Abstract
In this work, we re-examine the previously reported phenomenon of the creation of a superactive glutamate dehydrogenase by proteolytic modification by chymotrypsin and explore the various discrepancies that came to light during those studies. We find that superactivation is caused by cleavage at the N terminus of the protein and not the C-terminal allosteric site, as has previously been suggested. N-terminal sequencing reveals that TLCK-treated chymotrypsin cleaves bovine glutamate dehydrogenase at phenylalanine 10. We suggest that trypsin contamination in nontreated chymotrypsin may have led to the production of the larger 4–5 kDa digestion product, previously misinterpreted as having caused the activation. In line with some previous studies, we can confirm that GTP inhibition is attenuated to some extent by the proteolysis, while ADP activation is almost abolished. Utilizing the recently solved structures of bovine glutamate dehydrogenase, we illustrate the cleavage points.
Keywords: chymotrypsin, glutamate dehydrogenase, activation, guanosine triphosphate, adenosine triphosphate
The glutamate dehydrogenase of bovine liver, readily available commercially in a pure form from the 1950s onward, attracted the interest of enzymologists over many years because of its interesting and complex regulatory behavior (Olson and Anfinsen 1953; Talal and Tomkins 1964; Yielding et al. 1964; Engel and Dalziel 1969; Huang and Frieden 1969) cited by Monod et al. (1963) as one of the striking examples to establish the reality of allosteric control. Specifically, in addition to homotropic allosteric behavior with the oxidized and reduced coenzymes (Olson and Anfinsen 1953; Yielding et al. 1964; Huang and Frieden 1969), the enzyme also exhibited potent inhibition by GTP and activation by ADP (Wolff 1962; Hershko and Kindler 1966). This led to much experimentation and controversy over the number and location of nucleotide sites and the relationship between them (Markau et al. 1971; Koberstein and Sund 1973; Bayley and O'Neill 1980). Chemical modification experiments (Colman and Frieden 1966; Price and Radda 1969; Coffee et al. 1971) uniformly showed that the response to purine nucleotides was associated with the C-terminal 50 amino acids of the protein, a finding that correlated well with the finding that homologous GDHs of prokaryotes and fungi lacked both the 50-amino acid section and the regulation by GTP/ADP (Wootton et al. 1974; Mattaj et al. 1982). Latterly, the recent solution of the three-dimensional structure of bovine GDH (Peterson and Smith 1999) has revealed that these 50 amino acids form an “antenna” protruding from the main body of the hexameric structure and totally missing in various other solved GDH structures (Rice et al. 1987; Stillman et al. 1995). This antenna is suggested to have been “exapted” from its original function linking fatty acid oxidation to amino acid catabolism in Ciliates (Allen et al. 2004).
Against this background, we have re-examined a remarkable series of findings on the results of partial proteolysis of bovine GDH. Initially, Beynon and Kay (1976), using bovine GDH as one of a series of proteinase substrates in an exploration of differential intracellular protein turnover, were surprised to discover that treatment with chymotrypsin resulted in a gradual three- to fourfold activation, in parallel with a decrease of about 5 kDa in the size of the main protein band seen in SDS-PAGE. They reported that the proteolysed enzyme was desensitized to ADP. Although it apparently retained the GTP response, it appeared possible that a domain responsible for ADP activation was being physically cleaved off the enzyme molecule (Aitchison and Engel 1980).
Subsequent experiments by Aitchison and Engel (1983) showed that, despite the proteolytic clip revealed by SDS-PAGE, the native Mr was unchanged, so that the putative 5-kDa fragment evidently remained noncovalently attached. They also showed that the concept of desensitization was an oversimplification, and that the qualitative result depended critically on the conditions of assay—pH, coenzyme concentration, and use of NAD+ or NADP+, etc.
Beynon and Kay (1976) had also noted a slow decline in activity following the initial activation, correlating with evident extensive further proteolysis. In a subsequent study, Place and Beynon (1982) showed that this decline varied with the batch of chymotrypsin and could be completely eliminated by using TLCK-treated chymotrypsin, being apparently due to the combined action of chymotrypsin and small amounts of contaminating trypsin. They did not comment, however, on a puzzling difference in the Mr of the proteolytic fragments as compared with the original report (Beynon and Kay 1976): The new SDS-PAGE results appeared to show removal of a much smaller fragment (1–2 kDa) to give a protein with a major band at ∼53 kDa and still showing the kinetic activation. These authors concluded that “chymotryptic digestion generates a ‘superactive’ form that is quite stable to further digestion” (by chymotrypsin), but that the addition of small amounts of trypsin to this stable species results in rapid inactivation. The apparent effect of trypsin was also of interest, given earlier reports that this proteinase was responsible for cleavage of the N-terminal amino acids (Hucho et al. 1975), a complication in the purification of GDH (McCarthy et al. 1980).
