Abstract
Although protein Z (PZ) has a domain arrangement similar to the essential coagulation proteins FVII, FIX, FX, and protein C, its serine protease (SP)-like domain is incomplete and does not exhibit proteolytic activity. We have generated a trial sequence of putative activated protein Z (PZa) by identifying amino acid mutations in the SP-like domain that might reasonably resurrect the serine protease catalytic activity of PZ. The structure of the activated form was then modeled based on the proposed sequence using homology modeling and solvent-equilibrated molecular dynamics simulations. In silico docking of inhibitors of FVIIa and FXa to the putative active site of equilibrated PZa, along with structural comparison with its homologous proteins, suggest that the designed PZa can possibly act as a serine protease.
Keywords: protein Z (PZ), sequence design, active form of protein Z (PZa), homology modeling, molecular dynamics simulation, molecular docking
Protein Z (PZ) is a vitamin K-dependent (VKD) protein relatively abundant in humans, with a wide plasma concentration range (Miletich and Broze Jr. 1987). PZ circulates in plasma as a complex with protein Z-dependent protease inhibitor (ZPI) (Tabatabai et al. 2001). The most important known physiological function of PZ is its ability to enhance the inhibition of factor Xa by ZPI 1000-fold in the presence of phospholipid membrane and Ca2+ ions (Han et al. 1998; Al-Shanqeeti et al. 2005).
PZ is homologous to coagulation factors FVII, FIX, FX, and protein C (PC), and the domain assembly of PZ is identical to these proteins. It is composed of four domains: an N-terminal Gla domain, two epidermal growth factor (EGF)-like domains (EGF1 and EGF2 domains), and a serine protease (SP)-like domain (Ichinose et al. 1990; Sejima et al. 1990). However, PZ lacks the critical histidine and serine residues of the catalytic triad, and is therefore not a zymogen of a serine protease. Based on its homology with the other coagulation factors, we proposed a solvent-equilibrated model of human PZ using comparative modeling and molecular dynamics (MD) simulation (Lee et al. 2007). One of the striking features in this model was that even though PZ lacked two of the critical catalytic triad residues, the relative spatial arrangement of the putative active site region and the distances between these residues were similar to the other serine proteases after a long time simulation. The two β-barrel domains in PZ were juxtaposed similar to other chymotrypsin-like enzymes, even without an active catalytic triad bridging them. In addition, the residues Met309 and Val331, which correspond to a disulfide bond (Cys191–Cys220, chymotrypsinogen numbering) in other VKD SP domains, were close to disulfide bond-forming distance without distance or position constraints imposed. These observations raised questions about the evolution of PZ and led us to consider if PZ could have once been functionally active as a serine protease. Studies on serine protease evolution have shown that apart from protease domain sequences, the development and maintenance of the active site structure plays an important role in the evolution of proteolytic enzymes (Krem and Di Cera 2001, 2002). The preservation of the active site three-dimensional (3D) structure in PZ compared with other serine proteases indicates the operation of a possible divergent evolution mechanism within this structural family and raises the question of whether an active PZ was disfavored in evolution through sequence mutations in the protease domain. A possible reason for this evolutionary choice may have been due to negative evolutionary pressure brought about by undesirable function. Another possibility is that PZ might have been an ancestor of FVII, FIX, FX, and PC. This possibility, however, seems unlikely based on genomic evidence from a recent study that characterized blood coagulation proteins (using new genetic databases) in a jawless vertebrate system (lamprey) (Doolittle et al. 2008). It was found that homologs for FVII, FIX, FX, and PC exist in jawless vertebrates, whereas no matches are found for PZ. The earliest homolog of PZ is found only in jawed vertebrates like fugu and zebrafish, which indicates that PZ evolved later than the other coagulation serine proteases (Davidson et al. 2003).
In order to understand the evolutionary significance, it is desirable to model the 3D structure of the putative activated form of PZ (PZa). Apart from the evolutionary aspect, this in silico model building is also interesting as a protein design challenge to generate an activated form of a serine protease starting from an inactive form. A recent experimental work (Ortlund et al. 2007) has shown that it is possible to resurrect an ancestral protein by employing evolutionary relationship to sequence comparisons, dramatically leading to the X-ray crystal structure.
