Abstract
Protein self-association is critical to many biological functions. However, atomic-level structural characterization of these assemblies has remained elusive. In this report we present insights into the mechanistic details of the process of self-association of the 136-residue GTPase effector domain (GED) of the endocytic protein dynamin into a megadalton-sized soluble mass. Our approach is based on NMR monitoring of regulated folding and association through Gdn-HCl titration. The results suggest the evolution of a sequence–self-association paradigm. Equally significantly, the study demonstrates an elegant bottom-up strategy that can render large protein self-assemblies accessible to NMR investigations that have remained difficult to date.
Keywords: protein folding, self-assembly, Gdn-HCl, relaxation measurement, NMR
Most cellular functions are mediated through protein–protein association. Many of these are related to self-association. Details regarding the determinants of self-association and whether there exists an intrinsic relation between sequence and self-association can only be understood when there are enough structural details available for large protein assemblies. However, crystallization difficulties with many such assemblies render it impossible to study them using X-ray crystallography, and molecular weight, if not solubility, puts a limit on them being studied by NMR. Biomolecular NMR studies have largely been limited to structures with relative molecular mass (M r) below 100 kDa, with only a few studies done on larger molecules (Fiaux et al. 2002; Foster et al. 2007; Sprangers and Kay 2007), the largest assembly being GroEL–GroES complex (M r ≈ 900 kDa) (Fiaux et al. 2002); for most of these systems a fair amount of crystallographic information was previously available (Braig et al. 1994; Lowe et al. 1995; Hunt et al. 1996; Groll et al. 1997; Xu et al. 1997; Unno et al. 2002). The study of large assemblies of megadalton size by solution NMR spectroscopy has largely been hampered because of large rotational correlation times (τc) and/or inter-/intramolecular conformational exchange, which lead to extensive broadening of signals beyond detection.
Dynamin, during endocytosis, self-assembles into a helix around the neck of an invaginating clathrin-coated pit through the GTPase effector domain (GED) (Hinshaw 2000). Recombinant GED forms large soluble assemblies, and residue level information on this process would add to the information about its role in dynamin self-assembly, the high-resolution picture of which is still unknown. With this background, we report herein the NMR characterization of a large assembly produced by the GED of dynamin (Praefcke and McMahon 2004). A bottom-up approach was used wherein changes in the NMR and CD spectra were monitored as a function of denaturant concentration to obtain molecular snapshots of the stepwise folding and concomitant self-assembly process.
Similar denaturant-dependent approaches have been used earlier for different purposes. For example, unfolding of molten globules—which are generally inaccessible—formed by human (Schulman et al. 1997) and bovine (Wijesinha-Bettoni et al. 2001) α-lactalbumin has been investigated. Likewise, rare insights into the equilibrium folding transition at residue level detail were obtained for SUMO-1 (Kumar et al. 2006) and HIV-1 protease (Bhavesh et al. 2003; Chatterjee et al. 2005) by systematic monitoring of denaturant-dependent changes in the 2D NMR spectra.
Results and Discussion
GED self-assembles into a megadalton mass
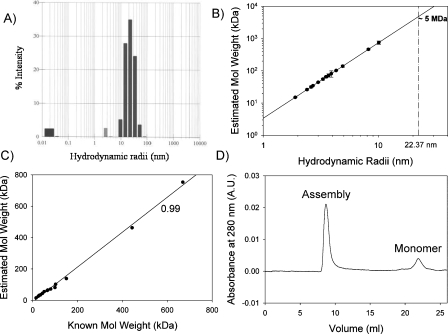
Hydrodynamic radii (R h), obtained from dynamic light-scattering (DLS) measurements, have been successfully used to derive molecular masses in several proteins (Claes et al. 1992). The average R h of GED using DLS was measured to be ∼22.37 nm (Fig. 1A; Chugh et al. 2006). The DLS-estimated molecular mass (M r) seems to have a good relationship with the average R h, and we observed that, for several proteins, the following empirical equation can be fitted (Fig. 1B):
Figure 1.
