Abstract
Dioxygenases catalyze dioxygen incorporation into various organic compounds and play a key role in the complex degradation pathway of mono- and polycyclic aromatic and hetero-aromatic compounds. Here we report the crystal structure of gentisate 1,2-dioxygenase from Silicibacter pomeroyi (GDOsp) at a 2.8 Å resolution. The enzyme possessed a conserved three-dimensional structure of the bicupin family, forming a homotetramerization. However, each subunit of GDOsp unusually contained two ferrous centers that were located in its two homologous cupin domains, respectively. Further mutagenic analysis indicated that the enzyme activity of GDOsp depends on the microenvironment in both metal-binding sites. Moreover, homologous structural comparison and functional study on GDOsp variants unveiled a group of functionally essential residues and suggested that the active site of the enzyme is located in the amino-terminal domain, but could be influenced by changes in the carboxyl domain. Therefore, GDOsp may provide a working model for studying long-distance communication within a protein (or its complex).
Keywords: gentisate 1,2-dioxygenase; extradiol dioxygenase; mononuclear metal center; bicupin folding; long-distance communication
Many bacteria degrade aromatic compounds through dioxygenases centered pathways. Dioxygenases catalyze dioxygen incorporation into various organic compounds and play a key role in the complex degradation pathway of mono- and polycyclic aromatic and hetero-aromatic compounds, including hydrocarbons recalcitrant to the environment and detrimental to human health. While aromatic hydrocarbons are diverse in the number of rings and modifications, the ring-fission dioxygenases are usually classified into two families, termed intradiol and extradiol. This classification is based on the cleavage site in ortho-dihydroxylated aromatic compounds, a group of common degradation intermediates of aromatic hydrocarbon. In particular, extradiol dioxygenases cleave at a point adjacent to one of the hydroxyl groups and yield α-hydroxymuconic semialdehyde adducts (Dagley et al. 1960; Fujisawa et al. 1972; Arciero et al. 1983; Whittaker et al. 1984; Hirano et al. 2007).
Bacterial ring-cleavage dioxygenases that have so far been isolated and characterized contain a variety of metal cations such as magnesium (Gibello et al. 1994), manganese (Boldt et al. 1995; Whiting et al. 1996), copper (Fusetti et al. 2002), and ferric and ferrous ions (Que et al. 1976; Adams et al. 2006; Liu et al. 2007) as the catalytic center. Primary sequence analysis of these dioxygenases shows that amino acid residues responsible for active-site metal binding are strongly conserved, which is further supported by structural studies on a subset of these enzymes. One of the common peptide folds of dioxygenases is called a bicupin structure, which is characterized by two homologous β-barrel (cupin) domains in each subunit (Jin et al. 2006). A functional cupin domain is characterized by two highly conservative sequence motifs of G(X)5HXH(X)3,4E(X)6G and G(X)5PXG(X)2H(X)3N (Dunwell et al. 2001). The two motifs together form one functional metal-binding site. In some cupin domains such motifs are lost, resulting in a conserved peptide fold, but lack of the metal-binding ability.
Gentisic acid (2,5-dihydroxybenzoate) serves as a focal point in the biodegradative pathways responsible for the metabolism of many abundant aromatic compounds (Chapman 1972; Bayly and Barbour 1984). Characteristics of the reaction catalyzed by a gentisate 1,2-dioxygenase (GDO, EC 1.13.11.4) appear to be similar to those of extradiol dioxygenases, although it is not an extradiol dioxygenase per se since the substrate is not a catechol. A GDO catalyzes the ring fission of gentisate between the carboxyl and proximal hydroxyl groups at positions 1 and 2 of the aromatic ring to form maleylpyruvate (Adams et al. 2006). Many known three-dimensional (3D) structures of GDO proteins belong to the bicupin family and contain one metal-binding site that is located in the amino (N)-terminal cupin domain (Adams et al. 2006), in which the conserved metal-binding motifs transform into two HXH-containing sequences (Supplemental Fig. S1). Despite extensive structural investigation of various members of this bicupin dioxygenase subfamily, the structural bases of their substrate specificity including the specificity of ring cleavage position remain elusive.