Given our 3D model for protein Z, an in silico PZa model was constructed by designing a primary sequence of PZa that might reasonably resurrect serine protease catalytic activity and then simulating a 3D solvent-equilibrated structure of the activated form. Further, a molecular docking study was employed to partially validate the predicted model by evaluating its ability to bind in silico to known serine protease inhibitors for FVIIa and FXa.
Results and Discussion
Model building
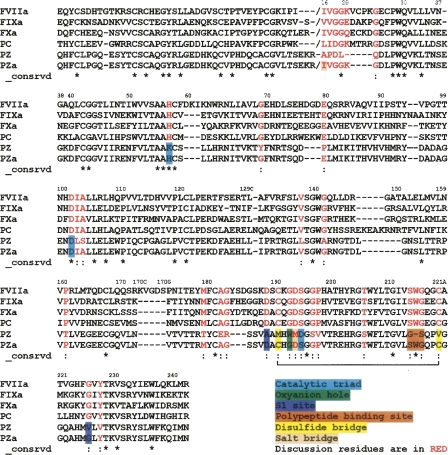
The proposed sequence of PZa has 36%–39% amino acid identities in the SP domain with the four structural templates used for model building (Fig. 1). This sequence identity is a substantial improvement from the PZ sequence for the SP domain (with all the active site residues conserved) and therefore offers a reasonable starting point for homology modeling. The root mean square deviations (RMSD) of the backbone atoms for our initial PZa model with the crystal structure of FVIIa (1DAN) and FXa (1XKA) in the SP domain were 0.93 and 0.73 Å for 824 and 868 atoms, respectively, using the program Swiss-PdbViewer (Guex and Peitsch 1997), while the corresponding value for FVIIa with FXa was 0.96 Å over 860 atoms. PROCHECK was used to calculate the φ-ψ angles for the Ramachandran plot, which shows that most of the φ-ψ angles (85.9%) are located in the core region of the plot, and only 0.3% were in the disallowed region (Supplemental Fig. S1). The bad contacts (steric clashes) present in the initial homology model have been eliminated after the MD simulation. The PZa model was also assessed using the Verify3D algorithm, and the 3D to 1D scores of our model are always positive (a criterion of proper folding) and are similar to those obtained with the template structures of FVIIa and FXa (Supplemental Fig. S2). The disulfide bonds were modeled based on homology with FVII, FIX, FX, and PC. In PZ (and in the PZa model), there is a possibility of an additional disulfide bond between residues Cys131 in the EGF2 domain and Cys234 in the SP domain (G.J. Broze, pers. comm.). After 11 ns of MD simulation, the Cα–Cα distance between these residues is 8.82 Å. Since these residues are present in nearby loop regions, there is a possibility that these cysteines could also be involved in disulfide bonding. We have not modeled in this disulfide in this work, since it does not appear in the other four homologs, although it represents a consideration in future work.
Figure 1.
Multiple sequence alignment of PZ with human FVIIa, FIXa, FXa, and PC. The PZa sequence in the alignment shows modifications of PZ to obtain the sequence of the activated form. The sequence modifications were carried out only in the SP-like domain of PZ. The chymotrypsinogen numbering is used to represent residues of the SP domain, with the N terminus Ile at position 16. The multiple sequence alignment for Gla, EGF1, and EGF2 domains is the same as our previous PZ paper (Lee et al. 2007) (part of EGF2 is shown in the alignment). The modified regions are color coded as shown in the legend. (*) Identity, (:) strongly similar.
Structural features of PZa
The objective of our study was to propose a reasonable sequence for an active PZ and to model the 3D structure of the active form based on the proposed sequence. Additionally, we wished to examine our equilibrated structure of the activated form for similarities with other active blood coagulation factors.