In the absence of protein chemical analysis, this series of observations left a puzzle as to the nature of these proteolytic changes, and in particular, as to the relationship of the reported small (1–2 kDa) and large (4–6 kDa) chymotryptic fragments. With the availability at last of a three-dimensional framework (Peterson and Smith 1999) to aid interpretation, it seemed desirable to establish at which end of the protein molecule the crucial cleavage occurs. With this information, it might now be possible to interpret the basis of the observed dramatic alteration in kinetic properties. In addition to the recent availability of the crystallographic structure, a recent revival of interest in the activity and allosteric properties of mammalian glutamate dehydrogenase has centered on the discovery that regulatory mutations of the glutamate dehydrogenase gene may cause hyperinsulinism and hyperammonemia in infants (Stanley et al. 1998). These disease-causing mutations impair enzyme sensitivity to allosteric inhibition by GTP and ATP, resulting in a gain of function (MacMullen et al. 2001). Haigis et al. (2006) also highlighted the importance of glutamate dehydrogenase and its down-regulation by ADP-ribosylation as important steps in the cell mechanism of regulating insulin excretion by pancreatic β cells.
Results and Discussion
Cleavage of GDH with TLCK-chymotrypsin
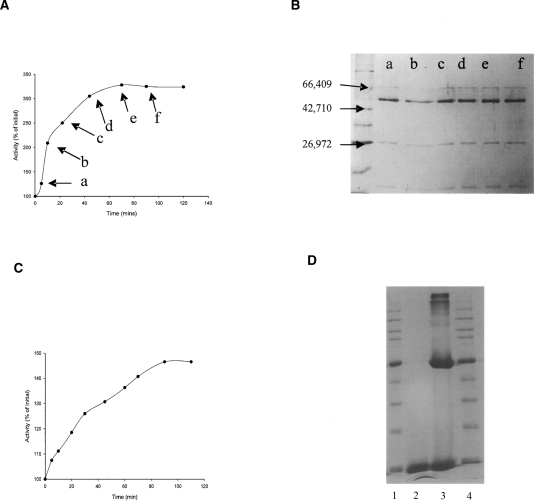
Figures 1 and 2 illustrate how chymotrypsin cleaves bovine glutamate dehydrogenase in such a fashion as to cause a rise in activity with NADP+ and NAD+ as coenzyme, but it is clear that the cleavage profiles differ depending on the grade of chymotrypsin utilized. Chymotrypsin is usually sold in two forms, one treated with TLCK, which will irreversibly inhibit trypsin, and also an untreated preparation, in which trace contamination by trypsin from the pancreatic source material might be expected. In both incubations there is a marked activation, over threefold in the NADP+ assay with TLCK-treated chymotrypsin. The distinguishing aspect with untreated chymotrypsin is that the activation is followed by a decrease in activity, accompanied by simultaneous formation of a GDH band at ∼51 kDa. There is no sign of this band in the incubation with TLCK-treated chymotrypsin, and Figure 1A suggests that the cleavage associated with the activation causes very little change in the size of the larger fragment, implying cleavage close to one end of the protein.
Figure 1.
Activation of bovine glutamate dehydrogenase by digestion with TLCK-treated chymotrypsin. GDH (1 mg/mL) was incubated with 0.2 mg/mL TLCK-treated chymotrypsin (Mr = 25,700) in 0.1 M phosphate buffer, pH 7.0 at 30°C. At the time points indicated in A by the labels a–f, samples were removed for assay of dehydrogenase activity (A) as described in Materials and Methods, utilizing NADP+ as the cofactor, and for SDS-PAGE analysis (B). For the latter purpose, 50-μL samples were treated with PMSF before being loaded onto a 12% SDS-PAGE gel. The commercial GDH (Mr = 55,600) also contains BSA (Mr = 66,400), added by the manufacturer as a stabilizing agent. Broad-range molecular weight markers were loaded in the first lane in B. Visible bands are rabbit muscle myosin (212,000), Escherichia coli MBP-β-galactosidase fusion (158,194), E. coli β-galactosidase (116,351), rabbit muscle phosphorylase b (97,184), BSA (66,409), bovine glutamic dehydrogenase (55,561), E. coli MBP2 (maltose binding protein) (42,710), E. coli thioredoxin reductase (34,622), E. coli triosephosphate isomerase (26,972), soybean trypsin inhibitor (20,100), and chicken egg-white lysozyme (14,313). Three marker sizes are shown with arrows. (C) Graph shows an activation time course similar to that in A, but in this case, activity was measured with glutamate and NAD+ at pH 8.5. (D) A control incubation of BSA with chymotrypsin under the same conditions. Lanes 1 and 4 contain broad-range markers, lane 2 contains just chymotrypsin, and lane 3 contains chymotrypsin incubated with BSA for 40 min.
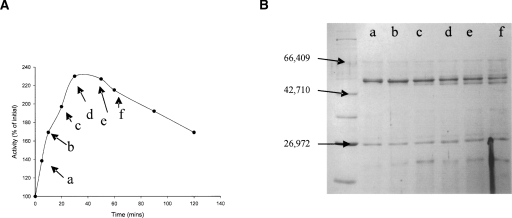
Figure 2.
Activation of bovine glutamate dehydrogenase by digestion with untreated chymotrypsin. GDH (Mr = 55,561) (0.5 mg/mL) was incubated with 0.2 mg/mL chymotrypsin (Mr = 25,700), in 0.1 M phosphate buffer (pH 7.0) at 30°C. Dehydrogenase activity was periodically assayed (A) as described in Materials and Methods, utilizing NADP+ as the cofactor. Samples (50 uL) were also removed for treatment with PMSF before being loaded onto 12% SDS-PAGE gels. (B) Broad-range molecular weight markers (as in Fig. 1) were loaded in the first lane.
