Abstract
Human glucose 6-phosphate dehydrogenase, purified after overexpression in E. coli, was shown to contain one molecule/subunit of acid-extractable “structural” NADP+ and no NADPH. This tightly bound NADP+ was reduced by G6P, presumably following migration to the catalytic site. Gel-filtration yielded apoenzyme, devoid of bound NADP+ but, surprisingly, still fully active. Mr of the main component of “stripped” enzyme by gel filtration was ∼100,000, suggesting a dimeric apoenzyme (subunit Mr = 59,000). Holoenzyme also contained tetramer molecules and, at high protein concentration, a dynamic equilibrium gave an apparent intermediate Mr of 150 kDa. Fluorescence titration of the stripped enzyme gave the K d for structural NADP+ as 37 nM, 200-fold lower than for “catalytic” NADP+. Structural NADP+ quenches 91% of protein fluorescence. At 37°C, stripped enzyme, much less stable than holoenzyme, inactivated irreversibly within 2 d. Inactivation at 4°C was partially reversed at room temperature, especially with added NADP+. Apoenzyme was immediately active, without any visible lag, in rapid-reaction studies. Human G6PD thus forms active dimer without structural NADP+. Apparently, the true role of the second, tightly bound NADP+ is to secure long-term stability. This fits the clinical pattern, G6PD deficiency affecting the long-lived non-nucleate erythrocyte. The K d values for two class I mutants, G488S and G488V, were 273 nM and 480 nM, respectively (seven- and 13-fold elevated), matching the structural prediction of weakened structural NADP+ binding, which would explain decreased stability and consequent disease. Preparation of native apoenzyme and measurement of K d constant for structural NADP+ will now allow quantitative assessment of this defect in clinical G6PD mutations.
Keywords: glucose 6-phosphate dehydrogenase, apoenzyme, “structural” NADP+, cofactor removal, enzyme stability, dissociation constant, G6PD deficiency
There have been many reports over the years that the subunits of human glucose 6-phosphate dehydrogenase (G6PD) [EC 1.1.1.49] possess a second NADP+-binding site in addition to that directly involved in catalysis (Kirkman 1962; Kirkman and Hendrickson 1962; Chung and Langdon 1963; Bonsignore et al. 1970; Yoshida 1973). The existence and possible role of this postulated “structural” NADP+ site remained controversial, however, not least because the first solved G6PD structure, that of the homologous enzyme of Leuconostoc mesenteroides (Lm), showed no sign of any coenzyme site other than the catalytic binding site (Rowland et al. 1994). The issue was finally and unambiguously resolved, however, by solution of the corresponding structure for human G6PD (the Canton mutant) (Au et al. 2000), which clearly shows bound NADP+ occupying a second site, quite distinct from the vacant catalytic NADP+ site. Considerable interest now attaches to the role of this tightly bound nucleotide molecule, since the crystallographic structure reveals that a high proportion of the clinical mutations associated with severe G6PD deficiency in man cluster around this structural NADP+ site (Au et al. 2000; Vulliamy and Luzzatto 2003). This overturns an earlier view that these mutations disrupt binding of the catalytic NADP+ molecule (Hirono et al. 1989). It leaves an open question, however, as to the true role of the auxiliary coenzyme molecule. It could be strictly required to achieve and maintain the active conformation of the enzyme protein and/or its quaternary structure. On the other hand, it might perhaps be only required for long-term stability.
The objectives of the present study, therefore, were to determine the stoichiometric content of bound NADP+; to discover whether it may be removed under gentle, non-denaturing conditions and, if so, whether it can be replaced and a dissociation constant determined; and finally to examine the properties of the enzyme protein in its absence. Taken together, these experiments, if successful, might afford a clearer view of the role of the structural NADP+ in human G6PD. They would also open the door to a more quantitative appraisal of the effects of many clinical mutations in this enzyme.
Results
Content of tightly bound NADP+
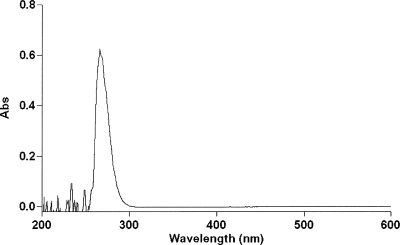
In order to release noncovalently bound nucleotide, 1 mL samples of freshly purified enzyme at 2 mg/mL were treated with trichloroacetic acid (TCA) to give final percentages between 5% (w/v) and 17.5% (w/v); 15% TCA was required to achieve complete precipitation of the protein, as judged by Bradford assays (Bradford 1976) on the supernatant after centrifugation, and this TCA concentration was adopted for subsequent measurements. A standard curve for NADP+ in 15% TCA gave an extinction coefficient at 260 nm of 18,700 M−1cm−1. Three separate G6PD samples were analyzed, and the released NADP+ was estimated as 1.14, 0.93, and 0.99 mol NADP+ per mol subunit. The average figure of 1.02 mol per mol subunit indicates full occupancy of an NADP+ site that, from crystallographic evidence (Au et al. 2000), can be assumed to be the second, structural site. A further sample at 3 mg/mL (600 μL, 50.8 μM, 30.5 nmol) was treated similarly and yielded 29.3 nmol NADP+ in the initial supernatant plus a further 2.6 nmol from a washing with 15% TCA for a total of 31.9 nmol. This again corresponds to a stoichiometry of 1.05 mol per subunit. In this case, however, the absorbance of the supernatant was scanned from 200 to 600 nm; the absence of any measurable absorbance beyond 300 nm (Fig. 1) shows that no NADPH is bound and is consistent with the assumption that the released ligand is entirely NADP+.
Figure 1.