(A) Histogram of distribution of hydrodynamic radii obtained from “regularization analysis” of data from dynamic light scattering experiments for 100 μM GED in 0.1 M phosphate buffer containing 1 mM EDTA, 150 mM NaCl at pH 5.7; average R h = 22.37 nm. (B) Plot of estimated molecular weight vs. hydrodynamic radii for 18 different proteins from literature (Claes et al. 1992); dashed line extrapolates the R h value of 22.37 nm on the fitted line (solid line) and corresponds to a molecular weight of ∼5 MDa. (C) Plot of estimated molecular weight vs. known molecular weight for the 18 proteins used in plot B; correlation coefficient between the two is found to be 0.99. (D) Size-exclusion chromatogram of GED (100 μM, 100 μL) in 0.1 M phosphate buffer containing 1 mM EDTA, 150 mM NaCl at pH 5.7, run on a Superose 6 10/300 GL column (GE Healthcare) using the BioLogic FPLC system (Bio-Rad), at a flow rate of 0.5 mL/min.
and on this basis the average R h of GED corresponds to an average molecular mass of ∼5 MDa for the GED assembly (vertical dashed line in Fig. 1B). Figure 1C shows the correlation between DLS-estimated and known molecular weights for the 18 proteins used in Figure 1B. The observed correlation coefficient of ∼0.99 indicates that the presently estimated average molecular mass appears to be a reliable estimate. However, the dispersion in the hydrodynamic radii from the DLS data (10–70 nm; Fig. 1A) indicates polydispersity of the aggregates. Accordingly, the molecular weight distribution could range from ∼0.7 to ∼70 MDa.
To further supplement the DLS data, the state of GED oligomer(s) in solution was monitored with size-exclusion chromatography using a Superose 6 10/300 GL column (GE Healthcare), the optimal separation range for which is 5 kDa–5 MDa. The majority of the GED fraction was found to elute in the void volume with a small peak corresponding to the monomer (Fig. 1D); the position corresponding to lysozyme (14.3 kDa) (data not shown). The presence of a major peak in the void volume thus hinted toward the presence of a megadalton-sized species (≥5 MDa) in solution. The monomer peak, when collected, concentrated, and reloaded on the column, produced a major peak in the void volume and a minor peak at the monomer position. This suggests the presence of equilibrium between multimer and monomer, with the equilibrium being mostly in favor of the multimer.
Additionally, the 1H-15N HSQC spectrum of GED (i.e., the fingerprint of all the amino acids in a protein) under native-like conditions displayed only 26 peaks, as against an expected 132 nonproline peaks (Fig. 2A). These peaks were assigned to the N-terminal residues using backbone-directed standard triple-resonance NMR methods (Supplemental Fig. S1). This indicated that these N-terminal residues did not participate in the formation of the core of the assembly and were flexible. Even TROSY-based NMR methods using perdeuterated proteins, which have shown much improvement over the standard triple-resonance methods in case of large-sized proteins (Salzmann et al. 1998; Riek et al. 2000, 2002), failed to produce any more peaks in the present case (data not shown). All this evidence taken together thus suggest that the GED molecule displays the property of self-assembly and exists as a conglomeration of megadalton-sized oligomers in solution.
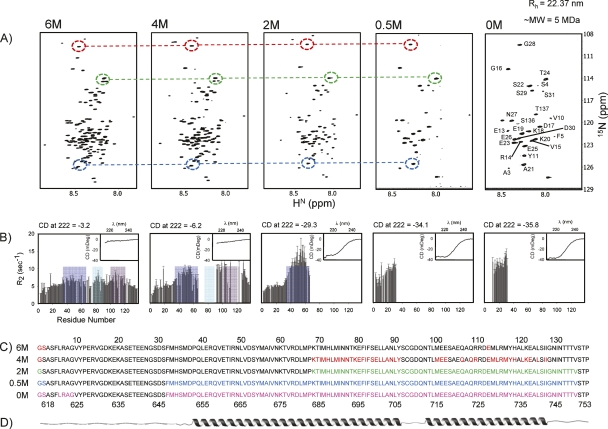
Figure 2.
Summary of the sequence of events that occur during the Gdn-HCl titration of GED as probed by NMR and CD. (A) 1H-15N HSQC spectra of GED at 800 MHz at varying Gdn-HCl concentrations; assignments at 0 M Gdn-HCl are marked; dashed circles and lines illustrate the similarity of the peak positions; there is a progressive disappearance of peaks as we proceed toward the folded state. (B) 15N transverse relaxation rates (R 2) measured at the corresponding Gdn-HCl concentrations as in A; blue, green, and purple patches correspond to the B, C, and D domains (see text) of restricted motion/conformational exchange; there is a progressive increase in the R 2 values followed by lack of data (due to peak disappearance in the HSQC spectra) in these domains; insets show far-UV CD spectra at the corresponding guanidine concentrations; ellipticity at 222 nm is shown at the top of each panel to indicate helicity changes; these are used to calculate molar ellipticity values and thereby the helical contents (see text). (C) Residues for which peaks disappeared in the HSQC spectra at different guanidine concentrations are marked in color on the sequence of the protein; residue numbers at the top are as per recombinant GED and those at the bottom are as per dynamin. (D) Predicted secondary structure along the sequence.