A plot of the backbone RMSD relative to the initial structure over 19 ns of MD simulation (Supplemental Fig. S3) shows that the model has equilibrated. Inspection of the final structures showed no bad contacts among residues. As expected, the model shows structural properties characteristic of the blood coagulation serine protease family and is structurally similar to FVIIa, FXa, and other serine proteases. The equilibrated structure of this protein reveals the presence of a His–Asp–Ser catalytic triad arrangement similar to that of other serine proteases. The H-bonding between His57 and Ser195 was, however, intermittent compared with other serine proteases, present only for ∼28% of over 7.5 ns of simulation time. A separate simulation of FVIIa using a comparable simulation protocol showed that the His57–Ser195 H-bonding was present for ∼60% of the simulation time. So, the H-bonding was intermittent in both the proteins, but was present relatively longer in FVIIa. The topology of the substrate-binding site adjacent to the catalytic triad is well preserved after a long time simulation. In PZa, the overall topology of the active site is similar to that of other serine proteases, as a majority of the binding site residues are conserved after sequence modification. The substitutions of Met191 and Val220 with Cys and the formation of a disulfide bond between these cysteine residues helped maintain an S1 specificity pocket in the PZa structure. The overall spatial architecture of the S1 pocket, which is built of two β-strands (residues 214–216, 226–228), loop residues 189–192, and the oxyanion hole (Gly193 and Ser195), is similar to other blood coagulation serine proteases. There are differences, however, in the distant structural elements that are often the determinants of S1 specificity and might lead to different ligand specificity. It was also observed from our simulation that the S1 site Asp189 residue forms a salt bridge interaction with Arg158, which causes a change in orientation of the Asp189 side chain in PZa compared with FVIIa and FXa, for which there is a Leu in place of Arg. We have carried out docking of known FVIIa and FXa inhibitors (below) to examine if the overall topology of the PZa active site region is similar to other blood coagulation serine proteases and also to study differences that might confer specificity to putative PZa.
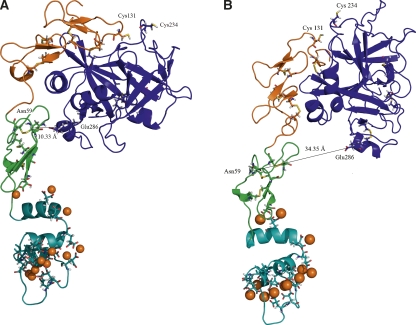
An unrestrained simulation of the modeled PZa revealed a bend in the structure of PZa in the N-terminal region of the EGF2 domain (Fig. 2A), which brings the orientation of the SP domain close to the EGF1 domain. This bend was also observed in our previous extended simulations of free FVIIa in the same region as PZa (not shown). A similar bend has been reported and discussed in the X-ray crystal structures of free FVIIa in this region (Kemball-Cook et al. 1999; Pike et al. 1999). In our PZa simulation, the bend occurs near Glu88 at the beginning of the α-helix between Glu88 and Thr94. As a result, the region between Gly285 and Gln290 in the protease domain contacts the EGF1 domain of PZa (with Asn59 and Glu286 forming a strong H-bond). The sequences of this bending region show no apparent similarities between PZa and FVIIa; however, this region responds similarly in both the proteins, with a more prominent bend in PZa. Another simulation of the PZa structure was carried out with the Gla and EGF domains held frozen so as to ensure a more upright orientation of the SP domain relative to the remainder of the PZa structure (Fig. 2B). It is our hypothesis that the upright form might approximate the effect of a putative cofactor (unknown)-bound form of PZa (similar to the tissue factor-bound form of FVIIa) with the bent form characteristic of free PZa. The resemblance of the simulated PZa structure to other members of the blood coagulation serine protease family indicates that the in silico design and modeling of PZa from PZ sequence has reasonably restored serine protease structural features.
Figure 2.
Cartoon representation of the solvent-equilibrated structural model of PZa. (Cyan) Gla domain, (green) EGF1 domain, (orange) EGF2 domain, (blue) SP domain, (orange spheres) Ca2+ ion, (sticks) Gla, Bha residues, and disulfide bonds. (A) Structure obtained from unconstrained simulation shows a bend of the SP domain. The Cα distance between Asn59 in the EGF1 domain and Glu286 in the SP domain is 10.33 Å. Residues Cys131 and Cys234 shown in the figure could be involved in disulfide bond formation, based on their distance after simulation (Cα distance 8.82 Å). (B) Structure obtained from keeping the Gla and EGF domains fixed, resulting in a more upright orientation of the SP domain. The figures were prepared using PyMOL (DeLano Scientific).