Absorption spectrum of coenzyme released from G6PD after TCA precipitation. Fifty percent (w/v) TCA was added to 3 mg/mL G6PD to final concentration of 15% (w/v). After being kept on ice for 1 h, the sample was centrifuged, and the supernatant containing released NADP+ was scanned between 200 and 600 nm.
Removal of tightly bound NADP+ without denaturation
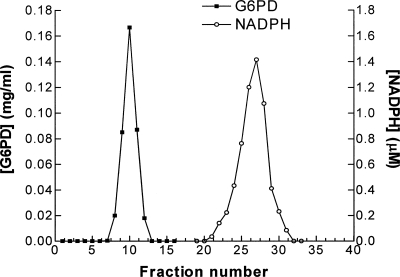
Two hundred microliters of G6PD at 1.5 mg/mL were mixed with 390 μL WHO buffer and 10 μL 60 mM G6P to give final concentrations of 8.47 μM G6PD and 1 mM G6P. Absorbance was monitored at 340 nm and rapidly increased by 0.051, corresponding (if due to NADPH) to 8.22 μM NADPH. This would suggest a reduction of 0.97 mol NADP+ per mol subunit. Gel filtration on a Sephadex G-75 column (1.5 × 13 cm) clearly separated two peaks (Fig. 2), an early one containing the protein and a later one absorbing at both 260 nm and 340 nm. The fractions under the second peak were measured fluorimetrically against a calibration with standard NADPH to give the quantitation shown in Figure 2. These data also allow a rough integration of protein and NADPH under the two peaks, yielding a ratio of 0.97 mol per mol. Enzyme thus treated was precipitated with 15% TCA and now showed no detectable nucleotide content.
Figure 2.
Displacement of tightly bound NADP+ by addition of glucose 6-phosphate. The purified G6PD enzyme, freed of any loosely bound NADP+ by serial dilution, was incubated at room temperature with glucose 6-phosphate at 1 mM (final concentration of 8.47 μM G6PD) for 30 min before passing down a Sephadex G-75 column pre-equilibrated and eluted with the 0.1 M Tris-HCl buffer at pH 7.6. The gel-filtration profile was monitored at 280 nm to locate the protein peak and at 340 nm to locate and quantitate the NADPH peak.
This result entirely fits with the rather surprising longstanding claim (Bonsignore et al. 1971a,b; De Flora et al. 1974) that the tightly bound NADP+ can be reduced by substrate, and also shows that, in this way, the cofactor can be quantitatively removed, since NADPH does not bind appreciably to the tight-binding structural NADP+ site.
Activity assay of stripped G6PD
In order to assess the role of the structural coenzyme molecule, the G6PD preparation, stripped of tightly bound nucleotide as described, was assayed for catalytic activity in the normal way (Betke et al. 1967) with 200 μM NADP+ and 600 μM glucose 6-phosphate. Unexpectedly, fresh preparations of the stripped enzyme showed 100% retention of catalytic activity without any obvious indication of a lag in the time course of reaction. This implies either that the enzyme is fully active in the absence of structural NADP+, or else that activity is fully restored in the period of ∼10 sec between mixing and the commencement of measurement by repopulation of the structural site by NADP+ from the reaction mixture.
Dissociation constant for structural NADP+
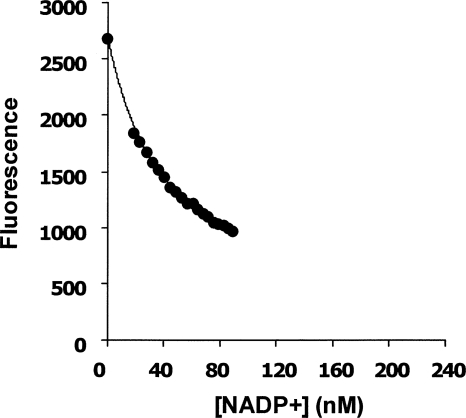
The freshly prepared stripped enzyme gives a fluorescence emission peak at 345 nm when excited with light at 290 nm. This emission is partially quenched by addition of NADP+. An initial Scatchard plot of the titration data (fluorescence quench/[NADP+] against fluorescence quench) gave a good straight line (data not shown), allowing rough estimation of the dissociation constant as 41.4 nM. However, this plot and calculation rest on the assumption that the ligand concentration is essentially unperturbed by the small decrement on binding to the enzyme, and this is not strictly true here, since at one end of the data set the ligand concentration was only nine times that of the enzyme. The data set also did not extend to high enough ligand concentrations to yield a direct measure of the fluorescence of the fully liganded enzyme. A computer, fit to the equation for a single binding site using GOSA software (Global Optimization by Simulated Annealing), gave a good fit to the data points for a K d of 37.0 nM and a quench of 90.7% (Fig. 3).
Figure 3.
Determination of the K d value of stripped G6PD for NADP+. A stripped G6PD sample (5 nM subunit concentration) was titrated with concentrated NADP+, and the fluorescence signal was recorded by fluorimeter after each addition. The resulting data were fitted to the equation for a single binding constant with the help of the GOSA software package (Global Optimization by Simulated Annealing) from Bio-Log. The calculated K d is 37.0 nM with uncertainty of 2.95 nM.