Bottom-up approach to follow the self-assembly process
Secondary-structure predictions of the GED sequence indicated earlier (Chugh et al. 2006) that the sequence had a tendency to assume a largely helical structure (∼50%) in the center, while the N terminus was mostly flexible. Two long helices along the length of the chain were predicted. The CD spectra of the assembly concurred largely with the predictions (Chugh et al. 2006), suggesting that the oligomer is an assembly of helical monomers. However, it is not possible to follow this association in a stepwise manner since it is not possible to obtain GED monomers under native conditions; gel filtration, DLS, and even NMR spectra recorded at 5 μM (SOFAST HMQC with 2048 scans [Schanda et al. 2005]) (data not shown) indicated that the protein formed this large oligomer even at such small concentrations. Therefore, we decided on a bottom-up approach wherein we followed the simultaneous folding and association process of the protein starting from a monomer created by using a denaturant (6 M Gdn-HCl), and the denaturant concentration was progressively decreased. To ensure the reversibility of Gdn-HCl–mediated denaturation, the CD and the HSQC spectra of the native and the refolded protein from the denatured state were compared and were found to be identical (data not shown).
In the 6 M guanidine-denatured state, characterized earlier (Chugh et al. 2007), 131 non-proline peaks were observed. The chemical shift dispersion of amide protons was very low (∼0.6 ppm), which suggested the absence of a persistent structure in this state. However, four regions (A, Glu13–Glu26; B, His34–Met66; C, Asn76–Leu90; and D, Met100–Ala123) of restricted motion/conformational exchange could be located in this state.
Next, in the graded folding exercise, described herein, the denaturant concentration was reduced in a stepwise manner (4, 2, and 0.5 M Gdn-HCl) and HSQC spectra at each step were recorded and individual assignments were obtained using a combination of HNN (Bhavesh et al. 2001; Panchal et al. 2001; Chugh et al. 2008), CBCANH (Ferentz and Wagner 2000), and TOCSY-HSQC (Fesik and Zuiderweg 1990) experiments; all the assigned HSQC spectra are included in the Supplemental material (Supplemental Figs. S2, S3, and S4 for 4, 2, and 0.5 M Gdn-HCl concentrations, respectively). To check for progressive change in the secondary structural characteristics, far-UV CD spectra were also recorded at each step. Simultaneously, the transverse relaxation rate, R 2, for the assigned residues at each Gdn-HCl concentration, was measured to look for changes in the regions of restricted motion/conformational exchange; since R 2 is largely sensitive to motions in milli- to microsecond timescale, it provides a good probe to monitor the changes in the regions of conformational exchange in a protein as it folds. A summary of all these results is depicted in Figure 2. A graded disappearance of peaks toward the middle of the sequence in the HSQC spectra, with a concomitant increase in the helicity (as seen from the far-UV CD spectra) is observed in the protein as the Gdn-HCl concentration is reduced (Fig. 2).
Folding and association go hand-in-hand
Gdn-HCl–induced unfolding (which includes dissociation and subsequent unfolding of the monomer) studied by far-UV CD (secondary structural probe) and fluorescence (tertiary structural probe) was found to be reversible and followed a two-state model with midpoints at 2.77 and 2.52 M, respectively (unpublished results). Thus at each of the 4, 2, and 0.5 M Gdn-HCl concentrations used herein, there are both oligomer and monomer populations. Besides, there will be different degrees of secondary- and tertiary-structure formations.