The initial reports of both Beynon and Kay (1976) and Aitchison and Engel (1980) correlated the activation seen in all studies with the appearance of a GDH band shortened by 4–5 kDa. However, the present result strongly indicates that the activation does not require that cleavage. Instead, the chymotryptic cleavage responsible for the activation causes only a small Mr change, and the 51-kDa fragment is only seen when contaminating trypsin is not inactivated, correlating, in fact, with the secondary inactivation also reported by Place and Beynon (1982). Although the latter authors do not comment on it, the fragment size visible in their gels is changed as compared with the earlier study, and is consistent with the small change seen here. In Figure 2, the maximum level of activation achieved is lower (2.4-fold), and this is presumably accounted for by the secondary inactivation. Figure 1C indicates that it is possible also to get an increase in activity with NAD+ as coenzyme, but this activation is only of the order of about 250%.
Comparison of effects of chymotrypsin and trypsin separately and sequentially
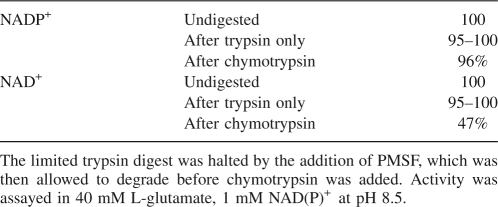
To highlight the differing individual effects of treatment with either protease, GDH was partially digested with trypsin and separately with chymotrypsin (TLCK treated) for 90 min before being run far down on a 10% SDS-PAGE gel (Fig. 3). This reveals firstly that the main enzyme band after digestion with trypsin (lane 4) is definitely smaller than after digestion with chymotrypsin alone (lane 2), and is 3–4 kDa smaller than undigested protein (lane 1). The size of the main large fragment in chymotrypsin-digested GDH seems to be as shown in the gels of Place and Beynon (1982), about 1 kDa smaller than the uncleaved protein subunit. This gel also suggests no further change in the size of the main band upon subsequent treatment of trypsin-digested GDH with chymotrypsin (lane 5). Limited trypsin digestion, in such a way as to produce the band in lane 4 of Figure 3, did not affect the activity of the enzyme with NAD+ or NADP+, and there was no observable effect on allosteric regulation. However, upon subsequent treatment with chymotrypsin, this trypsin-digested enzyme did not show activation, but instead lost activity of up to 50% with NAD+ within an hour (Table 1). The SDS-PAGE gel would suggest that this is not due to a further partial proteolysis, and it might instead reflect more extensive degradation of a portion of the enzyme, yielding small, inactive peptide fragments.
Figure 3.
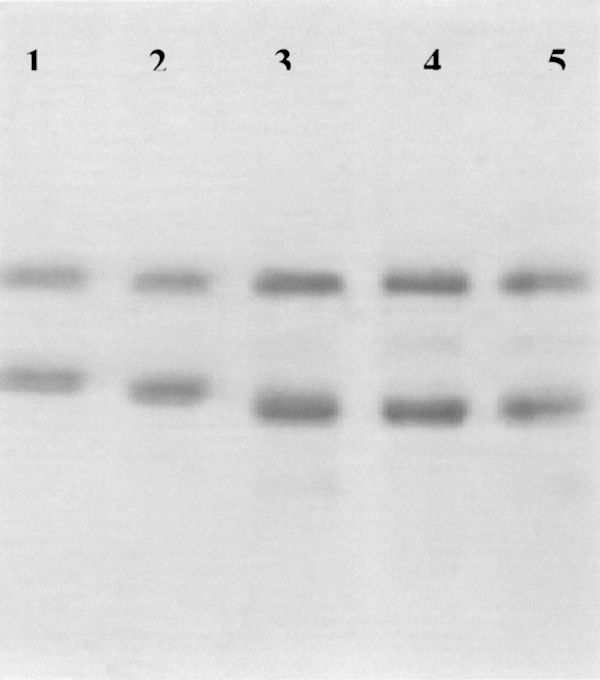
Ten percent SDS-PAGE displaying the effect of digestion by chymotrypsin and trypsin, separately and in combination, on bovine GDH. Lane 1 shows undigested GDH (55,561), lane 2 is GDH digested with chymotrypsin (200 μg/mL), lane 3 is GDH digested with chymotrypsin followed by trypsin (50 μg/mL), lane 4 shows GDH after trypsin digestion only, lane 5 is the result of trypsin digestion, followed by chymotrypsin. No attempt was made to remove BSA, the uppermost band (66,409) in all five tracks. The proteolytic enzymes were allowed to run off the gel.
Table 1.