Stability of stripped enzyme
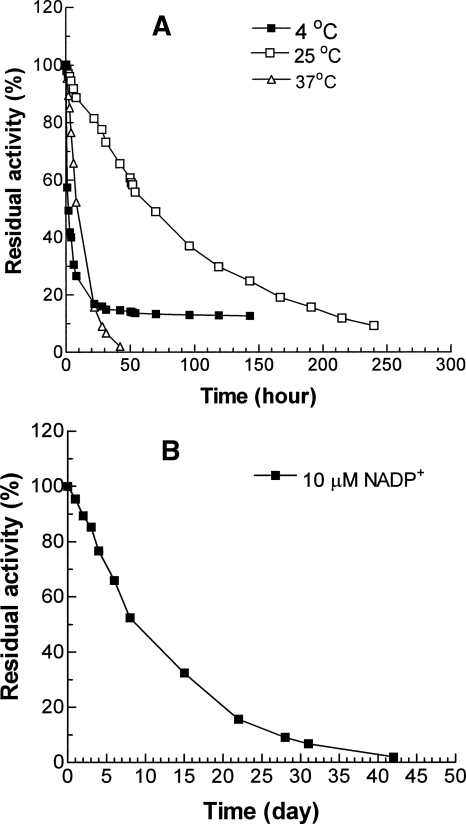
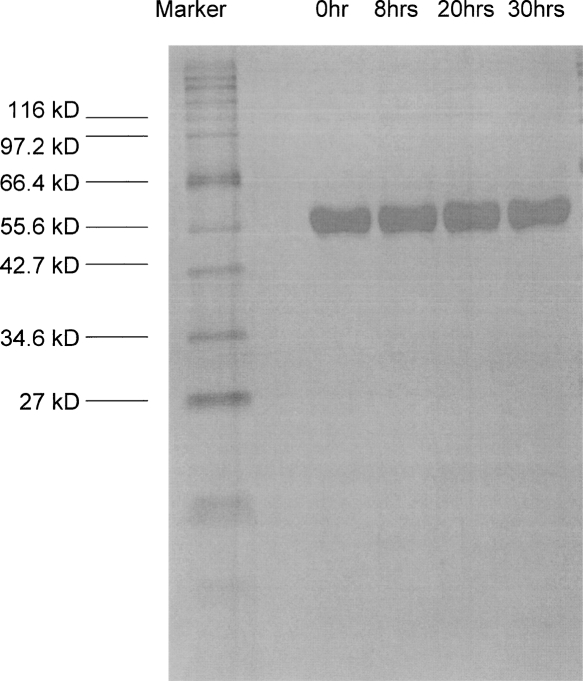
The activity measurements above suggest that the structural NADP+ is not essential, at least in the short term, for achieving catalysis, and this inevitably focuses attention on the question of whether it is needed for protein stability. Therefore three identical stripped G6PD samples at 50 μg/mL in Tris-HCl (pH 7.6) with 5 mM EDTA were incubated at the physiological temperature, 37°C, at the standard assay temperature, 25°C, and at 4°C, respectively. A fourth sample was also incubated at 37°C but with 10 μM NADP+ added. Loss of activity was most rapid at 4°C, with a decline of more than 70% in the first 10 h (Fig. 4A), suggesting that this apoenzyme is cold labile. However, the activity did not reach zero, and after 30 h, it reached a constant level of 14%. At 37°C, enzyme activity also declined, more slowly at first (Fig. 4), but in this case, the inactivation continued until after 2 d virtually all activity was lost. Contrasting with both, at 25°C the enzyme also gradually lost activity but much more slowly, so that, for example, after 2 d about 60% of activity was retained, and even after 10 d, the sample was still 10% active. In order to ensure that loss of activity truly reflected intrinsic stability properties and not, for example, degradation by traces of bacterial proteinases, samples were withdrawn at intervals from an incubation at 37°C and run on SDS-PAGE. Figure 5 shows that over 30 h, there was no change in the size of the main band and no sign of new bands corresponding to material of lower Mr.
Figure 4.
Stability properties of stripped G6PD. (A) Residual activity of G6PD without structural NADP+ incubated at 50 μg/mL in Tris-HCl (pH 7.6) with 5 mM EDTA at 4°C, 25°C, and 37°C. (B) Residual activity of an identical sample of G6PD incubated at 37°C after addition of 10 μM NADP+ to the buffer. In both cases, samples were withdrawn at intervals for assay under standard WHO conditions.
Figure 5.
SDS-PAGE of G6PD without structural NADP+ incubated at 37°C. Stripped G6PD enzyme at 50 μg/mL in Tris-HCl (pH 7.6) with 5 mM EDTA was incubated at 37°C. Samples were withdrawn at different time points and run on SDS-PAGE.
The addition of NADP+ at 10 μM stabilized the enzyme: In contrast to loss of 50% of activity in 8 h at 37°C without NADP+, the same loss in the presence of coenzyme required 8 d (Fig. 4B).
Reactivation of G6PD
In contrast to the apparently irreversible inactivation at 37°C, the enzyme incubated in the cold appeared to reach an equilibrium position, suggestive of a slow, but reversible conformational change. To test this, therefore, the enzyme at 14% residual activity was transferred to 25°C. Without any added coenzyme, the enzyme returned to a maximum of 36% activity within 30 min. The extent of recovery was enhanced by adding NADP+ at the same time as raising the temperature. With 10, 100, and 1000 μM NADP+, the activity rose to 43%, 54%, and 59%, respectively, in 30 min.
In assessing these results it has to be borne in mind that under similar conditions and over the same timescale, the intact holoenzyme (with structural NADP+ tightly bound, but no coenzyme bound at the catalytic site) would remain stable at all three temperatures (Kirkman 1962; Chung and Langdon 1963). It thus emerges clearly that the structural NADP+ is essential for the maintenance of activity of an enzyme that, in the intact red cell, is required to remain active over many weeks.