Docking of inhibitors
The PZa model was tested by docking known FVIIa and FXa inhibitors into the putative active site of PZa. The primary objective of this docking exercise was to validate to a reasonable extent the structural morphology of the PZa active site. AutoDock 3.0.5 (Morris et al. 1998) was used to perform the docking calculations. In order to test the scoring function of AutoDock and the adopted parameters for the inhibitors, we performed unbiased re-dockings with the unbound 3D structures of both the receptor (FVIIa, FXa) and the ligand (Supplemental Fig. S4) and used the known complex structure (X-ray crystal structure) for evaluating the performance. The idea behind this was to measure how well the docking protocol performs when tested against known partners used in this study. For both FVIIa and FXa, we identified a near-native structure (within 1.5 Å from the experimental structure) with the top prediction. The top prediction in each case was the lowest docked energy pose. The best docked pose for the FVIIa inhibitor had an RMSD of 0.76 Å relative to the crystal orientation with a docked energy of −12.06 kcal/mol, while the RMSD value for FXa inhibitor was 1.21 Å and the docked energy was −13.75 kcal/mol. Blind docking of the inhibitors from FVIIa and FXa to a region corresponding to the putative active site of PZa was then carried out. The top-ranking pose from each case was selected and analysis was done by visual comparison of the obtained coordinates relative to the referenced crystallographic coordinates.
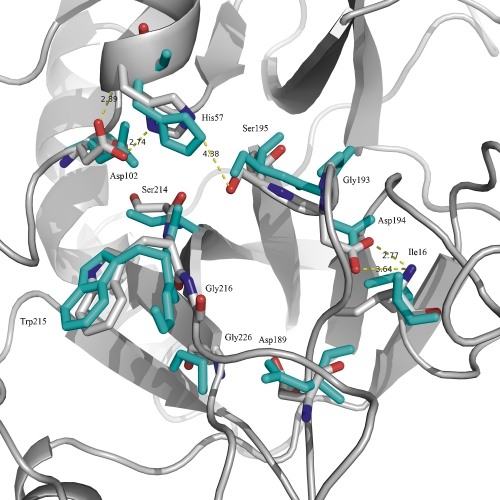
One of the important residues in the S1 pocket of serine proteases is Asp189. The negatively charged side chain participates in a salt bridge interaction with an arginine or lysine side chain from the substrate. Small molecule inhibitors exploit this interaction using an amidine or a guanidine group of the inhibitor to form an ion pair with the carboxylate of the aspartate. Initial docking of the inhibitor into the PZa active site showed that there were steric clashes that prevented the inhibitor from forming the salt bridge interaction with Asp189 at the bottom of the S1 pocket. Studies on serine protease specificity have shown that apart from the structure of the active site cleft, the ability of the enzyme to undergo conformational change to accommodate different substrates via an induced fit mechanism is also important for broad specificity (Post and Ray 1995). Several studies on a Met192 mutation in α-lytic protease have found that the S1 site especially can sample different conformations and facilitate hydrolysis of many different substrates (Davis and Agard 1998; Miller and Agard 1999; Ota and Agard 2001). Keeping this in mind, we performed an induced minimization of the SP domain of PZa with the FVIIa inhibitor in the S1 pocket so as to expand the pocket slightly and eliminate the steric clashes. Residues that line the S1 binding pocket in PZa are shown in Figure 3. The fixed structure of PZa after the induced minimization was then used to dock the FVIIa and FXa inhibitors.
Figure 3.
Catalytic triad region and the residues around the S1 pocket of solvent-equilibrated PZa (gray). The overall spatial architecture of this region is similar to FVIIa and FXa. SP domain of FVIIa is overlaid and the corresponding residues are shown (cyan).
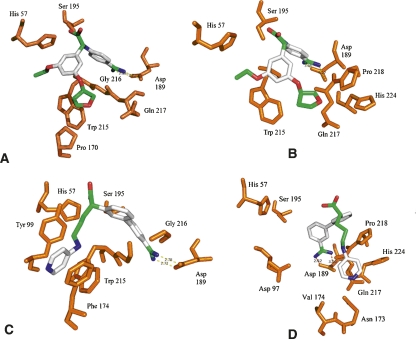
The docking of the FVIIa inhibitor to PZa (Fig. 4) shows that the inhibitor binds in a similar orientation to the PZa binding pocket as to the FVIIa pocket (with a docked energy of −9.12 kcal/mol), but the salt bridge interaction with the S1 site Asp189 is slightly weaker compared with FVIIa (interaction distances larger than for FVIIa). The inhibitor, however, sits in a similar overall conformation to FVIIa in the proximal hydrophobic S2 pocket and the distal S3 pocket. The docking of the FXa inhibitor to PZa shows that the amidine group of the inhibitor forms the expected salt bridge interaction with the carboxylate group of Asp189 in the S1 pocket (with a docked energy of −14.81 kcal/mol), but the overall binding conformation differs from the X-ray crystal conformation observed for FXa with its inhibitor. The S1 salt bridge interaction is stronger compared with the FVIIa inhibitor, but the distal hydrophobic interactions are different. Overall, it is encouraging that the inhibitor docking results indicate that several of the binding characteristics of FVIIa and FXa inhibitor binding are exhibited by the putative PZa model.