Percentage activity with both oxidized coenzymes after digestion with trypsin first, and then, subsequently, chymotrypsin
N-terminal sequencing analysis
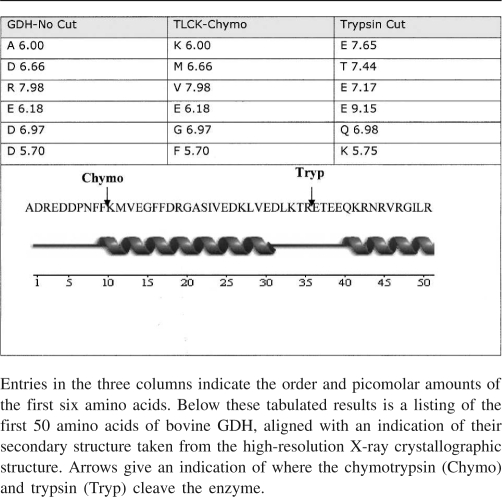
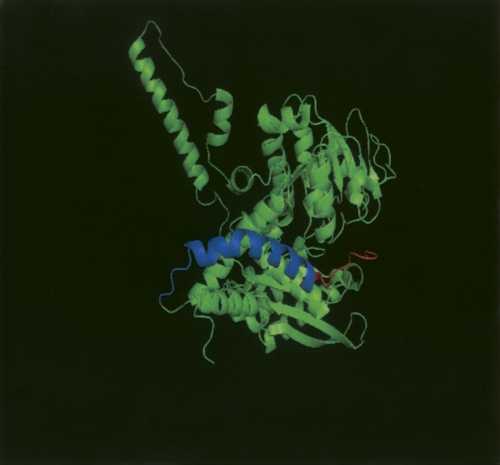
In the present study, Edman sequencing of a TLCK-chymotrypsin and trypsin digest was carried out to establish which of the termini was actually cleaved (Table 2). Undigested GDH was also N-terminally sequenced to first establish that the protein was indeed intact at the N-terminal, given that previous studies by McCarthy and Tipton (1985) revealed that the first four amino acids are lost in many commercial preparations. The first six amino acids of the GDH cleaved by chymotrypsin were revealed to be K-M-V-E-G-F, establishing that treatment of GDH with that protease causes cleavage of the enzymes at the –COOH end of phenylalanine 10. The loss of the first 10 amino acids of bovine GDH would represent a loss in MW of 1225.2 Da, which approximates to what is seen in SDS-PAGE, and the final residues of the C-terminal would seem to be inaccessible for cleavage (Banerjee et al. 2003). The Edman analysis also gave a clear indication of a clean N-terminal cut after limited trypsin treatment: The N-terminal sequence of E-T-E-E-Q-K-R places the tryptic cleavage as occurring between arginine 35 and glutamate 36, on an exposed loop after the first α-helix of the glutamate (substrate)-binding domain. This would reduce the relative molecular mass of the principal polypeptide of the bovine GDH subunit by 4090. Figure 4 illustrates where this proteolysis takes place in the three-dimensional structure. The fact that both cleavages occur at the N-terminal end of the protein accounts for the fact that chymotrypsin added after trypsin produces no further decrease in the size of the main GDH protein band.
Table 2.
Results of Edman degradation for TLCK chymotrypsin-cut, trypsin-cut, and intact GDH protein
Figure 4.
PyMOL (DeLano Scientific) cartoon representation of a monomer from beef GDH in its open domain conformation. Highlighted in red are the first 10 amino acids, which we have revealed to be the fragment cleaved by chymotrypsin. Highlighted in a blue color are the subsequent amino acids 11–35. Limited cleavage by trypsin produces a new N-terminal sequence beginning at amino acid 36.
Effect of chymotryptic cut on heterotropic allosteric effects
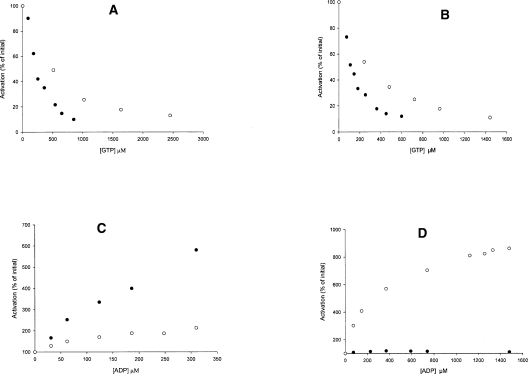
As stated earlier, the specific activity of GDH after chymotrypsin cleavage increased over threefold with NADP+, while a lesser increase was observed with NAD+. We have found that it is still possible to bring about an 80% inhibition of this superactivated enzyme by GTP, but the concentration of effector required to achieve such inhibition is increased two- to threefold (Fig. 5A,B). This holds true for activity assays carried out with both NAD+ and NADP+ at pH 8.5 and would seem to run contrary to earlier reports of Aitchison and Engel (1983), who stated that GTP inhibition of the superactive form was almost abolished at pH 8. Under the limited experimental conditions studied here, and as reported earlier (Place and Beynon 1983), ADP activation was much more greatly affected by the chymotryptic cleavage (Fig. 5C,D).
Figure 5.
Enzymes that have been treated with chymotrypsin to reveal an increase in activity at pH 8.5 also have an altered response to their allosteric effectors. A and B demonstrate the response of the uncleaved (filled circles) and cleaved (open circles) enzyme to GTP with (A) NAD+ and (B) NADP+ as the cofactor. (C,D) Results for the same enzyme samples assayed for ADP response with NAD+ and NADP+, respectively.