Measurement of molecular size
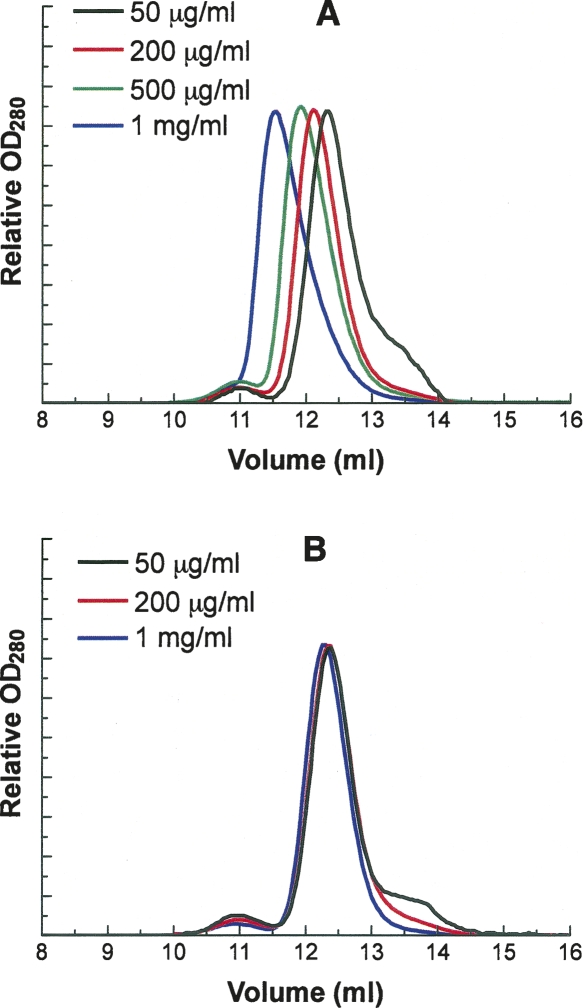
Native human G6PD can exist either as dimer or as tetramer, the two species being in an equilibrium strongly affected, among other things, by pH (Cohen and Rosemeyer 1969; Wrigley et al. 1972). The relation between quaternary structure and activity has remained a matter of dispute, and it was of interest to discover whether removal of the NADP+ affects the subunit structure. The apparent molecular weight of native, unstripped G6PD and the apoenzyme samples (100 μL at 0.05 to 1 mg/mL) was determined by FPLC. As to the intact holoenzyme (defined as above), at lower concentration of protein (0.05 mg/mL) there was one major peak and one small minor peak with elution volumes at 12.33 mL and 10.95 mL, corresponding, according to the calibration plot, to Mr values of 101.5 kDa and 203.6 kDa (Fig. 6A). These figures correspond roughly to the values for a dimer and tetramer. When the protein concentration increased, these two peaks converged and at 1 mg/mL merged into a single peak with Mr value of 150.5 kDa, corresponding closely to the value for a trimer. The symmetry of this protein makes it highly unlikely that the enzyme is trimeric, but the observed elution volume could readily be accounted for if dimers, once saturated with structural NADP+, are more likely to associate at high concentration, giving rise to a rapidly adjusting dimer–tetramer equilibrium and an apparent intermediate, average Mr from gel filtration, corresponding to neither species. In contrast, with the stripped enzyme, the main peak with apparent Mr of ∼100 kDa rather than 150 kDa could always be observed even at high protein concentration, indicating that in the absence of structural NADP+, the formation of tetramer is largely suppressed (Fig. 6B).
Figure 6.
Gel-elution profile of native G6PD and stripped G6PD by FPLC. One hundred microliter samples of different G6PD protein solutions were loaded on a Superdex 200 HR 10/30 column equilibrated with 0.1 M Tris-HCl, 150 mM NaCl (pH 7.6). To facilitate comparison, the absorbance of each protein peak is shown as a relative value rather than an absolute unit. (A) Apparent molecular weights of native G6PD peaks at different protein concentrations: 50 μg/mL, 101.5 kDa (12.33 mL) and 203.6 kDa (10.95 mL); 200 μg/mL, 112.9 kDa (12.12 mL) and 200.6 kDa (10.98 mL); 500 μg/mL, 124.9 kDa (11.92 mL) and 198.6 kDa (11.00 mL); and 1.0 mg/mL, 150.5 kDa (11.55 mL). (B) Apparent molecular weights of stripped G6PD peaks at different protein concentrations: 50 μg/mL, 100.01 kDa (12.36 mL) and 204.66 kDa (10.94 mL); 200 μg/mL, 100.52 kDa (12.35 mL) and 201.6 kDa (10.97 mL); and 1.0 mg/mL, 104.9 kDa (12.28 mL) and 200.6 kDa (10.98 mL).
Rapid-reaction experiments
As seen above, the freshly prepared apoenzyme appears to be fully active as judged by standard assays. However, since in such an assay there is a variable delay of ∼10 sec before the fully mixed solution comes under observation, it is at least possible that bound structural NADP+ is required to establish an active enzyme molecule but that during the 10-sec blind period the holoenzyme is fully reconstituted. This was tested by mixing the enzyme with the standard reactants in a stopped-flow apparatus with a dead time of ∼1 msec. Even on this timescale, a linear steady-state increase in A340 was observed, without any hint of a lag phase. This means either that the apoenzyme is fully active or that the structural NADP+ can be completely re-bound within 1 msec. However, a rough calculation shows that, given the molar concentrations of apo-G6PD (26–106 nM) and NADP+ (200 μM) used in this experiment, the second possibility would require a second-order “on” rate constant far in excess of the upper limit for a diffusion-controlled reaction.
Dissociation constants for two clinical G6PD mutants
A persistent belief in relation to the mutations responsible for G6PD deficiency in man has been that they destabilize the enzyme protein by weakening coenzyme binding. The methodology described here provides at last a way to put this idea to the test in a quantitatively rigorous fashion. The method was therefore applied to determine the dissociation constants of structural NADP+ for two severe class I naturally occurring G6PD mutants, G6PDCampinas (G488V) (Baronciani et al. 1993) and G6PDFukaya (G488S) (Vulliamy et al. 1997). The K d values for G488S and G488V were 273 nM and 480 nM, respectively (data not shown), about sevenfold and 13-fold higher than the figure for wild-type enzyme.