Here we present the crystal structure of a newly identified gentisate, 1,2-dioxygenase, from Silicibacter pomeroyi DSS-3 (GDOsp) (Liu et al. 2007). Structural analysis, extensive mutagenesis, and biochemistry characterizations demonstrated that each subunit of this homotetrameric enzyme has two metal-binding sites, one in each cupin domain. This observation is in sharp contrast to most of its bicupin homologs, which are believed to have only one metal-binding site per subunit. Importantly, both metal-binding sites are essential for the GDOsp activity. Based on available structural and biochemical data, a putative substrate-binding mode is proposed.
Results
GDOsp has a typical bicupin folding with a long N-terminal arm
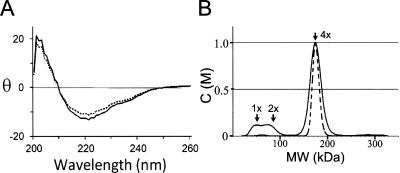
The recombinant protein of GDOsp (also called gtdA-2), a member of the gentisate 1,2-dioxygenase family, was expressed and purified from Escherichia coli following a previous protocol (Liu et al. 2007). The native protein consists of 374 amino acid residues. To characterize the recombinant full-length wild-type (WT) enzyme, we analyzed its secondary structure components using circular dichroism (CD) at 220 nm (Fig. 1). Thermal denaturation assay showed that the protein unfolded and lost its secondary structures above 70°C (data not shown), and this thermal denaturation process was irreversible because the protein visibly precipitated around 75°C. To demonstrate the enzymatic activity of the recombinant protein, we repeated the activity assay in the report by Liu et al. (2007), and the KM and kcat /KM values at pH 7.5 were determined to be 12 ± 3 μM and 0.24 ± 0.07 s−1 μM−1 (subunit), respectively.
Figure 1.
Properties of recombinant WT full-length GDOsp. (A) A representative wavelength scan of CD spectrum from 200 to 260 nm measured at 20°C. The dashed line is from the Fe-depleted sample. The unit of the vertical axis is degree mM−1cm−1. (B) Sedimentation coefficient distribution of GDOsp sample deduced from AUC sedimentation velocity analysis. The dashed line is from the Fe-depleted sample. The Y-axis is in an arbitrary unit. The positions of the monomer, dimer, and tetramer are marked as 1x, 2x, and 4x, respectively.
The recombinant protein of full-length GDOsp with an N-terminal tag was crystallized, and the crystal structure was determined at 2.8 Å resolution. The crystal form belongs to the R32 space group, with two protein molecules (named as A and B) per asymmetric unit and ∼45% solvent content. The model of GDOsp was refined to a final Rworking of 21% (Rfree of 25%) with good geometry statistics (Table 1). Overall, the two protein molecules were nearly identical; the root-mean-square deviation (RMSD) for all Cα atoms in residues 19−373 between molecules A and B was 0.7 Å. Four metal ions, two per peptide chain, were clearly visible at the predicted metal-binding sites of the conserved cupin motifs. Consistent with previous biochemistry data (Liu et al. 2007), homologous structures (Adams et al. 2006), and our own analysis (see below), all four metal ions were refined as ferrous with full occupancy and reasonable B-factors (Table 1). The two Fe2+ ions were 27.7 Å apart in the two domains of molecule A, and were 27.4 Å apart in molecule B. An intramolecular disulfide bond was observed between Cys100 and Cys323 in each GDOsp molecule that was shown to be important for the enzyme activity (see below).
Table 1.