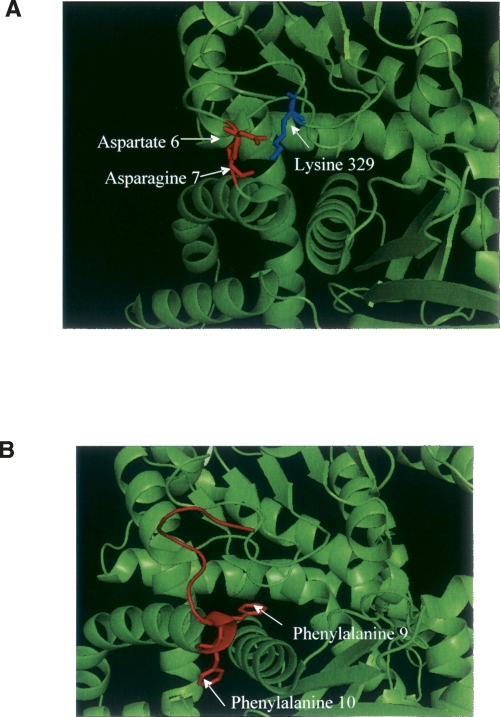
McCarthy and Tipton (1985) highlighted only minor kinetic effects on bovine GDH of losing the first four residues. The much greater effect of chymotryptic digestion may result from loss of critical contacts formed by some of the residues from 6 to10 in untreated GDH. Attributing particular significance to any one specific contact may be risky, but some of the most obvious interactions can be observed in Figure 6A. It appears possible that the N-terminal amino acids, which form part of the glutamate-binding domain, can make critical contact with overhanging loops of the coenzyme-binding domain. The crystal structure reveals interactions occurring between aspartate 6, asparagine 7, and lysine 329. It is also obvious that the two phenylalanine residues at 9 and 10 rest on either side of the third helix of the glutamate-binding domain (Fig. 6B), and it is just as plausible to believe that the loss of these might contribute to the reported kinetic effects.
Figure 6.
(A) Close-up of an N-terminal interaction in a domain from 1HWY, a domain with bound glutamate, NADH, and GTP. Highlighted in red are aspartate 6, and asparagine 7, and the prominent lysine 329 (in blue) of the coenzyme-binding domain, which are all within 3.5 Å of each other. (B) PyMOL (DeLano Scientific) cartoon displaying a close-up of the N-terminal of bovine GDH (1HWY), and in particular, amino acids 1–10 in red with the F9 and F10 in stick form. These phenylalanine residues lie on either side of the third helix of the glutamate-binding domain, and the peptide bond between residues 10 and 11 is the site of cleavage by chymotrypsin.
Aitchison and Engel (1983) reported that the 4-kDa fragment, now definitely identified as the first α-helix of bovine GDH, could not be separated on a gel-filtration column, and this may account for why limited trypsin digestion in the absence of chymotrypsin has no significant effect on enzyme functionality.
Possible structural basis of kinetic activation
In the oxidative deamination reaction at high pH and high substrate concentrations, glutamate is thought to bind to the enzyme before the NAD(P)H molecule dissociates from the catalytic cleft, thereby forming a tightly bound NAD(P)H-GLU abortive complex (George and Bell 1980). Under these conditions, hydride transfer is no longer rate-limiting, but rather the release of coenzyme from this abortive complex. In the reductive amination reaction, the analogous NAD(P)+ketoglutarate abortive complex is formed at high substrate concentrations and low pH (Bailey et al. 1982). It has been proposed that ADP and GTP realize their allosteric behavior on the basis of their interaction and effect with these abortive complexes (Pantaloni and Iwatsubo 1967; George and Bell 1980). In the case of GTP, a stabilizing effect seems to lead to an inhibition, while ADP, by binding behind the pivot helix of bovine GDH, will promote the disruption of the abortive complex, and thus, allow the enzyme rate to increase. Here, we raise the possibility that any increase in activity observed upon the chymotryptic digestion of the enzyme is also possibly due to disruption of the rate-limiting enzyme–substrate complex. It is interesting to note that Aitchison and Engel (1983) also found that there was no increase in kinetic activity in the situations where the coenzyme concentration was kept at 20 μM.
Conclusion
These studies show that previous suggestions, that activation of bovine glutamate dehydrogenase by partial proteolysis and concomitant decrease in, or loss of allosteric properties, were caused by cleavage/removal of an allosteric binding domain, are unfounded. Although such a domain is indeed formed by the C-terminal 50 residues, the key cleavage resulting in activation is at the other end of the GDH polypeptide chain. Further, it is now clear that the major kinetic effect on the enzyme is due to cleavage between residues 10 and 11, and not, as previously thought, to cleavage or removal of a much larger fragment. The formation of the latter fragment was clearly due instead to trace contamination with trypsin with the relative levels of the two proteinases, presumably accounting for the apparent dominance of a fragment 4–5 kDa smaller than the native subunit in the early studies (Beynon and Kay 1976; Aitchison and Engel 1983).
The present results are interesting in view of studies on two isozymes of human GDH (glud1 and glud2), which differ by just 15 amino acids (Shashidharan et al. 1994). In those studies it was found that a combination of mutations, many outside of the allosteric regions, seems to be responsible for the altered GTP and ADP sensitivity of the two enzymes (Plaitakis et al. 2003; Kanavouras et al. 2007).
