Abstract
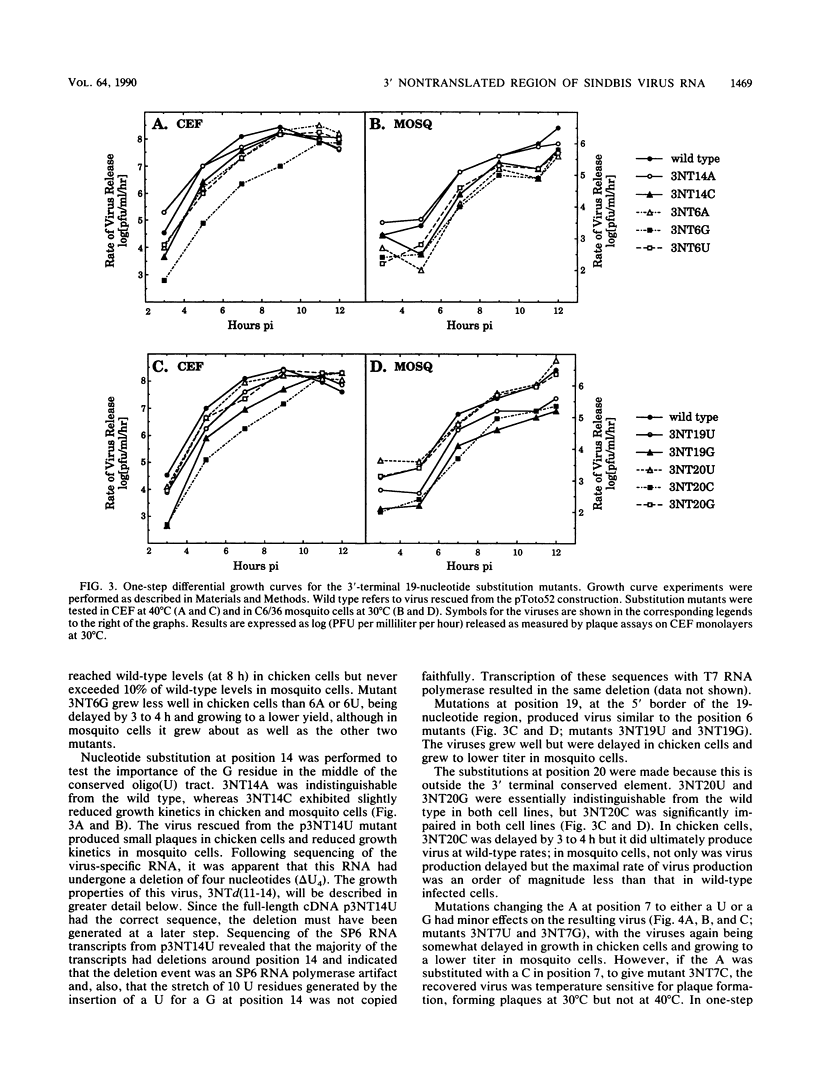
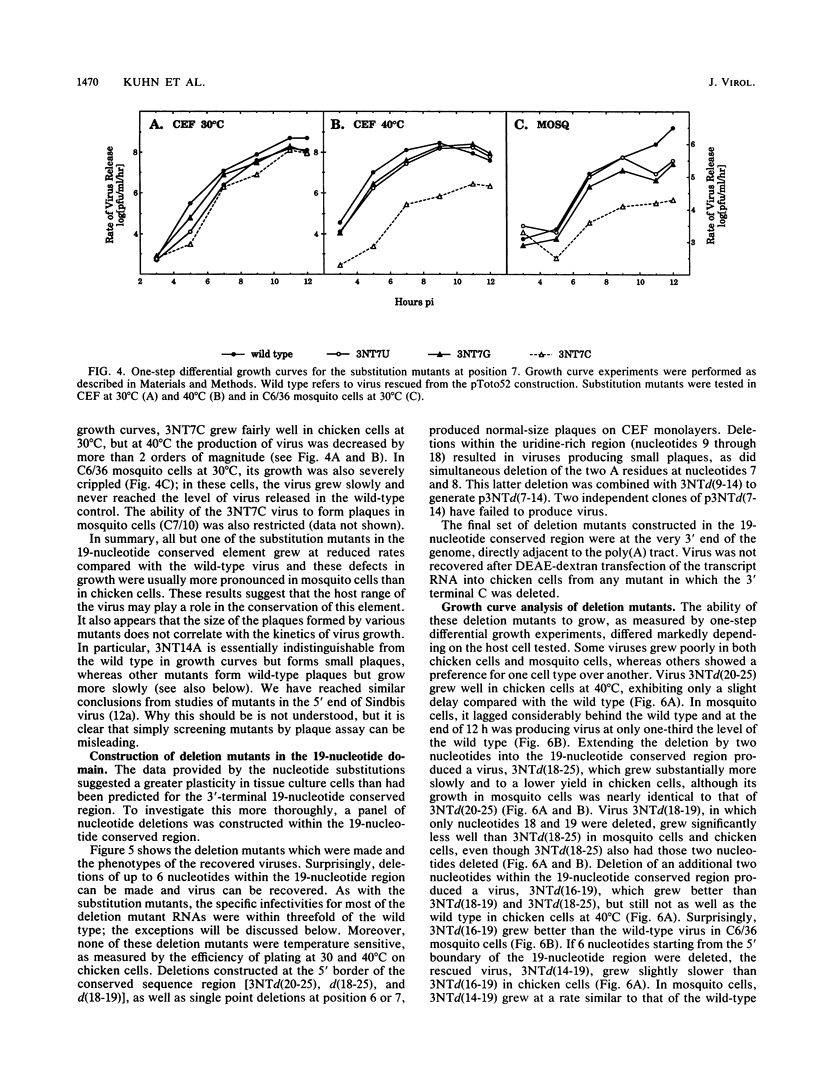
A cDNA clone from which infectious RNA can be transcribed was used to construct 42 site-specific mutations in the 3' nontranslated region of the Sindbis virus genome. The majority of these mutations were made in the 3'-terminal 19-nucleotide conserved sequence element and consisted of single nucleotide substitutions or of small (1 to 8) nucleotide deletions. An attempt was made to recover mutant viruses after transfection of SP6-transcribed RNA into chicken cells. In most cases, viable virus was recovered, but almost all mutants grew more poorly than wild-type virus when tested under a number of culture conditions. In the case of mutations having only a moderate effect, the virus grew as well as the wild type but was slightly delayed in growth. Mutations having a more severe effect led to lower virus yields. In many cases, virus growth was more severely impaired in mosquito cells than in chicken cells, but the opposite phenotype was also seen, in which the mutant grew as well as or better than the wild type in mosquito cells but more poorly in chicken cells. One substitution mutant, 3NT7C, was temperature sensitive for growth in chicken cells and severely crippled for growth in mosquito cells. Insertion mutations were also constructed which displaced the 19-nucleotide element by a few nucleotides relative to the poly(A) tail. These mutations had little effect on virus growth. Deletion of large regions (31 to 293 nucleotides long) of the 3' nontranslated region outside of the 19-nucleotide element resulted in viruses which were more severely crippled in mosquito cells than in chicken cells. From these results, the following principles emerge. (i) The entire 3' nontranslated region is important for efficient virus replication, although there is considerable plasticity in this region in that most nucleotide substitutions or deletions made resulted in viable virus and, in some cases, in virus that grew quite efficiently. Replication competence was particularly