Figure 4.
Lowest docked energy poses of FVIIa and FXa inhibitors. (A) Redocking of FVIIa inhibitor to FVIIa, (B) blind docking of FVIIa inhibitor to PZa, (C) redocking of FXa inhibitor to FXa, (D) blind docking of FXa inhibitor to PZa. (Orange) Protein side chains and backbones. Inhibitors are colored by atom type.
Conclusions
There currently exists a reasonable understanding of the relationship between sequence and function for blood coagulation serine proteases. In this study, by employing computational methods, we have attempted to use existing knowledge to design a sequence for an activated form of PZ that could function as a serine protease. Iterative sequence design, model building, and subsequent simulations have shown that apart from key structural features, distant structural elements and residues conserved across the family also play an important role in maintaining the structure of the protein. Experimental studies to express and investigate possible catalytic activity of this sequence could provide clues toward understanding how evolutionary changes in the sequence have affected the function of this protein.
Materials and Methods
Design of the PZa sequence
The blood coagulation serine proteases have conserved enzyme structures and share common structural features of their active sites. For substrate recognition and catalytic activity, the serine protease domain must have the following key characteristics (chymotrypsinogen numbering is used): the His57–Asp102–Ser195 catalytic triad, a selectivity S1 site composed of Asp189, Gly216, and Gly226, an oxyanion hole formed by the backbone NHs of Gly193 and Ser195, a proximal hydrophobic S2 pocket, a distal hydrophobic S3 pocket, and the salt bridge between Asp194 and N-terminal residue (Ile16) (Perona and Craik 1995; Czapinska and Otlewski 1999; Hedstrom 2002).
In order to identify a reasonable primary sequence for the activated form of PZ, the multiple sequence alignment of PZ with FVII, FIX, FX, and PC was used as the starting point (Lee et al. 2007). The amino acid residues in PZ corresponding to the key residues (above) that confer these serine protease characteristics were first modified based on the multiple sequence alignment. Multiple models were built based on this sequence, and the best model was subjected to a MD simulation (solvated) to evaluate the structure of the model, based on its similarity to SP domains from other blood coagulation factors. This model was not adequate. Iteration of this procedure, however, led to a sequence for which, apart from these key residues, all amino acid residues that were conserved in the other four proteins were also modified (Fig. 1). This additional sequence modification is consistent with earlier observations that found that even substitution of all of the amino acids in the S1 site of trypsin with their counterparts in chymotrypsin failed to transfer chymotrypsin-like characteristics to the variant enzyme (Hedstrom et al. 1992). Subsequent studies have pointed out that distal enzyme segments not contacting the substrate are important for serine protease specificity (Perona and Craik 1997). The N terminus of the SP domain was chosen based on the minimum sequence required to form the salt bridge with Asp194 and was assigned based on FVIIa, since there was no consensus for that peptide sequence among the coagulation proteins. A total of 32 residues were modified. This new sequence was used to build the model of the activated form of PZ.
Homology modeling
The overall procedure for the homology modeling was similar to that employed in a previous study (Lee et al. 2007). A multiple sequence alignment of human FVIIa, FIXa, FXa, and PC sequences was created with CLUSTALW, using the BLOSUM matrices for scoring the alignments (Thompson et al. 1994). Their corresponding X-ray structures ([PDB entry 1DAN] [Banner et al. 1996], [1RFN] [Hopfner et al. 1999], [1XKA] [Kamata et al. 1998], and [1AUT] [Mather et al. 1996], respectively) were used as structural templates for the homology modeling. The obtained alignments were checked for insertions and deletions in the structurally conserved regions and also fine-tuned manually. The best alignment was selected based on both the alignment score as well as the reciprocal positions of the conserved residues in the SP domain. The resulting alignment was used as an input to construct homology models of PZa using MODELLER 8v2 (Sali et al. 1995). Residue type and topology information were added for the two nonstandard residues, Gla and Bha. Three-dimensional models were built based on a distance restraint algorithm imposed from the multiple sequence alignment of the target sequence with the template structures, applying the CHARMM energy terms. An optimization method involving conjugate gradients and molecular dynamics with simulated annealing, available in MODELLER, was employed to minimize violations of spatial restraints. For the model-building process, default parameters included in the automodel class were used, and explicit manual restraints were specified for disulfide bridges. The model was built as a two-chain system with 10 disulfide bonds (Res. 18–23, 51–62, 56–71, 73–82, 89–101, 97–110, 112–125, 164–180, 288–302, and 310–337). To guarantee sufficient conformational sampling of each active site residue, an ensemble of 30 models was built, from which the best final model was selected based on evaluation of stereochemical values from PROCHECK (Laskowski et al. 1993), the objective function from MODELLER, and by visual inspection. The model was also assessed using the Verify3D algorithm (Bowie et al. 1991; Luethy et al. 1992) to identify any region of improper folding.
