). It is thought that His 19 and Trp 23 in BM2 have the same roles in BM2 channel function as they do in the A/M2 channel (6).
The activity of A/M2 ion channel is inhibited by the antiviral drug amantadine and its methyl derivative rimantadine (26, 27). The drug binds at both neutral (closed state) and acidic pH (open channel). Influenza A viruses that mutate and become drug resistant contain mutations in the A/M2 TM domain and naturally arising mutations mostly occur at TM domain residues Val 27, Ala 30, Ser 31, and Gly 34 (26). Neither influenza B virus growth nor BM2 ion channel activity are inhibited by the drugs.
Recently, Stouffer et al. (28) using X-ray crystallography published the atomic structure of the A/M2 TM domain with and without bound amantadine and Schnell and Chou (29), using NMR methods, copublished the structure of a somewhat larger A/M2 peptide (M2 residues 18–60) that included the TM domain and 17 residues of the cytoplasmic tail. As predicted from earlier biochemical and NMR studies (17, 30) the recent high-resolution structural data show the overall architecture of the TM domain is a four-helix bundle and the data confirm predictions that His 37 forms the pH sensor and Trp 41 the channel gate (25, 31). The Trp 41 indole rings are at van der Waals distance from each other, prohibiting passage of water or ions and the gate is further stabilized in the closed state by inter-subunit hydrogen bonds with Asp 44 (28, 29).
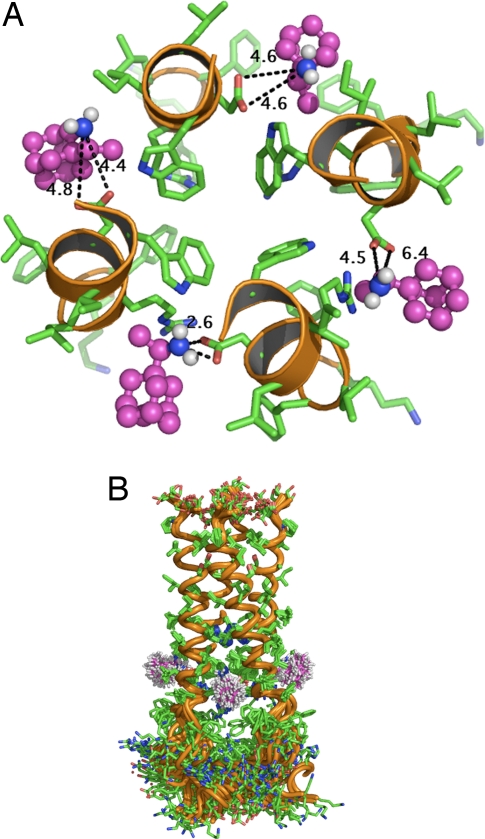
The crystal structures were solved at lower pH than the NMR structure, and the former structures are more open near the C-terminal His 37 than the NMR structure. However, the structures have a major difference in that the crystal structure data indicate a single amantadine molecule in the pore of the channel that is surrounded by Val 27, Ala 30, Ser 31, and Gly 34 (28). In contrast, from the NMR data obtained with the peptide in a solution of 40 mM rimantadine and ∼300 mM dihexanoyl-phosphatidyl-choline, four rimantadine molecules were found bound on the outside of the protein helix facing the lipid bilayer and located in the membrane environment at the end of the helix toward the cytoplasmic face of the channel. Drug binding includes interactions with residues 40–45 with a polar hydrogen bond between rimantadine and aspartic acid residue 44 (D44) that appears to be important (29) (Fig. 1). However, although the NMR structure is very well defined in the TM region, the drug-binding site around D44 is not well defined and rimantadine adopts many different conformations. Even within a single snapshot, each of the four rimantadine molecules within a tetramer display significant variability in their interactions. Fig. 1A illustrates the variability in geometries of rimantadine-N to Asp-carboxylate interactions (distances in angstroms) using data from frame 7 of 2RLF. Fig. 1B shows a structure in which the TM regions from the ensemble of NMR structures are aligned on the well-structured TM region (including the residues expected to form the binding site). The protein is well ordered in the TM region, except in the vicinity of the drug where both the side chains and the structure of the drug are variable.
Fig. 1.