Discussion
Over the past 40 years, many investigators have proposed that G6PD purified from erythrocytes might have a structural NADP+ moiety in addition to a separate site for the “catalytic” NADP+ (Kirkman 1962; Chung and Langdon 1963; Luzzatto and Allan 1965; Bonsignore et al. 1970; Yoshida 1973). The crystal structure of human G6PDCanton (Au et al. 2000), clearly showed a structural NADP+ molecule bound between the dimer interface and the C terminus, well separated from the catalytic coenzyme-binding site. The results here for purified recombinant human G6PD now unambiguously prove the stoichiometry of this binding site: Each monomer contains one tightly bound NADP+ molecule.
The method used here for making a native apoenzyme is at first sight surprising. The simplest interpretation of the original experiments from 30 years ago (Bonsignore et al. 1971a,b; De Flora et al. 1974) might have been that a proportion of the active sites bound the cofactor more tightly than the others. However, the crystallography shows no cofactor at the catalytic site in the enzyme as normally prepared, and the structural NADP+, on the other hand, is evidently very tightly bound, not being removed by the usual dialysis and chromatographic procedures. It might be assumed, therefore, that this structural NADP+, tightly bound at a non-catalytic site, would be unavailable for reduction by G6P. However, even though a tight binding constant imposes an upper limit on the kinetic “off” constant for ligand release, it is still entirely possible for such a ligand to be in reasonably rapid exchange with the surrounding medium. Conversion to NADPH in the presence of G6P indicates that the structural NADP+ is indeed in a dynamic equilibrium, since it must involve translocation to a catalytic site. This may be due to conformational changes of the enzyme induced on the binding of G6P. This suggestion is supported by the recently determined crystallographic results for human ΔG6PD-G6P, a mutant with a deletion of 25 residues from the N-terminal (Kotaka et al. 2005). Based on the structure, although the G6P binding site is ∼15 Å distant from the structural NADP+-binding site, the presence of G6P could affect the latter site through movements of one or two amino acid residues from two small stretches of the primary sequence (363–366 and 393–395), each of which spans from the substrate site to the structural NADP+-binding site to the other (Kotaka et al. 2005). In the binary complex ΔG6PD-G6P, 10 residues in the C terminus are disordered and NADP+ occupancy at the structural site is also low. It is possible that binding of the sugar phosphate triggers the conformational change at the C terminus, decreasing the affinity for the structural NADP+, and thus allowing it to migrate to the catalytic site. An alternative sequence, in which the disordered C terminus is a consequence of prior migration of the structural NADP+, cannot be excluded, however.
The results also imply that NADPH binds to the structural site very weakly, if at all, since, once reduced, the cofactor is readily removed by gel filtration and since the cofactor removed by TCA treatment is entirely in the oxidized form. It is therefore important to note that the intracellular NADP+/NADPH pool is largely in the reduced form (Kirkman and Gaetani 1986), and accordingly control of the oxidation–reduction balance of this coenzyme might have an important bearing on long-term stability of G6PD.
Previous studies demonstrated that G6PD stripped of structural NADP+ seemed to be inactive and monomeric (Bonsignore et al. 1971b; Wrigley et al. 1972; Cancedda et al. 1973). However, the fact that the enzyme can be totally stripped of NADP+ in a gentle fashion by catalytic turnover would appear to imply that it must remain active even after the coenzyme has vacated the structural site: Initially, NADP+ leaving one G6PD subunit might be reduced at the catalytic site of a second, still-intact subunit, but this would not easily account for rapid total stripping of the entire population of enzyme molecules. Indeed, in our study, G6PD carefully stripped of its structural NADP+ was found to be both active and dimeric. The discrepancy between this and earlier results could be due to the G6PD samples, semi-purified from different patients' blood in previous studies, or possibly to differences in the timing of experiments.
Thus, the proposed notion that the structural NADP+ is necessary for dimerization of G6PD seems to be inaccurate. On the other hand, our results indicate that the second dimerization to form a tetrameric molecule is promoted by the presence of structural NADP+. Accordingly, it may also readily be deduced that, contrary to some earlier assertions, the tetramer state is not required for catalytic activity. Further experiments, however, need to be carried out to investigate the quaternary structure of G6PD with and without structural NADP+ in greater depth.
The structural and catalytic NADP+ sites differ markedly in their affinity for the coenzyme: The K d value for structural NADP+ determined here at 25°C is 37 nM, ∼200-fold lower than 7.8 μM for catalytic NADP+ (Wang et al. 2005). This, interestingly, correlates with unpublished estimates, cited many years ago by Yoshida (1973), of values of 170 nM at 37°C and 1.5 nM at 4°C for the K d for NADP+ obtained by equilibrium dialysis. Yoshida also drew attention to the fact that these values were far lower than those inferred from the concentration dependence of the catalytic reaction, suggesting, therefore, that there might be a second type of binding site for the coenzyme (Yoshida 1973). From the results obtained here, this tightly bound NADP+ seems to be crucial, not for activity, but rather for the long-term stability of the enzyme at the physiological temperature of 37°C. Scopes et al. (1998), without being specifically aware of the role of a structural NADP+ site, also noted the connection between NADP+ concentration and stabilization of the dimer state. Enzyme stability is particularly crucial in mature red blood cells, which lack a nucleus and the machinery to synthesize proteins. It is striking to find that more than 40 severe class I mutations are located in a distinct cluster between amino acids 360 and 450, in and around exon 10, which is close to the structural NADP+ site (Hirono et al. 1989; Au et al. 2000; Vulliamy and Luzzatto 2003; Kotaka et al. 2005; Mason et al. 2007). It seems likely that these mutations affect the binding capacity for structural NADP+ and thus decrease enzyme stability, causing the severe disease phenotype. Detailed investigation of such mutants may further clarify the significance of structural NADP+.