Molecular dynamics (MD) simulation
A stepwise structure refinement approach for the homology model was performed through MD simulation, to obtain a solvent-equilibrated model and to remove bad contacts. All MD simulations were performed using PMEMD9 in the AMBER9 (Case et al. 2006) suite of molecular modeling programs. Force field parameters used were taken from the ff99SB force field included with the AMBER9 molecular dynamics package. The EGF and Gla domains are based on the PZ model from our previous study (Lee et al. 2007). The seven conserved Ca2+ ions in the Gla domain were placed based on FVIIa X-ray structure, and the additional Ca2+ ions were placed with malonate coordination (both carboxylates of Gla involved). The total system was composed of 85,023 atoms including 12 Ca2+ ions, five Na+ counterions for neutralizing the system, and 26,502 TIP3P (Jorgensen et al. 1983) water molecules (solvated in a 12.5 Å layer of water molecules).
Prior to structural equilibration, the model was subjected to several stages of energy minimization and relaxation. In the first step, belly dynamics were performed on all the water molecules and counterions for 25 ps. Belly dynamics involved allowing motional freedom to the water molecules and the counterions to relax their positions, while the protein atoms were kept fixed. This was followed by an energy minimization of all the water molecules and counterions in 10,000 conjugate gradient steps to remove steric clashes, while the protein was held fixed. The whole system was then subjected to minimization, where in the initial stages of minimization and NPT equilibration phase, constraints were applied on the backbone atoms of the model and the H-bond-forming atoms of His57 and Ser195 in the catalytic triad. A stepwise heating procedure was implemented over a 35-ps period to bring the system to 300K. In the later stages of equilibration, the constraints over the backbone atoms were removed and side chain atom constraints for the catalytic triad were gradually removed. Equilibration was concluded by 3.5-ns simulations, and these were followed by an unconstrained production run. Long-range interactions were treated using the particle mesh Ewald (PME) method (Darden et al. 1993; Essmann et al. 1995), and a time step of 1.5 fs was used in all of the molecular dynamics calculations. The stability of the system and the state of equilibration were followed by monitoring the backbone RMSD and the potential energy of the system. The final 7.5-ns unconstrained trajectories were used for analysis.
Docking of inhibitors
The flexible ligand docking program AutoDock 3.0.5 (Morris et al. 1998) was used to dock two known inhibitors, one from FVIIa (2BZ6) (Groebke et al. 2006) and another from FXa (1XKA) (Kamata et al. 1998; Supplemental Fig. S1), into the binding site of modeled PZa, which was fixed. The coordinates of the protein atoms were obtained by building an average structure from the final 100 ps of the simulation. The Lamarckian genetic algorithm (GA) was used as the search method, and grid maps with 61 × 61 × 61 points centered on the ligand with a grid point spacing of 0.375 Å were computed. The scoring function in AutoDock is a regression-based method, consisting of a van der Waals, an electrostatic, a hydrogen-bonding, and a desolvation energy term, in addition to an entropy term that accounts for the loss of torsional degrees of freedom upon binding. For each inhibitor, 50 docking runs were performed with an initial population of 300, with those differing by <2.0 Å in positional RMSD being clustered together. Step sizes of 2 Å for translation and 50° for rotation were chosen, and a maximum number of 250,000 energy evaluations and 27,000 generations were considered for each system.