Structure of the rimantadine binding site at the boundary of the A/M2 TM domain with the cytoplasmic tail. (A). A ribbon diagram showing four TM domain α-helices and the interactions between rimantadine (purple) and D44. Drawn from frame 7 of Protein Data Bank accession no. 2RLF. The variability in rimantadine-N to Asp-carboxylate geometries (distances in angstroms) are indicated. (B) A ribbon diagram showing a structure in which the TM regions from the ensemble of NMR structures are aligned on the well-structured TM region (including the residues expected to form the drug-binding site). The protein is well ordered in the TM region, except in the vicinity of the drug where both the side-chains and the drug are variable. Drawn from Protein Data Bank accession no. 2RLF from the data of Schnell and Chou (29).
These two distinct drug-binding sites led to two incompatible drug inhibition mechanisms (32). Stouffer et al. (28) proposed that amantadine physically occludes the pore of the channel restricting passage of protons and the naturally arising drug-resistant mutations (TM domain residues 27, 30, 31, and 34) are close to the proposed channel pore drug-binding site. The Hill coefficient of 1 for amantadine inhibition is consistent with one amantadine molecule blocking the channel (33) although other drug-binding models are possible. The finding of electron density in the pore of the channel in the presence of amantadine and the absence of density without amantadine is consistent with the drug being present in the pore of the channel (28), nonetheless the caveat has to be added that at 3.5 Å resolution it cannot be proven the density represents amantadine. Schnell and Chou (29) proposed channel inhibition occurred by an allosteric mechanism with external drug binding stabilizing the closed state making it more difficult to move the four helices and open the channel. Important in this regard is the notion of long-range helical effects because of the observation that an amantadine-resistant mutation maps to Leu 38 (L38F) (33) and it was suggested the mutation probably disturbs helix–helix packing and lowers the energetic cost of opening the channel (29).
We mutagenized D44 and R45 to alanine as these mutations would be expected to interfere with the observed rimantadine interaction and may lead to a drug insensitive channel, but the channel remained drug sensitive. We also substituted A/M2 pore residues 24–36 into BM2 (in place of BM2 pore residues 6–18) and this chimeric protein yielded a pH-activated ion channel that was partially inhibited by amantadine. Thus, our functional data suggest that amantadine/rimantadine binding outside of the channel pore is not the primary site associated with the pharmacological inhibition of the A/M2 ion channel.
Results and Discussion
Ion Channel Activity and Sensitivity to Amantadine Inhibition of M2 Mutants.
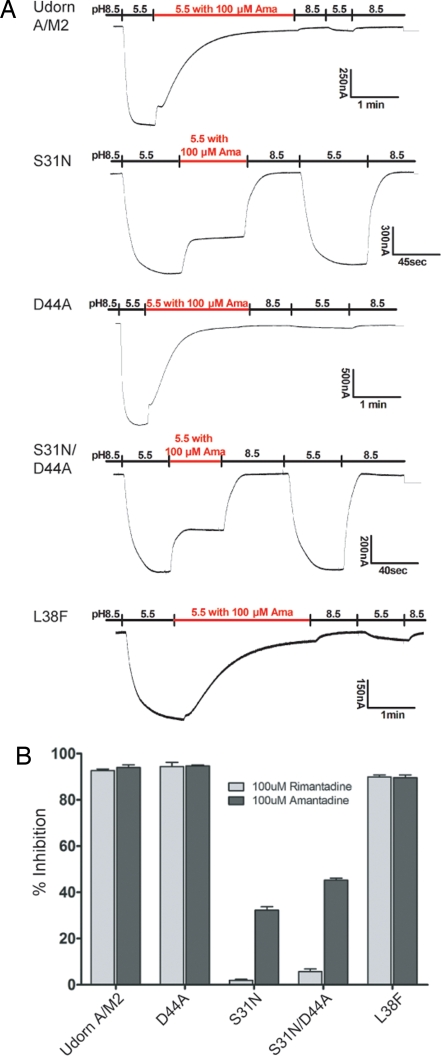
To investigate the interaction between A/M2 ion channel pore residue Asp 44 and amantadine/rimantadine, we mutated Asp 44 to alanine and examined the ion channel activity and its sensitivity to amantadine in oocytes of Xenopus laevis (Fig. 2A). We also examined the ion channel activity of a known amantadine-resistant mutant, S31N and a double mutant S31N/D44A. The levels of cell-surface expression of the WT and mutant channels in oocytes were equivalent (data not shown). The oocytes were initially bathed at pH 8.5 to keep the M2 channel in the closed state and then the bathing solution was changed to pH 5.5 to open the channel (4). Whereas, WT M2 and D44A ion channel currents were sensitive to amantadine, S31N and S31N/D44A channel currents were largely resistant to the drug (Fig. 2A). For WT A/M2 and D44A the amantadine inhibition of current remains even after the oocytes were returned to pH 8.5 and then bathed in pH 5.5 because amantadine inhibition