sensitive to changes involving the C at position 1, the A at position 7, and a stretch of 9 U residues punctuated by a G at position 14. (ii) The panel of mutants examined collectively deleted the entire 3' nontranslated region. Only mutants in which 8 nucleotides in the 3' terminal 19 nucleotides had been deleted or in which the 3' terminal C was deleted were nonviable. Although the 3' terminal C was essential for replication, it could be displaced by at least 7 nucleotides from its 3' terminal position adjacent to the poly(A) tract.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barton D. J., Sawicki S. G., Sawicki D. L. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J Virol. 1988 Oct;62(10):3597–3602. doi: 10.1128/jvi.62.10.3597-3602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Condreay L. D., Adams R. H., Edwards J., Brown D. T. Effect of actinomycin D and cycloheximide on replication of Sindbis virus in Aedes albopictus (mosquito) cells. J Virol. 1988 Aug;62(8):2629–2635. doi: 10.1128/jvi.62.8.2629-2635.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgarno L., Rice C. M., Strauss J. H. Ross River virus 26 s RNA: complete nucleotide sequence and deduced sequence of the encoded structural proteins. Virology. 1983 Aug;129(1):170–187. doi: 10.1016/0042-6822(83)90404-x. [DOI] [PubMed] [Google Scholar]
- Faragher S. G., Dalgarno L. Regions of conservation and divergence in the 3' untranslated sequences of genomic RNA from Ross River virus isolates. J Mol Biol. 1986 Jul 20;190(2):141–148. doi: 10.1016/0022-2836(86)90287-1. [DOI] [PubMed] [Google Scholar]
- Hahn C. S., Strauss E. G., Strauss J. H. Dideoxy sequencing of RNA using reverse transcriptase. Methods Enzymol. 1989;180:121–130. doi: 10.1016/0076-6879(89)80097-7. [DOI] [PubMed] [Google Scholar]