From 131 peaks in 6 M Gdn-HCl (Fig. 2A), the HSQC spectrum simplified to 90 peaks at 4 M Gdn-HCl (Fig. 2A, Supplemental Fig. S2). The CD value at 222 nm increased to −3.10 × 105 deg cm2 dmol−1 from −1.60 × 105 deg cm2 dmol−1 (Fig. 2B, inset), indicating a rise in helicity (0% at 6 M and 1.4% at 4 M Gdn-HCl) in the polypeptide chain as denaturant concentration is reduced. Sequence-specific resonance assignment revealed that a continuous stretch of residues, Lys68–Tyr91, and a discontinuous stretch between residues, Met100 and Ile127, belonged to the missing signals in the 4 M Gdn-HCl HSQC spectrum (Fig. 2C). In addition to a one-to-one correlation between the 90 peaks present at 4 M Gdn-HCl and the corresponding peaks at 6 M Gdn-HCl (Fig. 2A), the sequence-corrected Hα secondary chemical shifts and cumulative (Cα, Cβ, and CO) secondary chemical shifts (Supplemental Fig. S5A) failed to show any structure formation for these common residues. Moreover, the amide proton temperature coefficients with values more negative than −4.5 ppb/K (Supplemental Fig. S6) (Baxter and Williamson 1997) signified the absence of any hydrogen bonding among the observed 90 residues. Thus, the increased CD value must be attributed to formation of transient helices in the stretches Lys68–Tyr91 and Met100–Ile127. It is important to note here that these stretches belong, respectively, to the “C” (Asn76–Leu90) and “D” (Met100–Ala123) regions displaying significant conformational transitions on the milli- to microsecond timescale observed in the presence of 6 M Gdn-HCl (Chugh et al. 2007). The disappearance of these peaks in 4 M Gdn-HCl could therefore be ascribed to slowing down of conformational transitions to intermediate exchange (millisecond timescale), which, in turn, must be due to initiation of the folding and transient association at these sites. Further, an increase in R 2 values was observed for the “B” region (His34–Met66) and for the remaining residues of the “C” and “D” regions. This indicated enhancement in conformational transitions (milli- to microsecond timescale) and suggested an increased tendency toward structure formation in those areas on reduction of Gdn-HCl concentration.
As the Gdn-HCl concentration was further reduced to 2 M, the number of peaks observed in the HSQC spectra decreased to 65 (Fig. 2A, Supplemental Fig. S3) with the CD value at 222 nm increasing to −1.46 × 106 deg cm2 dmol−1 (Fig. 2B, inset), indicating a significant increase in the helicity (11.5%) along the chain. Strikingly, the stretches for which the peaks disappeared extended further and covered the C-terminal half of the protein, Lys68–Val135 (Fig. 2C). This indicated that the entire C-terminal half upon folding participated in the self-association process. Also, the “B” region (His34–Met66) showed further enhancement in the conformational exchange, as shown by a considerable increase in the R 2 values at this Gdn-HCl concentration. However, the sequence-corrected Hα secondary chemical shifts, cumulative (Cα, Cβ, and CO) secondary chemical shifts (Supplemental Fig. S5B), and amide proton temperature coefficients (Supplemental Fig. S6) still suggested a lack of structure for these residues; also, these peaks could be directly correlated to the corresponding peaks at 6 M Gdn-HCl (Fig. 2A). Thus, the disappearance of peaks in the HSQC spectrum with the increase in helicity must again be credited to helix formation and transient association.
A further decrease in Gdn-HCl concentration to 0.5 M led to the disappearance of the “B” region peaks (showing high R 2 values at 2 M Gdn-HCl) from the HSQC spectrum, thereby reducing the number of observed peaks to a mere 30 (Fig. 2A, Supplemental Fig. S4). Interestingly, these peaks for Ala3–Ser31 and Ser136–Thr137 constituted the N- and C-terminal stretches (Fig. 2C), respectively, and correlated well with the corresponding peaks at 6 M Gdn-HCl. The amide proton temperature coefficients (Supplemental Fig. S6) also suggested the absence of any structure in this region. The CD value at 222 nm increased to −1.71 × 106 deg cm2 dmol−1 (Fig. 2B, inset), suggesting structure formation in the “B” region as well (global helicity = 35.8%). It can also be seen that the spectra in 0.5 and 0 M guanidine are nearly identical (Fig. 2A), indicating that the folding process is nearly complete at 0.5 M itself, and all of these folded units participate in transient association. The R 2 values in the latter case were, however, higher (Fig. 2B), suggesting a greater degree of millisecond to microsecond timescale motions in the N terminus of the native assembly. The CD value at 222 nm further increased to −1.79 × 106 deg cm2 dmol−1 and global helicity to 39.4% (Fig. 2B, inset) in the native state. As discussed earlier, an equilibrium between associated and monomeric states also occurs in the native state, with the equilibrium being mostly in favor of the associated state.