is effectively irreversible on this time scale: the reverse reaction rate constant of amantadine block is 3 × 10−4s−1 (33). The expression and amantadine sensitivity of WT M2, S31N, D44A, and S31/D44A was also examined in mammalian cells (CHO-K1 cells) and very similar drug sensitivity to that found in oocytes was observed [supporting information (SI) Fig. S1]. The inward currents of CHO-K1 cells expressing the D44A and S31N/D44A mutants were transient in nature when the cells were exposed to solutions of low pH (Fig. S1), possibly indicating altered gating, consistent with the interactions observed between the Trp-41 gate and Asp44 in the NMR structure (27, 28). It has also been noted previously in a study of the differences between the ion channel activities of influenza virus strains A/chicken/Germany/34 (H7N1, Rostock strain) and A/chicken/Germany/27 (H7N7, Weybridge strain) that substitution of aspartic acid residue 44 for asparagine yields an amantadine sensitive channel activity (34).
Fig. 2.
Influenza virus M2 mutant ion channels D44A and L38F are sensitive to amantadine inhibition. Amantadine and rimantadine sensitivity of WT M2 and M2 mutant ion channels D44A, S31N, D44A/S31N, and L38F. The mRNAs encoding the WT and mutant M2 ion channels were expressed in oocytes of Xenopus laevis and surface currents measured using a two-electode voltage-clamp apparatus (4). (A) Representative recording traces WT M2 and mutant channels. Amantadine sensitivity of these A/M2 variants was determined by bathing the oocytes in pH 5.5 Barth's solution that contain 100 μM amantadine when the oocytes displayed maximal inward current (downward deflection). (B) The percentage of inhibition of A/M2 inward current in the presence of either 100 μM amantadine or 100 μM rimantadine (n = a minimum of 5).
In the external drug-binding model (29) the previously reported amantadine resistance of mutant L38F (33) was of importance because it was the only known amantadine resistance mutation C-terminal to those found between M2 residues 26 and 34. The existence of L38F as amantadine resistant bolstered the argument for long-range effects of drug binding stabilizing the four-helix bundle in the closed state whereas the mutant would destabilize the helices and allow the channel to open. Thus, we constructed the L38F mutation into the identical M2 sequence used for all other mutants used here (M2 sequence derived from influenza A/Udorn/72) and investigated channel properties of M2 mutant L38F. As shown in Fig. 2A mutant L38F had ion channel properties indistinguishable from WT M2 and was completely sensitive to amantadine. It is possible that the earlier finding of amantadine insensitivity of the L38F mutant occurred because less time was allowed for inhibition to occur, and we conclude there are no known examples of mutations that lead to M2 amantadine resistance more C-terminal than residue 34. It can also be deduced from the data shown in Betakova et al., (32) that M2 of Rostock influenza virus that contains the L38F mutation is amantadine sensitive.
Amantadine and its methyl derivative rimantadine both inhibit the M2 ion channel activity and influenza virus replication (5, 26, 33). Because in the X-ray structure amantadine was modeled (28) and in the NMR structure rimantadine was modeled (29) it was important to determine that there was no major difference in the inhibitory action of the two drugs. As shown in Fig. 2B the ion channel activity of WT M2 and mutants D44A and L38F was completely sensitive to both drugs. Mutants S31N and S31N/D44A were resistant to the drug but mutants containing S31N were more resistant to rimantadine than amantadine. This may be related to the fact that several drug-resistant mutations in M2 pore-lining residues retain the ability to bind drug (35) and rimantadine may bind less well to the S31N protein than amantadine.