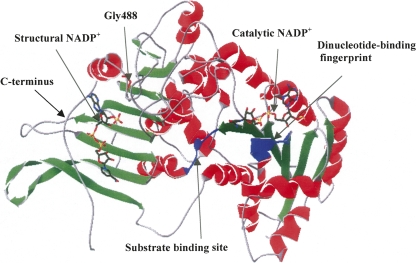
The methodology presented here now offers the possibility of direct comparison of structural NADP+ dissociation constants for native G6PD and such clinical mutants. In the current study, the K d values of another two severe class I mutants, G488S and G488V, were also determined, giving figures of 273 nM and 480 nM, respectively, about sevenfold to 13-fold higher than the value for the wild-type enzyme. Based on the three-dimensional structure of human G6PD (Fig. 7), Gly488 is located in the βO-αo turn of the C terminus, important in positioning Arg 487 to interact with the adenine and the 2′-phosphate of structural NADP+ directly. If Gly488 was mutated to Ser or Val, the larger side chains of these two amino acid residues could affect the binding of structural NADP+. The results here clearly showed that these mutations do indeed decrease binding affinity for structural NADP+. We have also recently applied the same approach to two class I mutations at position 393, in which Arg has been replaced, respectively, by His (G6PDNashville) and Gly (G6PDWisconsin) (Wang et al. 2006). The analysis produced an unambiguous differentiation in that the binding of structural NADP+ was weakened 15- to 20-fold in the first mutant and hardly at all in the second. Therefore, the determination of K d values for the binding of the structural NADP+ molecule seems likely to be of wide applicability in the assessment of mutational defects in G6PD deficiency. Moreover, the function of structural NADP+ in oligomerization and refolding as previously reported (Beutler and Collins 1965; Yoshida et al. 1967; Gomez-Gallego et al. 1996) also now needs to be further investigated.
Figure 7.
The human G6PDCanton monomer (PDB no. 2bh9). Helices, sheet strands, and coils of the subunit are shown in red, green, and gray, respectively. Substrate and catalytic NADP+-binding sites are shown in blue. Gly488 is highlighted in red.
Materials and Methods
Coenzymes, NADP+ (98% purity), and NADPH (100% purity) were from Roche; glucose 6-phosphate and TCA were from Sigma Chemical Co.; and Sephadex G-75 was from Pharmacia. FPLC column, Superdex 200 (10 × 30 cm), was also purchased from Pharmacia. Spectrophotometric measurements were made throughout with a Cary Bio50. Protein fluorescence was measured with a Hitachi F-4500 fluorimeter. Rapid-reaction measurements were carried out with a SX.18 MV-R stopped-flow reaction analyzer from Applied Photophysics, Ltd.
The expression plasmids of two naturally occurring mutants, G6PDCampinas (G488V) (Baronciani et al. 1993) and G6PDFukaya (G488S) (Vulliamy et al. 1997), were constructed according to a previous report (Wang et al. 2006). In brief, the desired mutations in G6PD G488S and G488V were created by megaprimer-based site-directed mutagenesis (Landt et al. 1990; Sarkar and Sommer 1990). The two flanking primers (below) were IMPACT NDES, generating an NdeI restriction site at the 5′ end of the gene, and G6PD-pQE-SalI-R, creating a SalI restriction site at the 3′ end. The internal mutagenic primers were G6PD-G488S and G6PD-G488V, with the mutagenic base change highlighted in bold in each case.
Primer IMPACT NDES: 5′pCAGGAAACAGCATATGGCAGAGC-3′
Primer G6PD-pQE-SalI-R: 5′pTAGGGCGTCGACTCAGAGCTTGTGGGGGTTCAC-3′
Mutagenic primer G6PD-G488S: 5′-ATGGCAGCCGAAGCCCCACGGAG-3′
Mutagenic primer G6PD-G488V: 5′ATGGCAGCCGAGTCCCCACGGAGGC-3′
The recombinant G6PD wild type and mutants G488S and G488V were overexpressed, purified, and stored as previously described (Wang et al. 2002, 2005), showing a similar specific activity of 180 IU/mg. The buffer throughout purification and subsequent procedures was 0.1 M Tris-HCl (pH 7.6), with 5 mM EDTA. Seventy-five μM NADP+ was added to the buffer, both to achieve elution from the affinity column and to ensure long-term stability of the purified enzyme. For these experiments, however, excess NADP+ was rapidly removed by repeated (8×) centrifugal filtration through a Centricon YM-50 cone and dilution in fresh buffer. The starting sample was 400 μL G6PD (1–2 mg/mL). In each filtration cycle, the enzyme was diluted fivefold to 2 mL and reduced again by filtration to ∼400 μL.
In order to release noncovalently bound nucleotide, G6PD was precipitated by adding 50% (w/v) TCA to give various final concentrations. Samples were centrifuged after 1 h on ice, and the white protein precipitate was resuspended in the same acidic solution to wash out any trapped nucleotide. The protein concentration of the combined supernatants was estimated (Bradford 1976), and the released nucleotide was estimated by measuring A260 and employing a directly determined extinction coefficient.
G6PD enzyme activities were estimated by using the standard WHO assay (Betke et al. 1967).
To remove tightly bound NADP+ without denaturation of the enzyme protein, we followed the observation of Bonsignore et al. (1971a,b) and De Flora et al. (1974), who reported that it could be reduced and released by adding the substrate glucose 6-phosphate. This method, though initially surprising, nevertheless proved effective in our hands. The purified enzyme, from which any loosely bound NADP+ had previously been removed by serial dilution, was incubated at room temperature with glucose 6-phosphate at a high concentration (1 mM) for 30 min before passing down a Sephadex G-75 column pre-equilibrated and eluted with the usual 0.1 M Tris-HCl buffer at pH 7.6.