Electronic supplemental material
The following supplemental material is available for this article:
Figure S1: Ramachandran plot of the best homology model obtained from Modeller;
Figure S2: Verify3D plot of the SP domain coordinates of the homology model and the corresponding coordinates from the crystal structures of FVIIa and Fxa;
Figure S3: Plot of backbone RMSD of each domain and all residues of PZa over 19-nsec MD simulation;
Figure S4: Inhibitors used for docking.
Acknowledgments
This work was supported by the National Institutes of Health (HL-06350) and NSF (ITP/APS-0121361). We acknowledge the use of the computational resources provided by ITS at UNC-CH and the Biomedical Unit of the PSC. We thank our colleagues at UNC-CH for helpful conversations.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Lee G. Pedersen, University of North Carolina at Chapel Hill, Chem CB 3290, Chapel Hill, NC 27599, USA; e-mail: lee_pedersen@unc.edu; fax: 919-962-2388.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.034801.108.
References
- Al-Shanqeeti, A., Vlieg, A.H., Berntorp, E., Rosendaal, F.R., Broze G.J., Jr Protein Z and protein Z-dependent protease inhibitor. Thromb. Haemost. 2005;93:411–413. doi: 10.1160/TH04-11-0715. [DOI] [PubMed] [Google Scholar]
- Banner, D.W., D'Arcy, A., Chene, C., Winkler, F.K., Guha, A., Konigsberg, W.H., Nemerson, Y., Kirchhofer, D. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- Bowie, J.U., Luethy, R., Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Case, D.A., Darden, T.A., Cheatham T.E., III, Simmerling, C.L., Wang, J., Duke, R.E., Luo, R., Merz, K.M., Pearlman, D.A., Crowley, M., et al. University of California; San Francisco, CA: 2006. AMBER 9. [Google Scholar]
- Czapinska, H., Otlewski, J. Structural and energetic determinants of the S1-site specificity in serine proteases. Eur. J. Biochem. 1999;260:671–695. doi: 10.1046/j.1432-1327.1999.00160.x. [DOI] [PubMed] [Google Scholar]
- Darden, T.A., York, D., Pedersen, L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- Davidson, C.J., Tuddenham, E.G., McVey, J.H. 450 million years of hemostasis. J. Thromb. Haemost. 2003;1:1487–1494. doi: 10.1046/j.1538-7836.2003.00334.x. [DOI] [PubMed] [Google Scholar]
- Davis, J.H., Agard, D.A. Relationship between enzyme specificity and the backbone dynamics of free and inhibited α-lytic protease. Biochemistry. 1998;37:7696–7707. doi: 10.1021/bi972963p. [DOI] [PubMed] [Google Scholar]
- Doolittle, R.F., Jiang, Y., Nand, J. Genomic evidence for a simpler clotting scheme in jawless vertebrates. J. Mol. Evol. 2008;66:185–196. doi: 10.1007/s00239-008-9074-8. [DOI] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M.L., Darden, T., Lee, H., Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- Groebke, Z.K., Banner, D.W., Hilpert, K., Himber, J., Lave, T., Riederer, M.A., Stahl, M., Tschopp, T.B., Obst-Sander, U. Dose-dependent antithrombotic activity of an orally active tissue factor/factor VIIa inhibitor without concomitant enhancement of bleeding propensity. Bioorg. Med. Chem. 2006;14:5357–5369. doi: 10.1016/j.bmc.2006.03.042. [DOI] [PubMed] [Google Scholar]
- Guex, N., Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Han, X., Fiehler, R., Broze G.J., Jr Isolation of a protein Z-dependent plasma protease inhibitor. Proc. Natl. Acad. Sci. 1998;95:9250–9255. doi: 10.1073/pnas.95.16.9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom, L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Hedstrom, L., Szilagyi, L., Rutter, W.J. Converting trypsin to chymotrypsin: The role of surface loops. Science. 1992;255:1249–1253. doi: 10.1126/science.1546324. [DOI] [PubMed] [Google Scholar]
- Hopfner, K.P., Lang, A., Karcher, A., Sichler, K., Kopetzki, E., Brandstetter, H., Huber, R., Bode, W., Engh, R.A. Coagulation factor IXa: The relaxed conformation of Tyr99 blocks substrate binding. Structure. 1999;7:989–996. doi: 10.1016/s0969-2126(99)80125-7. [DOI] [PubMed] [Google Scholar]