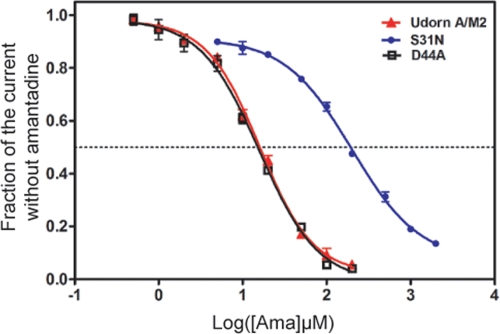
To be sure that amantadine was equally efficacious in inhibiting D44A as it is WT, a dose–response curve was determined by measuring inhibition after 2 min of drug application for each of many concentrations of the drug. It was found that WT A/M2 and D44A had virtually identical amantadine sensitivities (Fig. 3).
Fig. 3.
Amantadine isochronic inhibition curves for WT A/M2, D44A and S31N ion channels. Nine different concentrations of amantadine were applied to oocytes expressing WT, D44A, and S31N ion channels for 2 min at pH 5.5 and the current was measured before and after drug addition. Three oocytes were used for each drug concentration. The results were fit into the dose-response curve in GraphPad Prism 5. The best fit IC50 values for WT A/M2, D44A, and S31N were 16.0 ± 1.1, 15.8 ± 1.1, and 199.9 ± 1.1 μM, respectively.
Growth and Drug Sensitivity of Influenza Viruses Containing M2 Mutations.
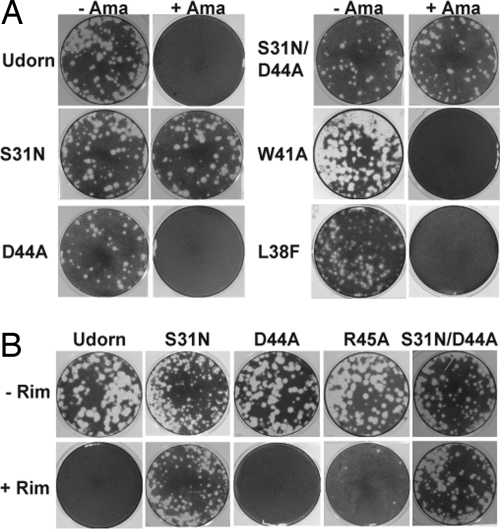
To examine the effect of M2 mutations D44A, S31N, S31N/D44A, and L38F in the context of an influenza virus infection, the mutations were introduced into the influenza A virus genome using reverse genetics procedures (36, 37) and the viruses recovered. Because of the involvement of M2 residue W41 (the channel gate) with D44 and R45, and in the NMR model the interaction of rimantadine with all three residues, two additional M2 mutant viruses W41A and R45A were constructed and recovered. As shown in Fig. 4, all of the viruses had equivalent plaque sizes. Although D44A showed a similar plaque size to WT virus, single-step growth curves of D44A virus showed slightly slower kinetics than WT virus and D44A reached a maximum titer of 1 log lower than WT virus cells (data not shown). Nonetheless the D44A mutant virus apparently contained an ion channel activity sufficient for growth in tissue culture. Plaque formation of WT virus and M2 mutant viruses D44A, L38F, W41A, and R45A was sensitive to amantadine/rimantadine (Fig. 4 A and B) and plaque formation of S31N and S31N/D44A was insensitive to either drug treatment. The drug sensitivity/insensitivity of viruses containing the M2 mutations is completely consistent with the electrophysiology recordings of the mutant protein ion channel activities. It is interesting to note that even though W41 is a key residue in channel function (the gate) and D44 and R45 interact with W41 (Fig. 1), the M2 channel in the virus can tolerate these changes and remains sensitive to amantadine/rimantadine.
Fig. 4.