The size exclusion column (Superdex 200 HR 10/30) for FPLC experiments was equilibrated with 0.1 M Tris-HCl, 150 mM NaCl, pH 7.6 and calibrated with ferritin (450 kDa), β-amylase (200 kDa), bovine serum albumin (68 kDa), hen ovalbumin (45 kDa), cytochrome c (12.5 kDa), and aprotinin (6.5 kDa).
In rapid-reaction studies, one of the two drive syringes contained the enzyme preparation at 6.25, 3.13, or 1.56 μg/mL in buffer and the other contained buffer with 400 μM NADP+ and 1.2 mM G6P. For each reaction, 150 μL each of the two solutions was rapidly mixed, and the increase in A340 was monitored.
In the stability test, G6PD lacking structural NADP+ was incubated at 4°C, 25°C, and 37°C, and the residual enzyme activity was followed. A further sample was monitored at 37°C after adding 10 μM NADP+.
In order to determine the dissociation constant for binding of NADP+ at the structural site, a centrifuged solution of G6PD (5 nM subunits) was titrated with a concentrated solution of NADP+ added with a Hamilton syringe. Protein fluorescence was monitored by exciting at 290 nm and measuring emission at 345 nm. The power setting was 950 W and the slit 10/10 nm. The resulting data were fitted to the equation for a single binding constant with the help of the GOSA software package from Bio-Log.
Acknowledgments
We dedicate this article to the memory of our coauthor V.M.S.L., who was tragically lost in Thailand in the “tsunami” disaster of December 26, 2004. This work was made possible by a grant to the late V.M.S.L. from the Research Grants Council of Hong Kong and has been completed under a Science Foundation Ireland Fellowship grant to P.C.E. We also thank Jess L.C. Chan for curating the G6PD mutants and supplying the clones.
Footnotes
Reprint requests to: Paul C. Engel, School of Biomolecular and Biomedical Science, Conway Institute, University College Dublin, Belfield, Dublin 4, Ireland; e-mail: paul.engel@ucd.ie; fax: 353-1-283-7211.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035352.108.
References
- Au, S.W.N., Gover, S., Lam, V.M.S., Adams, M.J. Human glucose-6-phosphate dehydrogenase: The crystal structure reveals a structural NADP+ molecule and provides insights into enzyme deficiency. Structure. 2000;8:293–303. doi: 10.1016/s0969-2126(00)00104-0. [DOI] [PubMed] [Google Scholar]
- Baronciani, L., Tricta, F., Beutler, E. G6PD “Campinas”: A deficient enzyme with a mutation at the far 3′ end of the gene. Hum. Mutat. 1993;2:77–78. doi: 10.1002/humu.1380020115. [DOI] [PubMed] [Google Scholar]
- Betke, K., Beutler, E., Brewer, G.J., Kirkman, H.N., Luzzatto, L., Motulsky, A.G., Ramot, B., Siniscolo, M. Standardisation of procedures for the study of glucose-6-phosphate dehydrogenase. Report of a WHO scientific group. WHO Tech. Rep. Ser. 1967;366:1–53. [PubMed] [Google Scholar]
- Beutler, E., Collins, Z. Hybridization of glucose-6-phosphate dehydrogenase from rat and human erythrocytes. Science. 1965;150:1306–1307. doi: 10.1126/science.150.3701.1306. [DOI] [PubMed] [Google Scholar]
- Bonsignore, A., Lorenzoni, I., Cancedda, R., De Flora, A. Distinctive patterns of NADP binding to dimeric and tetrameric glucose 6-phosphate dehydrogenase from human red cells. Biochem. Biophys. Res. Commun. 1970;39:142–148. doi: 10.1016/0006-291x(70)90769-2. [DOI] [PubMed] [Google Scholar]
- Bonsignore, A., Cancedda, R., Nicolini, A., Damiani, G., De Flora, A. Metabolism of human erythrocyte glucose-6-phosphate dehydrogenase. VI. Interconversion of multiple molecular forms. Arch. Biochem. Biophys. 1971a;147:493–501. doi: 10.1016/0003-9861(71)90406-1. [DOI] [PubMed] [Google Scholar]
- Bonsignore, A., Cancedda, R., Lorenzoni, I., Cosulich, M.E., De Flora, A. Human erythrocyte glucose 6-phosphate dehydrogenase. Physical properties. Biochem. Biophys. Res. Commun. 1971b;43:94–101. doi: 10.1016/s0006-291x(71)80091-8. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cancedda, R., Ogunmola, G., Luzzatto, L. Genetic variants of human erythrocyte glucose-6-phosphate dehydrogenase. Discrete conformational states stabilized by NADP+ and NADPH. Eur. J. Biochem. 1973;34:199–204. doi: 10.1111/j.1432-1033.1973.tb02746.x. [DOI] [PubMed] [Google Scholar]
- Chung, A.E., Langdon, R.G. Human erythrocyte glucose 6-phosphate dehydrogenase. II. Enzyme-coenzyme interrelationship. J. Biol. Chem. 1963;238:2317–2324. [PubMed] [Google Scholar]
- Cohen, P., Rosemeyer, M.A. Subunit interactions of human glucose 6-phosphate dehydrogenase from human erythrocytes. Eur. J. Biochem. 1969;8:8–15. doi: 10.1111/j.1432-1033.1969.tb00488.x. [DOI] [PubMed] [Google Scholar]
- De Flora, A., Morelli, A., Giuliano, F. Human glucose 6-phosphate dehydrogenase: Content of bound coenzyme. Biochem. Biophys. Res. Commun. 1974;59:406–413. doi: 10.1016/s0006-291x(74)80221-4. [DOI] [PubMed] [Google Scholar]