Data collection and refinement statistics
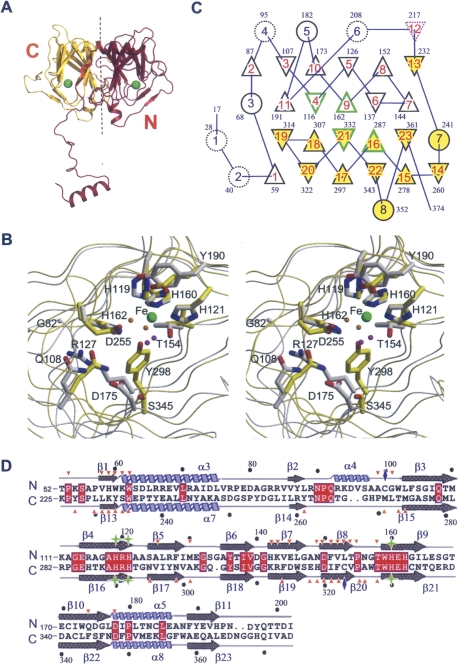
The GDOsp molecule contained an N-terminal arm ∼50 residues in length and a bicupin core formed by two structurally similar cupin domains positioned back-to-back around an internal dyad symmetry axis (Fig. 2A). No electron density was apparent for residues 1–18 of molecule A and residues 1−16 of molecule B in addition to the invisible N-terminal tag, indicating a flexible N terminus in either case. Each cupin domain mainly contained a seashell-shaped, β-barrel, with one metal-binding site located inside the barrel and close to the opening. The β-barrel is formed by two antiparallel β-sheets, smaller and larger (Fig. 2C,D; Supplemental Fig. S1).
Figure 2.
Structural comparison of GDOsp N and C domains. (A) Ribbon diagram of the GDOsp subunit. The N-terminal arm and N-terminal cupin domain are colored burgundy, and the C domain is colored gold. The pseudo-dyad axis is represented by the black dashed line. The two Fe2+-binding sites are marked as green spheres. (B) Stereoview of the superposition of the N- and C-domain metal-binding sites. The view looks from the entrance into the cupin cavity. The superposition was based on Cα atoms of three Fe2+-ligand His residues from the two domains. The backbone trace and corresponding carbon atoms of the N domain are colored gray, and those from the C domain are colored yellow. Oxygen, nitrogen, and iron are colored red, blue, and green, respectively. Water molecules associated with the N domain are colored magenta, and those from the C domain are colored orange. The two Fe2+ ions are superimposed perfectly. Selected side chains are labeled. (C) Topology diagram of the secondary structure of the GDOsp monomer. α-Helices are presented as circles, and β-strands as triangles. Each type of subject is labeled by numbers inside the symbol. Their location in the primary sequence are indicated by the number of residues in that subject. Subjects of the N and C domains are filled with white and yellow colors, respectively. Subjects following internal dyad symmetry have solid line borders; otherwise, dashed lines. Subjects containing Fe2+-binding triads have green borders; β-strands that are involved in oligomerization have magenta borders; otherwise, borders are black. Note that N and C domains swap their β1 and β13 strands in the 3D structure. (D) Structure-based sequence alignment of GDOsp N and C domains. Conserved residues are highlighted in red. Major secondary structural elements from the two domains are marked on the top and the bottom of alignment, respectively. Selected residue numbers are labeled. Residues involved in domain–domain interface (<3.5 Å) are marked with orange triangles, the interdomain disulfide bonded cysteine residues are marked with blue diamonds, and residues involved in metal binding are marked with green stars. This figure was prepared with the program Espript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi/).
Although the homology is low (identity <20%) between the primary sequence of the GDOsp N domain (residues 52–222) and the carboxy (C) domain (223–374) (Fig. 2D), the two domains clearly share similar folding. The RMSD between the N- and C-terminal halves was 1.45 Å for 128 Cα atom pairs (using a 3.0 Å cutoff). Such a structural superposition based on peptide Cα atoms of the two domains also resulted in an excellent overlay of their corresponding metal-binding sites, including the iron ion and three coordinating His residues (Fig. 2B).