Effect of amantadine and rimantadine on influenza A virus plaque formation. WT influenza virus (A/Udorn/72) and influenza virus containing mutations in the M2 TM domain were recovered from cloned DNA (36) and assayed for plaque formation on MDCK cells in the presence or absence of 5 μM amantadine (Ama) (A) or 5 μM rimantadine (Rim) (B). Monolayers of MDCK cells were preincubated with or without amantadine (5 μM) or rimantadine (5 μM) for 30 min and then infected with the appropriate virus in the presence or absence of the drug. After 1 h of incubation, monolayers were washed with PBS (with or without drug) and overlaid with 0.6% Avicel microcrystalline cellulose-DMEM supplemented with 1.0 μg/ml of N-acetyl trypsin, with or without amantadine (5 μM) or rimantadine (5 μM). After 2–3 days of incubation, monolayers were fixed and stained with naphthalene black dye solution. The equivalent dilution is shown for each matched pair of monolayers with and without drug (−Ama, +Ama; −Rim, +Rim).
An A/M2 and BM2 Chimeric TM Domain Renders the BM2 Ion Channel Amantadine Sensitive.
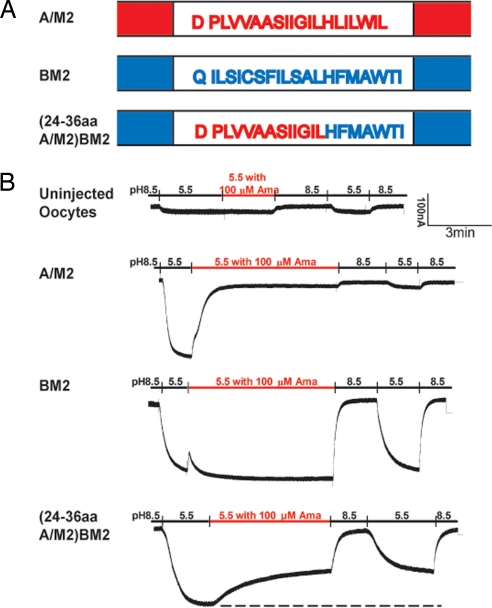
To provide another line of evidence for the notion that amantadine inhibits the A/M2 ion channel by binding to the pore between residues 26 and 34, a hybrid channel was constructed between the two proton-selective ion channels A/M2 (amantadine sensitive) and BM2 (amantadine insensitive). BM2 TM domain residues 6–18 were replaced with A/M2 residues 24–36. The chimeric channel, (24–36 aa A/M2)BM2 was expressed in oocytes and its channel activity measured in comparison to both A/M2 and BM2 ion channel activities. Like A/M2 and BM2 the chimeric channel was activated by pH 5.5 (Fig. 5). The current of the A/M2 channel was inhibited by the addition of 100 μM amantadine (94.0% ± 0.9% SE, n = 8) and 100 μM rimantadine (92.7% ± 0.7% SE; n = 8) whereas in contrast the current of BM2 was not inhibited by the addition of 100 μM amantadine (−1.4% ± 0.2%, n = 5). Importantly, the current of the chimeric channel was partially inhibited by amantadine (49.1 ± 1.7% SE, n = 8) and 100 μM rimantadine (50.2 ± 1.6% SE, n = 3). The time constant (τ) (in seconds) of inhibition was determined by fitting the inhibition curves to a standard exponential decay equation and for A/M2 channel τ = 34 ± 2.3 sec. SE (n = 8), and for the chimeric (24–36 aa A/M2)BM2 channel τ = 108.4 ± 2.8 sec SE (n = 8). Given that single side-chain substitutions at residues 27, 30, 31, and 34 lead to amantadine resistance, the sensitivity of the chimeric channel to amantadine is especially noteworthy, particularly as this chimeric pore may well be altered in structure in subtle ways from the WT M2 pore. Thus, the simplest interpretation of the data is that amantadine/rimantadine interacts with A/M2 residues 24–36 to occlude the channel pore and that these residues can be transferred into the BM2 pore to render BM2 partially amantadine sensitive.
Fig. 5.