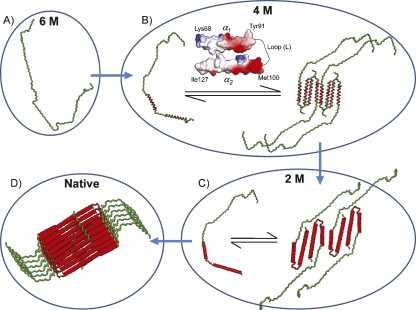
The above results provide valuable insights into the stepwise progression of the folding process of the GED, assembly domain of dynamin. The observed loss of signals maps to the predicted regions of helical structure (Fig. 2D). While on one hand this provides experimental support to the theoretical predictions, it also suggests that helix formation might create a surface that is conducive for association. Figure 3A depicts one of the several possible topologies at 6 M Gdn-HCl, wherein the polypeptide chain is completely denatured and is a monomer. At 4 M Gdn-HCl, the transient helices occur in the stretches Lys68–Tyr91 (part of α1) and Met100–Ile127 (part of α2). Importantly, the electrostatic potential surface of these two helices showed oppositely charged surfaces and also a fair amount of neutral surface (Fig. 3B). Therefore it is conceivable that both these factors promote interaction of these helices. The fact that the intervening loop remains flexible suggests that the two helices might interact via intramolecular folding; the loop could act as a hinge to bring the transient helices in close proximity. However, the possibility of an intermolecular association of these helices can also not be excluded. The surface of this folded structure has again a fair degree of hydrophobicity. On further reducing the denaturant concentration to 2 M, the transient helices stabilize to form more permanent helices, α2 further extends toward the C terminus, and the loop (L) also becomes rigid (Fig. 3C). On going further down to 0.5 M Gdn-HCl, α1 also extends toward the N terminus, thereby engaging all the helices into self-association. The ∼30 N-terminal residues for which the peaks are visible under native conditions remain largely unfolded.
Figure 3.
Proposed folding and association pathway of GED as probed by varying Gdn-HCl concentrations. (A) One of the several possible topologies at 6 M Gdn-HCl, the polypeptide chain is completely denatured and is a monomer. (B) GED monomer and a transiently formed assembly at 4 M Gdn-HCl are in equilibrium; transient helices are formed in the stretches Lys68–Tyr91 and Met100–Ile127 (marked in red along the polypeptide chain); electrostatic surface potential of part of α1, and part of α2, is shown to emphasize the type of interactions that lead to closeness between the two transiently formed helices; (C) at 2 M Gdn-HCl, the α2 helix extends toward the C terminus and covers the stretch Met100–Val135; helices α1 and α2 are more stable, and the loop (L) region also becomes rigid; parts of α1, L, and α2 are shown in red along the chain as the corresponding peaks are absent in the HSQC spectra. (D) Conceptual helical bundle of GED: α1 extends toward the N terminus and covers Phe32–Tyr91; ∼30 residues at the N terminus are free and flexible and not involved in assembly formation; the geometric arrangement is purely arbitrary and hypothesis-driven.
Considering all the above observations, different models can be built for the GED self-assembly. One such possibility involving intramolecular as well as intermolecular association is schematically shown in Figure 3D. The geometrical arrangements presented in the figure are only suggestive and are by no means experimentally established. Details may differ within the overall picture presented in the figure.
Conclusions
Using multidimensional NMR, residue-level insights have been obtained for the self-association process of GED of dynamin. The structural and motional characteristics at different guanidine concentrations provided stepwise information regarding folding and concomitant self-association. The results presented herein have profound implications. Firstly, these demonstrate the possibility of probing large molecular assemblies using NMR by adopting innovative strategies. Secondly, the so-called “sequence–structure paradigm” can possibly be extended to include the “sequence–self-association paradigm.”
Materials and Methods
Protein expression and purification
GED was expressed and purified as described elsewhere (Chugh et al. 2007). An isotopically enriched (15N or 13C and 15N, or 13C, 15N, and 2H, as required) GED sample (∼1 mM) was equilibrated with different concentrations of denaturant (6 M, 4 M, 2 M and 0.5 M Gdn-HCl, in 10 mM acetate buffer at pH 5.0 containing 1 mM EDTA, 150 mM NaCl, and 1 mM DTT). Equilibrium was tested with series of HSQC spectra recorded at different time points.