Finally, since virtually all of the voluminous literature on bovine GDH has been based on studies of commercially prepared enzyme, and since different commercial preparations do not behave identically in proteolysis experiments, there is clearly a need to check some of the key observations using enzyme that can be shown to be homogeneous at the N terminus.
Materials and Methods
Materials
The study was initially complicated by the realization that responses to partial proteolysis differed between GDH preparations from different commercial sources, and that even from a single source, they varied over a period of years, possibly reflecting changes in production or storage procedures. For the main study reported here, preparations of bovine liver glutamate dehydrogenase were obtained from Biozyme Laboratories, Ltd., in lyophilized form and stabilized with ADP and BSA. Sigma currently also provides enzyme precipitated in ammonium sulphate and free of BSA, but this was found not to be inhibited to the same extent by GTP, nor activated by chymotryptic cleavage in the same manner as lyophilized forms. This is possibly unsurprising, given that proteolytic cleavage of GDH stored in ammonium sulphate was a problem highlighted in previous work (Place and Beynon 1983). L-glutamate, α-chymotrypsin (TLCK treated and untreated), DPCC-treated trypsin, and the protease inhibitor, phenylmethylsulphonyl fluoride, were obtained from Sigma-Aldrich. NAD+ and NADP+ at >98% purity were obtained from Apollo Scientific. Wide-range protein molecular weight markers were from New England BioLabs.
Assay of glutamate dehydrogenase
The activity of glutamate dehydrogenase was measured with 40 mM L-glutamate and 1 mM NAD(P)+ in 0.1 M pyrophosphate buffer, pH 8.5, using a Cary 50 Spec uv-vis spectrophotometer (Varian, Inc.)
Digestion of glutamate dehydrogenase and BSA
Glutamate dehydrogenase was first separated from ADP present in the commercial preparation by the use of a PD-10 desalting column (Amersham Sciences). Before most experiments, much of the BSA was also removed by binding the GDH to a Q-Sepharose column equilibrated with 20 mM Tris buffer at pH 7.4, washing with 20 mM Tris, and then eluting the GDH with a salt gradient of 0–0.5 mM NaCl. The eluate was precipitated with 60% ammonium sulphate, but dialyzed against phosphate buffer within 2 d. This step did not remove all of the BSA, but made it easier to identify the relevant bands after proteolytic cleavage. Proteolytic incubations were conducted at 30°C in 500 μL of 0.1 M potassium phosphate buffer (pH 7.0). GDH concentrations were ca. 500 μg/mL, with chymotrypsin concentration set to about 200 μg/mL. BSA was also incubated for 40 min with an equal concentration of chymotrypsin as a control (Fig. 1D). The DPCC-trypsin digest was carried out with a concentration of 50 μg/mL protease and was typically allowed to proceed for 1 h. In all of the proteolytic incubations, samples were taken out at intervals and treated with PMSF (0.5 mM), a potent inhibitor of both chymotrypsin and trypsin, and then immediately assayed for activity and kept for SDS-PAGE analysis. In situations where a trypsin digest was subsequently treated with chymotrypsin, at least 6 h were allowed to pass to allow excess PMSF to hydrolyze. Absorption coefficients (A280 1%) of 15.4 and 20.0 were used to calculate the concentrations of trypsin and chymotrypsin, respectively. Protein fragments to undergo N-terminal sequencing were electroblotted from an SDS-PAGE gel onto PVDF membrane and stained according to the method of Matsudaira (1987).
Acknowledgments
This work was supported by a Fellowship grant to P.C.E. from Science Foundation Ireland, whose assistance is gratefully recognized. We are grateful to Dr. Arthur Moir, University of Sheffield, for protein sequence analysis.
Footnotes
Reprint requests to: Paul C. Engel, School of Biomolecular and Biomedical Science, Conway Institute, University College Dublin, Belfield, Dublin 4, Ireland; e-mail: Paul.Engel@ucd.ie; fax: 353-1-2837211.
Abbreviations: GDH, L-glutamate dehydrogenase [EC 1.4.1.2-4]; BSA, bovine serum albumin; TLCK, tosyl-L-lysine chloromethyl ketone; PMSF, phenylmethanesulphonylfluoride; DPCC, diphenylcarbamyl chloride; NAD+, nicotinamide adenine dinucleotide; NADP+, nicotinamide adenine dinucleotide phosphate; GTP, guanosine triphosphate; ADP, adenosine triphosphate.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.034785.108.