Generation of a chimeric A/M2 and BM2 ion channel that is amantadine sensitive. (A) Schematic diagram showing the TM domain residues of A/M2 (derived from sequence of influenza virus A/Udorn/72 M2) (45) and BM2 (derived from sequence of influenza virus B/Lee/40 BM2) (46), and a chimeric BM2 protein with BM2 TM domain residues 6–18 aa substituted with A/M2 TM domain residues 24–36. (B) Ion channel activities of A/M2, BM2, and the chimeric ion channel (24–36 aa A/M2)BM2. Oocytes expressing the chimera ion channel displayed robust pH-activated inward current as compared to uninjected oocytes. Amantadine (100 μM) inhibited A/M2 current (94.0 ± 0.9%; n = 8). BM2 ion channel activity was insensitive to 100 μM amantadine. The chimeric BM2 chimera ion channel was partially inhibited (49.1% ± 1.7%; n = 8) by 100 μM amantadine and partially inhibited by 100 μM rimantadine (50.2% ± 1.6%; n = 3) (data not shown). Like WT M2 channel the drug inhibition was not reversible over a short time period.
Summary.
Our functional studies on the influenza A virus A/M2 proton-selective ion channel and its inhibition by amantadine or rimantadine suggest that amantadine/rimantadine binding outside of the channel pore is not the primary site associated with the pharmacological inhibition of the A/M2 ion channel. First, mutation of D44 and R45 to alanine did not alter the sensitivity of the channels to the drug, suggesting that the interactions of rimantadine with D44 and R45 are not important for drug inhibition. Second, the original finding that mutating L38 to phenylalanine caused amantadine resistance appears to be incorrect and thus there is no evidence for long-range effects of amantadine resistance, which was proposed in the allosteric mechanism for rimantadine inhibition from the cytoplasmic side of the four-helix bundle. Third, the chimeric channel (24–36 aa A/M2)BM2 channel is activated by low pH and partially inhibited by amantadine. The interpretation of these data that we prefer is that we transferred the drug site from A/M2 to the normally drug-insensitive BM2 ion channel. It seems unlikely amantadine stabilizes the closed state of the chimeric channel by an allosteric mechanism and involving M2 residues 24–36, when this chimeric protein lacks the D44 and R45 residues that interact with rimantadine in the NMR model.
There are many examples of second substrate or ligand-binding sites for proteins but the biological significance of these second sites is often unclear. For influenza virus, a second sialic acid binding site was identified on the hemagglutinin (38) and the avian N9 neuraminidase contains a second sialic acid binding site (39, 40). However, neither of these second sites appears to be required for virus replication. Amantadine partitions into lipid bilayers (41–43) giving the drug access to the cytoplasmic side of the four-helix bundle. Whether the external rimantadine binding site, that we consider to be a second drug-binding site, has another role in properties of A/M2 function that are not directly associated with inhibition of channel activity remains to be determined.
Materials and Methods
Cells, Viruses, and Plasmids.
293-T and Madin–Darby canine kidney (MDCK) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% FBS. Influenza A/Udorn/72 virus (WT) and mutant viruses were propagated in MDCK cells overlaid with serum-free DMEM containing 3.0 μg/ml N-acetyl trypsin (NAT; Sigma-Aldrich, St. Louis, MO) at 37°C. WT and mutant viruses were generated by using reverse genetics from cDNAs essentially as described previously (36). The eight genome-sense (pHH21) plasmids and four protein-expressing (pcDNA3.1) plasmids used to generate influenza virus by reverse genetics have been described previously (36). Mutations into the M2 gene in pHH21 and pGEM3 vectors were generated using Quick Change mutagenesis (Stratagene, La Jolla, CA). 293-T cells were transfected using TransIT-LT1 (Mirus, Madison, WI) according to the manufacturer protocols. Virus stocks were propagated in MDCK cells, and the virus titers were determined by plaque assay on MDCK cells. For determination of viral genome sequences viral RNA was extracted by using the QIAamp viral RNA kit (Qiagen, Valencia, CA), followed by Super Reverse Transcriptase (Molecular Genetic Resources, Tampa, FL), using genome-specific primers, and amplified with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA). The complete nucleotide sequences of the M genes were determined using a 3100-Avant genetic analyzer (Applied Biosystems).
For additional information, see SI Materials and Methods.
Plaque Assays.