- Gomez-Gallego, F., Garrido-Pertierra, A., Mason, P.J., Bautista, J.M. Unproductive folding of the human G6PD-deficient variant A- FASEB J. 1996;10:153–158. doi: 10.1096/fasebj.10.1.8566536. [DOI] [PubMed] [Google Scholar]
- Hirono, A., Kuhl, W., Gelbart, T., Forman, L., Fairbanks, V.F., Beutler, E. Identification of the binding domain for NADP+ of human glucose-6-phosphate dehydrogenase by sequence analysis of mutants. Proc. Natl. Acad. Sci. 1989;86:10015–10017. doi: 10.1073/pnas.86.24.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman, H.N. Glucose 6-phosphate dehydrogenase from human erythrocytes. I. Further purification and characterization. J. Biol. Chem. 1962;237:2364–2370. [PubMed] [Google Scholar]
- Kirkman, H.N., Gaetani, G.F. Regulation of glucose-6-phosphate dehydrogenase in human erythrocytes. J. Biol. Chem. 1986;261:4033–4038. [PubMed] [Google Scholar]
- Kirkman, H.N., Hendrickson, E.M. Glucose 6-phosphate dehydrogenase from human erythrocytes. II. Subactive states of the enzyme from normal persons. J. Biol. Chem. 1962;237:2371–2376. [PubMed] [Google Scholar]
- Kotaka, M., Gover, S., Vandeputte-Rutten, L., Au, S.W.N., Lam, V.M.S., Adams, M.J. Structural studies of glucose-6-phosphate and NADP+ binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr. D Biol. Crystallogr. 2005;61:495–504. doi: 10.1107/S0907444905002350. [DOI] [PubMed] [Google Scholar]
- Landt, O., Grunert, H.P., Hahn, U.A. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Luzzatto, L., Allan, N.C. Different properties of glucose 6-phosphate dehydrogenase from human erythrocytes with normal and abnormal enzyme levels. Biochem. Biophys. Res. Commun. 1965;21:547–554. doi: 10.1016/0006-291x(65)90520-6. [DOI] [PubMed] [Google Scholar]
- Mason, P.J., Bautista, J.M., Gilsanz, F. G6PD deficiency: The genotype-phenotype association. Blood Rev. 2007;21:267–283. doi: 10.1016/j.blre.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Rowland, P., Basak, A.K., Gover, S., Levy, H.R., Adams, M.J. The three-dimensional structure of glucose 6-phosphate dehydrogenase from Leuconostoc mesenteroides refined at 2.0 Å resolution. Structure. 1994;2:1073–1087. doi: 10.1016/s0969-2126(94)00110-3. [DOI] [PubMed] [Google Scholar]
- Sarkar, G., Sommer, S.S. The “megaprimer” method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- Scopes, D.A., Bautista, J.M., Naylor, C.E., Adams, M.J., Mason, P.J. Amino acid substitutions at the dimer interface of human glucose-6-phosphate dehydrogenase that increase thermostability and reduce the stabilising effect of NADP. Eur. J. Biochem. 1998;251:382–388. doi: 10.1046/j.1432-1327.1998.2510382.x. [DOI] [PubMed] [Google Scholar]
- Vulliamy, T.J., Luzzatto, L. Glucose-6-phosphate dehydrogenase deficiency and related disorders. In: Handin R.I., et al., editors. Blood: Principles and practice of hematology. Lippincott Williams and Wilkins; Philadelphia, PA: 2003. pp. 1921–1950. [Google Scholar]
- Vulliamy, T., Luzzatto, L., Hirono, A., Beutler, E. Hematologically important mutations: glucose-6-phosphate dehydrogenase. Blood Cells Mol. Dis. 1997;23:302–313. doi: 10.1006/bcmd.1997.0147. [DOI] [PubMed] [Google Scholar]
- Wang, X.T., Au, S.W.N., Lam, V.M.S., Engel, P.C. Recombinant human glucose-6-phosphate dehydrogenase. Evidence for a rapid-equilibrium random-order mechanism. Eur. J. Biochem. 2002;269:3417–3424. doi: 10.1046/j.1432-1033.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- Wang, X.T., Lam, V.M.S., Engel, P.C. Marked decrease in specific activity contributes to disease phenotype in two human glucose 6-phosphate dehydrogenase mutants, G6PD(Union) and G6PD(Andalus) Hum. Mutat. 2005;26:284. doi: 10.1002/humu.9367. [DOI] [PubMed] [Google Scholar]
- Wang, X.T., Lam, V.M.S., Engel, P.C. Functional properties of two mutants of human glucose 6-phosphate dehydrogenase, R393G and R393H, corresponding to the clinical variants G6PD Wisconsin and Nashville. Biochim. Biophys. Acta. 2006;1762:767–774. doi: 10.1016/j.bbadis.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Wrigley, N.G., Heather, J.V., Bonsignore, A., De Flora, A. Human erythrocyte glucose 6-phosphate dehydrogenase: Electron microscope studies on structure and interconversion of tetramers, dimers and monomers. J. Mol. Biol. 1972;68:483–499. doi: 10.1016/0022-2836(72)90101-5. [DOI] [PubMed] [Google Scholar]
- Yoshida, A. Hemolytic anemia and G6PD deficiency. Science. 1973;179:532–537. doi: 10.1126/science.179.4073.532. [DOI] [PubMed] [Google Scholar]
- Yoshida, A., Steinmann, L., Harbert, P. In vitro hybridization of normal and variant human glucose-6-phosphate dehydrogenase. Nature. 1967;216:275–276. doi: 10.1038/216275a0. [DOI] [PubMed] [Google Scholar]