- Ichinose, A., Takeya, H., Espling, E., Iwanaga, S., Kisiel, W., Davie, E.W. Amino acid sequence of human protein Z, a vitamin K-dependent plasma glycoprotein. Biochem. Biophys. Res. Commun. 1990;172:1139–1144. doi: 10.1016/0006-291x(90)91566-b. [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impey, R.W. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Kamata, K., Kawamoto, H., Honma, T., Iwama, T., Kim, S.H. Structural basis for chemical inhibition of human blood coagulation factor Xa. Proc. Natl. Acad. Sci. 1998;95:6630–6635. doi: 10.1073/pnas.95.12.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemball-Cook, G., Johnson, D.J., Tuddenham, E.G., Harlos, K. Crytsal structure of active site-inhibited human coagulation factor VIIa (des-Gla) J. Struct. Biol. 1999;127:213–223. doi: 10.1006/jsbi.1999.4158. [DOI] [PubMed] [Google Scholar]
- Krem, M., Di Cera, E. Molecular markers of serine protease evolution. EMBO J. 2001;20:3036–3045. doi: 10.1093/emboj/20.12.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krem, M., Di Cera, E. Evolution of enzyme cascadses from embryonic development to blood coagulation. Trends Biochem. Sci. 2002;27:67–74. doi: 10.1016/s0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lee, C.J., Chandrasekaran, V., Duke, R.E., Perera, L., Pedersen, L.G. A proposed structural model of human protein Z (PZ) J. Thromb. Haemost. 2007;5:1558–1561. doi: 10.1111/j.1538-7836.2007.02597.x. [DOI] [PubMed] [Google Scholar]
- Luethy, R., Bowie, J.U., Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Mather, T., Oganessyan, V., Hof, P., Huber, R., Foundling, S., Esmon, C., Bode, W. The 2.8 Å crystal structure of Gla-domainless activated protein C. EMBO J. 1996;15:6822–6831. [PMC free article] [PubMed] [Google Scholar]
- Miletich, J.P., Broze G.J., Jr Human plasma protein Z antigen: Range in normal subjects and effect of warfarin therapy. Blood. 1987;69:1580–1586. [PubMed] [Google Scholar]
- Miller, D.W., Agard, D.A. Enzyme specificity under dynamic control: A normal mode analysis of α-lytic protease. J. Mol. Biol. 1999;286:267–278. doi: 10.1006/jmbi.1998.2445. [DOI] [PubMed] [Google Scholar]
- Morris, G.M., Goodsell, D.S., Halliday, R.S., Huey, R., Hart, W.E., Belew, R.K., Olson, A.J. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- Ortlund, E.A., Bridgham, J.T., Redinbo, M.R., Thornton, J.W. Crystal structure of an ancient protein: Evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, N., Agard, D.A. Enzyme specificity under dynamic control II: Principal component analysis of α-lytic protease using global and local solvent boundary conditions. Protein Sci. 2001;10:1403–1414. doi: 10.1110/ps.800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona, J.J., Craik, C.S. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona, J.J., Craik, C.S. Evolutionary divergence of substrate specificity within the chymotrypsin-like seine protease fold. J. Biol. Chem. 1997;272:29987–29990. doi: 10.1074/jbc.272.48.29987. [DOI] [PubMed] [Google Scholar]
- Pike, A.C.W., Brzozowski, A.M., Roberts, S.M., Olsen, O.H., Person, E. Structure of human factor VIIa and its implications for the triggering of blood coagulation. Proc. Natl. Acad. Sci. 1999;96:8925–8930. doi: 10.1073/pnas.96.16.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post, C.B., Ray, J.W.J. Reexamination of induced fit as a determinant of substrate specificity in enzymic reactions. Biochemistry. 1995;34:15881–15885. doi: 10.1021/bi00049a001. [DOI] [PubMed] [Google Scholar]
- Sali, A., Potterton, L., Yuan, F., van Vlijmen, H., Karplus, M. Evaluation of comparative protein modelling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- Sejima, H., Hayashi, T., Deyashiki, Y., Nishioka, J., Suzuki, K. Primary structure of vitamin K-dependent human protein Z. Biochem. Biophys. Res. Commun. 1990;171:661–668. doi: 10.1016/0006-291x(90)91197-z. [DOI] [PubMed] [Google Scholar]
- Tabatabai, A., Fiehler, R., Broze G.J., Jr Protein Z circulates in plasma in a complex with protein Z-dependent protease inhibitor. Thromb. Haemost. 2001;85:655–660. [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]