Confluent monolayers of MDCK cells were incubated with 10-fold serially diluted virus samples in DMEM 1% BSA for 1 h at 37°C. The inoculums were removed, and the cells were washed with phosphate-buffered saline (PBS). The cells were then overlaid with DMEM containing 0.6% Avicel microcrystalline cellulose (FMC BioPolymer, Philadelphia, PA) (44) and NAT (1.0 μg/ml). To examine the effect of the drug (amantadine or rimantadine) on plaque formation, monolayers were preincubated with DMEM supplemented with the drug (5 μM) at 37°C for 30 min, and virus samples were preincubated with DMEM 1% BSA with the drug (5 μM) at 4°C for 30 min before infection. At 2–3 days after infection, the monolayers were fixed and stained with naphthalene black dye solution (0.1% naphthalene black, 6% glacial acetic acid, 1.36% anhydrous sodium acetate).
mRNA Synthesis, Culture, and Microinjection of Oocytes.
WT A/M2-pGEM3 cDNA and mutant DNAs (D44A/S31N/D44A,S31N/L38F) were linearized using HindIII and in vitro transcription reactions were performed on the linearized cDNA using a T7 mMESSAGE mMACHINE transcription kit (Ambion, Austin, TX) and injected into oocytes as described previously (4).
Two-Electrode Voltage-Clamp Analysis.
Whole-cell two-electrode voltage-clamp currents were measured 48–72 h after injection of oocytes as described previously (4). Currents were acquired and analyzed using the pCLAMP 10.0 software package (Axon Instruments, Sunnyvale, CA).
Supplementary Material
Acknowledgments.
The research was supported by the National Institutes of Health research grants R01 AI-20201 (R.A.L.) and R01 Al-57363 (L.H.P.). R.A.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804958105/DCSupplemental.
References
- 1.Lamb RA, Krug RM. In: Fields Virology. 4th Ed. Knipe D M, Howley PM, editors. Philadelphia: Lippincott, Williams and Wilkins; 2001. pp. 1487–1531. [Google Scholar]
- 2.Lamb RA, Zebedee SL, Richardson CD. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 3.Paterson RG, Takeda M, Ohigashi Y, Pinto LH, Lamb RA. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology. 2003;306:7–17. doi: 10.1016/s0042-6822(02)00083-1. [DOI] [PubMed] [Google Scholar]
- 4.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 5.Chizhmakov IV, et al. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mould JA, et al. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 7.Zhirnov OP. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology. 1990;176:274–279. doi: 10.1016/0042-6822(90)90253-n. [DOI] [PubMed] [Google Scholar]
- 8.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 9.Ciampor F, et al. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans Golgi compartment. Virology. 1992;188:14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- 10.Grambas S, Hay AJ. Maturation of influenza A virus hemagglutinin–estimates of the pH encountered during transport and its regulation by the M2 protein. Virology. 1992;190:11–18. doi: 10.1016/0042-6822(92)91187-y. [DOI] [PubMed] [Google Scholar]
- 11.Lamb RA, Pinto LH. In: Contemporary Topics in Influenza Virology. Kawaoka Y, editor. Wymondham, Norfolk, UK: Caister Academic; 2005. pp. 65–93. [Google Scholar]
- 12.Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 13.Sugrue RJ, Hay AJ. Structural characteristics of the M2 protein of the influenza A viruses: Evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holsinger LJ, Shaughnessy MA, Micko A, Pinto LH, Lamb RA. Analysis of the posttranslational modifications of the influenza virus M2 protein. J Virol. 1995;69:1219–1225. doi: 10.1128/jvi.69.2.1219-1225.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castrucci MR, et al. The cysteine residues of the M2 protein are not required for influenza A virus replication. Virology. 1997;238:128–134. doi: 10.1006/viro.1997.8809. [DOI] [PubMed] [Google Scholar]
- 16.Sakaguchi T, Tu Q, Pinto LH, Lamb RA. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc Natl Acad Sci USA. 1997;94:5000–5004. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto LH, et al. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer CM, Pinto LH, Cross TA, Lamb RA. The influenza virus M2 ion channel protein: Probing the structure of the transmembrane domain in intact cells by using engineered disulfide cross-linking. Virology. 1999;254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- 19.Shuck K, Lamb RA, Pinto LH. Analysis of the pore structure of the influenza A virus M2 ion channel by the substituted-cysteine accessibility method. J Virol. 2000;74:7755–7761. doi: 10.1128/jvi.74.17.7755-7761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balannik V, Lamb RA, Pinto LH. The oligomeric state of the active BM2 ion channel protein of influenza B virus. J Biol Chem. 2008;283:4895–4904. doi: 10.1074/jbc.M709433200. [DOI] [PubMed] [Google Scholar]