Circular dichroism
All the far-UV CD spectra were recorded as described elsewhere (Chugh et al. 2007) on a Jasco J-810 spectropolarimeter. Protein (20 μM) was equilibrated with different Gdn-HCl concentrations for at least 12 h before the experiments.
Size-exclusion chromatography
Size-exclusion chromatography was performed using Superose 6 10/300 GL column (GE Healthcare) with 0.1 M phosphate buffer containing 1 mM EDTA, 150 mM NaCl at pH 5.7, at a flow rate of 0.5 mL/min with absorbance monitored at 280 nm using a BioLogic FPLC system (Bio-Rad). Recombinant GED (100 μM, 100 μL) was centrifuged at 15,400g at 4°C for 10 min, and the supernatant was applied to the column.
NMR spectroscopy
All NMR experiments were performed on a Bruker 800-MHz Avance spectrometer, equipped with a triple-resonance CryoProbe. The experiments were performed at 15°C, processed using Felix (Accerlys Software Inc.), and analyzed using Felix and CARA (Keller 2004). Backbone assignment was obtained using standard strategy based on heteronuclear triple-resonance experiments: HNN, CBCANH, CBCA(CO)NH, HNCO, and HN(CA)CO. TOCSY-HSQC and NOESY-HSQC were used to confirm the spin systems. Relaxation measurements were carried out as described elsewhere (Chugh et al. 2007). Amide proton temperature coefficients were measured by recording HSQC spectra from 15°C to 39°C at steps of 3°C. SOFAST HMQC was performed on 5 μM 15N-labeled GED sample with a delay of 100 ms and 2048 scans.
Electronic supplemental material
The Supplemental material contains the annotated HSQC for native, 4 M, 2 M, and 0.5 M Gdn-HCl–denatured GED; secondary chemical shifts; and amide proton temperature coefficient data.
Acknowledgments
We thank Dr. Rohit Mittal for the GED clone and his critical comments and discussion. The authors acknowledge Abijar Bhori and Professor Arvind Lali at UDCT, Mumbai, for help with the gel-filtration experiments. We thank the Government of India for providing financial support to the National Facility for High-Field NMR at TIFR. J.C. is the recipient of a TIFR Alumni Association fellowship for career development through the years 2002–2005.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Ramakrishna V. Hosur, Department of Chemical Sciences, Tata Institute of Fundamental Research, 1 Homi Bhabha Road, Colaba, Mumbai 400005, India; e-mail: hosur@tifr.res.in; fax: +91-22-2280-4610.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035840.108.
References
- Baxter, N.J., Williamson, M.P. Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR. 1997;9:359–369. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- Bhavesh, N.S., Panchal, S.C., Hosur, R.V. An efficient high-throughput resonance assignment procedure for structural genomics and protein folding research by NMR. Biochemistry. 2001;40:14727–14735. doi: 10.1021/bi015683p. [DOI] [PubMed] [Google Scholar]
- Bhavesh, N.S., Sinha, R., Mohan, P.M., Hosur, R.V. NMR elucidation of early folding hierarchy in HIV-1 protease. J. Biol. Chem. 2003;278:19980–19985. doi: 10.1074/jbc.M301615200. [DOI] [PubMed] [Google Scholar]
- Braig, K., Otwinowski, Z., Hegde, R., Boisvert, D.C., Joachimiak, A., Horwich, A.L., Sigler, P.B. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- Chatterjee, A., Mridula, P., Mishra, R.K., Mittal, R., Hosur, R.V. Folding regulates autoprocessing of HIV-1 protease precursor. J. Biol. Chem. 2005;280:11369–11378. doi: 10.1074/jbc.M412603200. [DOI] [PubMed] [Google Scholar]
- Chugh, J., Chatterjee, A., Kumar, A., Mishra, R.K., Mittal, R., Hosur, R.V. Structural characterization of the large soluble oligomers of the GTPase effector domain of dynamin. FEBS J. 2006;273:388–397. doi: 10.1111/j.1742-4658.2005.05072.x. [DOI] [PubMed] [Google Scholar]
- Chugh, J., Sharma, S., Hosur, R.V. Pockets of short-range transient order and restricted topological heterogeneity in the guanidine-denatured state ensemble of GED of dynamin. Biochemistry. 2007;46:11819–11832. doi: 10.1021/bi701280p. [DOI] [PubMed] [Google Scholar]