References
- Aitchison, M.J., Engel, P.C. The effects of digestion by bovine pancreas α-chymotrypsin on native bovine glutamate dehydrogenase. Biochem. Soc. Trans. 1980;8:649. doi: 10.1042/bst0080649. [DOI] [PubMed] [Google Scholar]
- Aitchison, M.J., Engel, P.C. Beef liver glutamate dehydrogenase: Effects of partial proteolysis with chymotrypsin. Int. J. Biochem. 1983;15:79–85. doi: 10.1016/0020-711x(83)90014-9. [DOI] [PubMed] [Google Scholar]
- Allen, A., Kwagh, J., Fang, J., Stanley, C.A., Smith, T.J. Evolution of glutamate dehydrogenase regulation of insulin homeostasis is an example of molecular exaptation. Biochemistry. 2004;43:14431–14443. doi: 10.1021/bi048817i. [DOI] [PubMed] [Google Scholar]
- Bailey, J., Bell, E.T., Bell, J.E. Regulation of bovine glutamate dehydrogenase. The effects of pH and ADP. J. Biol. Chem. 1982;257:5579–5583. [PubMed] [Google Scholar]
- Banerjee, S., Schmidt, T., Fang, J., Stanley, C.A., Smith, T.J. Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation. Biochemistry. 2003;42:3446–3456. doi: 10.1021/bi0206917. [DOI] [PubMed] [Google Scholar]
- Bayley, P.M., O'Neill, K.T.J. The binding of oxidixed coenzyme to bovine-liver glutamate dehydrogenase studied by circular-dichroism spectroscopy. Eur. J. Biochem. 1980;112:521–531. doi: 10.1111/j.1432-1033.1980.tb06115.x. [DOI] [PubMed] [Google Scholar]
- Beynon, R.J., Kay, J. The chymotrypsin catalyzed activation of glutamate dehydrogenase. Int. J. Biochem. 1976;7:449–453. [Google Scholar]
- Coffee, C.J., Bradshaw, R.A., Goldin, B.R., Frieden, C. Identification of the sites of modification of bovine liver glutamate dehydrogenase reacted with trinitrobenzenesulfonate. Biochemistry. 1971;10:3516–3526. doi: 10.1021/bi00795a005. [DOI] [PubMed] [Google Scholar]
- Colman, R.F., Frieden, C. On the role of amino groups in the structure and function of glutamate dehydrogenase. I. Effect of acetylation on catalytic and regulatory properties. J. Biol. Chem. 1966;241:3652–3660. [PubMed] [Google Scholar]
- Engel, P.C., Dalziel, K. Kinetic studies of glutamate dehydrogenase with glutamate and norvaline as substrates. Coenzyme activation and negative homotropic interactions in allosteric enzymes. Biochem. J. 1969;115:621–631. doi: 10.1042/bj1150621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, A., Bell, J.E. Effects of adenosine 5′-diphosphate on bovine glutamate dehydrogenase: Diethyl pyrocarbonate modification. Biochemistry. 1980;19:6057–6061. doi: 10.1021/bi00567a017. [DOI] [PubMed] [Google Scholar]
- Haigis, M.C., Mostoslavsky, R., Haigis, K.M., Fahie, K., Christodoulou, D.C., Murphy, A.J., Valenzuela, D.M., Yancopoulos, G.D., Karow, M., Blander, G., et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hershko, A., Kindler, S.H. Mode of interaction of purine nucleotides and amino acids with glutamate dehydrogenase. Biochem. J. 1966;101:661–664. doi: 10.1042/bj1010661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.Y., Frieden, C. Rates of GDP-induced and GTP-induced depolymerization of glutamate dehydrogenase: A possible factor in metabolic regulation. Proc. Natl. Acad. Sci. 1969;64:338–344. doi: 10.1073/pnas.64.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucho, F., Rasched, I., Sund, H. Studies of glutamate dehydrogenase: Analysis of functional areas and functional groups. Eur. J. Biochem. 1975;52:221–230. doi: 10.1111/j.1432-1033.1975.tb03990.x. [DOI] [PubMed] [Google Scholar]
- Kanavouras, K., Mastorodemos, V., Borompokas, N., Spanaki, C., Plaitakis, A. Properties and molecular evolution of human GLUD2 (neural and testicular tissue-specific) glutamate dehydrogenase. J. Neurosci. Res. 2007;85:1101–1109. doi: 10.1002/jnr.21197. [DOI] [PubMed] [Google Scholar]
- Koberstein, R., Sund, H. Studies of glutamate dehydrogenase. The influence of ADP, GTP, and L-glutamate on the binding of the reduced coenzyme to beef-liver glutamate dehydrogenase. Eur. J. Biochem. 1973;36:545–552. doi: 10.1111/j.1432-1033.1973.tb02942.x. [DOI] [PubMed] [Google Scholar]
- MacMullen, C., Fang, J., Hsu, B.Y., Kelly, A., de Lonlay-Debeney, P., Saudubray, J.M., Ganguly, A., Smith, T.J., Stanley, C.A. Hyperinsulinism/hyperammonemia syndrome in children with regulatory mutations in the inhibitory guanosine triphosphate-binding domain of glutamate dehydrogenase. J. Clin. Endocrinol. Metab. 2001;86:1782–1787. doi: 10.1210/jcem.86.4.7414. [DOI] [PubMed] [Google Scholar]
- Markau, K., Schneider, J., Sund, H. Studies of glutamate dehydrogenase. The mechanism of the association-dissociation equilibrium of beef-liver glutamate dehydrogenase. Eur. J. Biochem. 1971;24:393–400. doi: 10.1111/j.1432-1033.1971.tb19698.x. [DOI] [PubMed] [Google Scholar]