- 21.Ma C, et al. Identification of the pore-lining residues of the BM2 ion channel protein of influenza B virus. J Biol Chem. 2008 doi: 10.1074/jbc.M710302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mould JA, et al. Permeation and activation of the M2 ion channel of influenza A virus. J Biol Chem. 2000;275:31038–31050. doi: 10.1074/jbc.M003663200. [DOI] [PubMed] [Google Scholar]
- 23.Hu J, et al. Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci USA. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkataraman P, Lamb RA, Pinto LH. Chemical rescue of histidine selectivity filter mutants of the M2 ion channel of influenza A virus. J Biol Chem. 2005;280:21463–21472. doi: 10.1074/jbc.M412406200. [DOI] [PubMed] [Google Scholar]
- 25.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 26.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamb RA, Holsinger LJ, Pinto LH. In: Receptor-Mediated Virus Entry into Cells. Wimmer E, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 303–321. [Google Scholar]
- 28.Stouffer AL, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovacs FA, Cross TA. Transmembrane four-helix bundle of influenza A M2 protein channel: Structural implications from helix tilt and orientation. Biophys J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 32.Miller M. Coughing up flu's proton channels (News and Views) Nature. 2008;451:532–533. doi: 10.1038/451532a. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: Characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betakova T, Ciampor F, Hay AJ. Influence of residue 44 on the activity of the M2 proton channel of influenza A virus. J Gen Virol. 2005;86:181–184. doi: 10.1099/vir.0.80358-0. [DOI] [PubMed] [Google Scholar]
- 35.Astrahan P, Kass I, Cooper MA, Arkin IT. A novel method of resistance for influenza against a channel-blocking antiviral drug. Proteins. 2004;55:251–257. doi: 10.1002/prot.20018. [DOI] [PubMed] [Google Scholar]
- 36.Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J Virol. 2002;76:1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neumann G, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauter NK, et al. Crystallographic detection of a second ligand binding site in influenza virus hemagglutinin. Proc Natl Acad Sci USA. 1992;89:324–328. doi: 10.1073/pnas.89.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Air GM, Ritchie LR, Laver WG, Colman PM. Gene and protein sequence of an influenza neuraminidase with hemagglutinin activity. Virology. 1985;145:1–7. doi: 10.1016/0042-6822(85)90206-5. [DOI] [PubMed] [Google Scholar]
- 40.Varghese JN, et al. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc Natl Acad Sci USA. 1997;94:11808–11812. doi: 10.1073/pnas.94.22.11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epand RM, Epand RF, McKenzie RC. Effects of viral chemotherapeutic agents on membrane properties: Studies of cyclosporin A, benzlooxycarbonyl-D-Phe-L-Phe-Gly and amantadine. J Biol Chem. 1987;262:1526–1529. [PubMed] [Google Scholar]
- 42.Subczynski WK, Wojas J, Pezeshk V, Pezeshk A. Partitioning and localization of spin-labeled amantadine in lipid bilayers: An epr study. J Pharmaceut Sci. 2000;87:1249–1254. doi: 10.1021/js970381n. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Schnell JR, Chou JJ. Amantadine partition and localization in phospholipid membrane: A solution NMR study. Biochem Biophys Res Commun. 2004;324:212–217. doi: 10.1016/j.bbrc.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 44.Matrosovich M, Matrosovich T, Garten W, Klenk HD. New low-viscosity overlay medium for viral plaque assays. Virol. J. 2006:3.63. doi: 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamb RA, Lai C-J. Conservation of the influenza virus membrane protein (M1) amino acid sequence and an open reading frame of RNA segment 7 encoding a second protein (M2) in H1N1 and H3N2 strains. Virology. 1981;112:746–751. doi: 10.1016/0042-6822(81)90319-6. [DOI] [PubMed] [Google Scholar]
- 46.Briedis DJ, Lamb RA, Choppin PW. Sequence of RNA segment 7 of the influenza B virus genome: Partial amino acid homology between the membrane proteins (M1) of influenza A and B viruses and conservation of a second open reading frame. Virology. 1982;116:581–588. doi: 10.1016/0042-6822(82)90150-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.