- Hahn Y. S., Grakoui A., Rice C. M., Strauss E. G., Strauss J. H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J Virol. 1989 Mar;63(3):1194–1202. doi: 10.1128/jvi.63.3.1194-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Strauss E. G., Strauss J. H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J Virol. 1989 Jul;63(7):3142–3150. doi: 10.1128/jvi.63.7.3142-3150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. J., Kinney R. M., Kost C. L., Trent D. W. Molecular determinants of alphavirus neurovirulence: nucleotide and deduced protein sequence changes during attenuation of Venezuelan equine encephalitis virus. J Gen Virol. 1986 Sep;67(Pt 9):1951–1960. doi: 10.1099/0022-1317-67-9-1951. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Weiss B. G., Tsiang M., Huang H., Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986 Jan 17;44(1):137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- Niesters H. G., Strauss J. H. Mutagenesis of the conserved 51-nucleotide region of Sindbis virus. J Virol. 1990 Apr;64(4):1639–1647. doi: 10.1128/jvi.64.4.1639-1647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J. H., Rice C. M., Dalgarno L., Strauss E. G., Strauss J. H. Sequence studies of several alphavirus genomic RNAs in the region containing the start of the subgenomic RNA. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5235–5239. doi: 10.1073/pnas.79.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J. H., Strauss E. G., Strauss J. H. Comparative studies of the 3'-terminal sequences of several alpha virus RNAs. Virology. 1981 Mar;109(2):281–289. doi: 10.1016/0042-6822(81)90499-2. [DOI] [PubMed] [Google Scholar]
- Ou J. H., Trent D. W., Strauss J. H. The 3'-non-coding regions of alphavirus RNAs contain repeating sequences. J Mol Biol. 1982 Apr 25;156(4):719–730. doi: 10.1016/0022-2836(82)90138-3. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Synthesis, cleavage and sequence analysis of DNA complementary to the 26 S messenger RNA of Sindbis virus. J Mol Biol. 1981 Aug 15;150(3):315–340. doi: 10.1016/0022-2836(81)90550-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. G., Lenches E. M., Strauss J. H. Mutants of sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology. 1976 Oct 1;74(1):154–168. doi: 10.1016/0042-6822(76)90137-9. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]