- Mattaj, I.W., McPherson, M.J., Wootton, J.C. Localization of a strongly conserved section of coding sequence in glutamate dehydrogenase genes. FEBS Lett. 1982;147:21–25. doi: 10.1016/0014-5793(82)81003-x. [DOI] [PubMed] [Google Scholar]
- McCarthy, A.D., Tipton, K.F. Ox glutamate dehydrogenase. Comparison of the kinetic properties of native and proteolysed preparations. Biochem. J. 1985;230:95–99. doi: 10.1042/bj2300095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, A.D., Walker, J.M., Tipton, K.F. Purification of glutamate dehydrogenase from ox brain and liver. Evidence that commercially available preparations of the enzyme from ox liver have suffered proteolytic cleavage. Biochem. J. 1980;191:605–611. doi: 10.1042/bj1910605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira, P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- Monod, J., Changeux, J.P., Jacob, F. Allosteric proteins and cellular control systems. J. Mol. Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- Olson, J.A., Anfinsen, C.B. Kinetic and equilibrium studies on crystalline 1-glutamic acid dehydrogenase. J. Biol. Chem. 1953;202:841–856. [PubMed] [Google Scholar]
- Pantaloni, D., Iwatsubo, M. Binding of ADP, NADH and NADPH on glutamatedehydrogenase, determined spectrophotometrically. Biochim. Biophys. Acta. 1967;132:217–220. doi: 10.1016/0005-2744(67)90217-3. [DOI] [PubMed] [Google Scholar]
- Peterson, P.E., Smith, T.J. The structure of bovine glutamate dehydrogenase provides insights into the mechanism of allostery. Structure. 1999;7:769–782. doi: 10.1016/s0969-2126(99)80101-4. [DOI] [PubMed] [Google Scholar]
- Place, G.A., Beynon, R.J. The chymotrypsin-catalysed activation of bovine liver glutamate dehydrogenase. Biochem. J. 1982;205:75–80. doi: 10.1042/bj2050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place, G.A., Beynon, R.J. Chymotryptic activation of glutamate dehydrogenase. Biochim. Biophys. Acta. 1983;747:26–31. doi: 10.1016/0167-4838(83)90116-4. [DOI] [PubMed] [Google Scholar]
- Plaitakis, A., Spanaki, C., Mastorodemos, V., Zaganas, I. Study of structure-function relationships in human glutamate dehydrogenases reveals novel molecular mechanisms for the regulation of the nerve tissue-specific (GLUD2) isoenzyme. Neurochem. Int. 2003;43:401–410. doi: 10.1016/s0197-0186(03)00028-7. [DOI] [PubMed] [Google Scholar]
- Price, N.C., Radda, G.K. Desensitization of glutamate dehydrogenase by reaction of tyrosine residues. Biochem. J. 1969;114:419–427. doi: 10.1042/bj1140419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, D.W., Baker, P.J., Farrants, G.W., Hornby, D.P. The crystal structure of glutamate dehydrogenase from Clostridium symbiosum at 0.6 nm resolution. Biochem. J. 1987;242:789–795. doi: 10.1042/bj2420789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidharan, P., Michaelidis, T.M., Robakis, N.K., Kresovali, A., Papamatheakis, J., Plaitakis, A. Novel human glutamate dehydrogenase expressed in neural and testicular tissues and encoded by an X-linked intronless gene. J. Biol. Chem. 1994;269:16971–16976. [PubMed] [Google Scholar]
- Stanley, C.A., Lieu, Y.K., Hsu, B.Y., Burlina, A.B., Greenberg, C.R., Hopwood, N.J., Perlman, K., Rich, B.H., Zammarchi, E., Poncz, M. Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N. Engl. J. Med. 1998;338:1352–1357. doi: 10.1056/NEJM199805073381904. [DOI] [PubMed] [Google Scholar]
- Stillman, T.J., Yip, K.S., Rice, D.W., Fuentes, A.M., Connerton, I. Crystallization and preliminary X-ray analysis of the NADP-specific glutamate dehydrogenase from Neurospora crassa . Acta Crystallogr. 1995;51:837–839. doi: 10.1107/S0907444995000904. [DOI] [PubMed] [Google Scholar]
- Talal, N., Tomkins, G.M. Allosteric properties ofglutamate dehydrogenases from different sources. Science. 1964;146:1309–1311. doi: 10.1126/science.146.3649.1309. [DOI] [PubMed] [Google Scholar]
- Wolff, J. The effect of thyroxine on isolated dehydrogenases. II. Sedimentation changes in glutamic dehydrogenase. J. Biol. Chem. 1962;237:230–235. [PubMed] [Google Scholar]
- Wootton, J.C., Chambers, G.K., Holder, A.A., Baron, A.J., Taylor, J.G., Fincham, J.R., Blumenthal, K.M., Moon, K., Smith, E.L. Amino-acid sequence of NADP-specific glutamate dehydrogenase of Neurospora crassa . Proc. Natl. Acad. Sci. 1974;71:4361–4365. doi: 10.1073/pnas.71.11.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yielding, K.L., Tomkins, G.M., Trundle, D.S. On the mechanism of inhibition of glutamate dehydrogenase by NADH and NADPH. Biochim. Biophys. Acta. 1964;85:342–345. doi: 10.1016/0926-6569(64)90257-3. [DOI] [PubMed] [Google Scholar]