- Chugh, J., Kumar, D., Hosur, R.V. Tuning the HNN experiment: Generation of serine–threonine check points. J. Biomol. NMR. 2008;40:145–152. doi: 10.1007/s10858-007-9217-z. [DOI] [PubMed] [Google Scholar]
- Claes, P., Dunford, M., Kenney, A., Vardy, P. An on-line dynamic light scattering instrument for macromolecular instrumentation. In: Harding S.E., Sattelle D.B., Bloomfield V.A., editors. Laser light scattering in biochemistry. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 66–76. [Google Scholar]
- Ferentz, A.E., Wagner, G. NMR spectroscopy: A multifaceted approach to macromolecular structure. Q. Rev. Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- Fesik, S.W., Zuiderweg, E.R. Heteronuclear three-dimensional NMR spectroscopy of isotopically labelled biological macromolecules. Q. Rev. Biophys. 1990;23:97–131. doi: 10.1017/s0033583500005515. [DOI] [PubMed] [Google Scholar]
- Fiaux, J., Bertelsen, E.B., Horwich, A.L., Wuthrich, K. NMR analysis of a 900K GroEL–GroES complex. Nature. 2002;418:207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- Foster, M.P., McElroy, C.A., Amero, C.D. Solution NMR of large molecules and assemblies. Biochemistry. 2007;46:331–340. doi: 10.1021/bi0621314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll, M., Ditzel, L., Lowe, J., Stock, D., Bochtler, M., Bartunik, H.D., Huber, R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Hinshaw, J.E. Dynamin and its role in membrane fission. Annu. Rev. Cell Dev. Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, J.F., Weaver, A.J., Landry, S.J., Gierasch, L., Deisenhofer, J. The crystal structure of the GroES co-chaperonin at 2.8 Å resolution. Nature. 1996;379:37–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- Keller, R. The computer-aided resonance assignment tutorial. Cantina Verlag; Goldau, Switzerland: 2004. [Google Scholar]
- Kumar, A., Srivastava, S., Kumar, M.R., Mittal, R., Hosur, R.V. Residue-level NMR view of the urea-driven equilibrium folding transition of SUMO-1 (1-97): Native preferences do not increase monotonously. J. Mol. Biol. 2006;361:180–194. doi: 10.1016/j.jmb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Lowe, J., Stock, D., Jap, B., Zwickl, P., Baumeister, W., Huber, R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Panchal, S.C., Bhavesh, N.S., Hosur, R.V. Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: Application to unfolded proteins. J. Biomol. NMR. 2001;20:135–147. doi: 10.1023/a:1011239023422. [DOI] [PubMed] [Google Scholar]
- Praefcke, G.J., McMahon, H.T. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Riek, R., Pervushin, K., Wuthrich, K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem. Sci. 2000;25:462–468. doi: 10.1016/s0968-0004(00)01665-0. [DOI] [PubMed] [Google Scholar]
- Riek, R., Fiaux, J., Bertelsen, E.B., Horwich, A.L., Wuthrich, K. Solution NMR techniques for large molecular and supramolecular structures. J. Am. Chem. Soc. 2002;124:12144–12153. doi: 10.1021/ja026763z. [DOI] [PubMed] [Google Scholar]
- Salzmann, M., Pervushin, K., Wider, G., Senn, H., Wuthrich, K. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanda, P., Kupce, E., Brutscher, B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- Schulman, B.A., Kim, P.S., Dobson, C.M., Redfield, C. A residue-specific NMR view of the non-cooperative unfolding of a molten globule. Nat. Struct. Biol. 1997;4:630–634. doi: 10.1038/nsb0897-630. [DOI] [PubMed] [Google Scholar]
- Sprangers, R., Kay, L.E. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- Unno, M., Mizushima, T., Morimoto, Y., Tomisugi, Y., Tanaka, K., Yasuoka, N., Tsukihara, T. The structure of the mammalian 20S proteasome at 2.75 Å resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- Wijesinha-Bettoni, R., Dobson, C.M., Redfield, C. Comparison of the denaturant-induced unfolding of the bovine and human α-lactalbumin molten globules. J. Mol. Biol. 2001;312:261–273. doi: 10.1006/jmbi.2001.4927. [DOI] [PubMed] [Google Scholar]
- Xu, Z., Horwich, A.L., Sigler, P.B. The crystal structure of the asymmetric GroEL–GroES–(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]