GDOsp forms homotetramers both in solution and in the crystal
Since functional GDO proteins are often observed in homotetramers (Harpel and Lipscomb 1990a; Suarez et al. 1996; Werwath et al. 1998; Adams et al. 2006), the oligomeric form of GDOsp was studied in solution. Using analytical ultracentrifugation (AUC) under submicromolar concentrations of GDOsp subunits, we demonstrated that GDOsp forms stable homotetramers in solution. The apparent molecular weight was 169 kDa for the N-terminal His-tagged full-length WT GDOsp (Fig. 1B). Consistent with this solution observation, GDOsp was also found to form a symmetric tetramer in the crystal, which is similar to previously described GDO proteins (Adams et al. 2006).
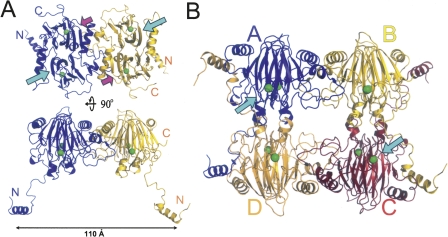
First, the two GDOsp molecules in an asymmetric unit, A and B, formed a homodimer with a non-crystallographic dyad symmetry (Fig. 3A). The dyad axis of this A–B dimer was parallel to the pseudo-dyad axis between the N and C domains in each subunit. Upon dimer formation, the Fe2+-binding site in the N-terminal domain is positioned away from the dimer interface and is solvent exposed, while the C-terminal domain was positioned in proximity to the interface. The dimer interface was extensive. Buried SAS (solvent accessible surface) in the A–B dimer interface was 1500 Å2 from each subunit.
Figure 3.
Ribbon diagram of the GDOsp oligomer crystal structure. (A) GDOsp dimer in an asymmetric unit in two views differing roughly by 90°. The two subunits are colored blue and gold, respectively. Their N and C termini are labeled. Ferrous ions are depicted as green spheres. The N- and C-domain cupin entrances of both subunits are marked in the top view with cyan and magenta arrows, respectively. (B) GDOsp tetramer formed by two neighboring asymmetric units. The four subunits are labeled as A−D. The blue-yellow dimer is in the same orientation as in the front view (bottom) in A. The N-domain cupin entrances of subunits A and C are located in the front and are marked with cyan arrows. This figure was prepared with the program PyMOL (DeLano Scientific).
The GDOsp dimer, A–B, further formed a dimer of dimers with a neighboring symmetry mate. This tetramer had approximate dimensions of 110 × 85 × 65 Å (Fig. 3B). Overall, 5200 Å2 (28%) of the total 18,300 Å2 SAS of each GDOsp subunit was buried upon the formation of the tetramer. This tetramer is the only one having a local 2-2-2 symmetry and assumes the most extensive protein–protein interaction in the crystal packing, supporting the notion that the same tetramer also exists in solution. In particular, the extended N-terminal arm (i.e., residues 1−52) wrapped across the dimer–dimer interface. This arm formed a helix bundle between its α1 and the α3 helix from another subunit. Each N-terminal arm contributed 1400 Å2 buried SAS from itself and another 1250 Å2 buried SAS from its counterpart to the tetramerization.
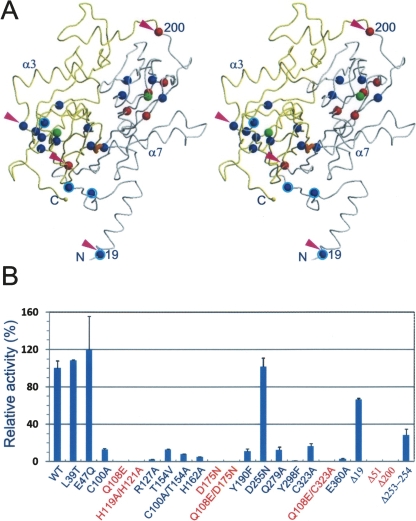
To test the importance of the N-terminal arm in both structure and function, three truncation mutants were constructed by the deletion of N-terminal 19, 51, and 58 residues, respectively (Fig. 4A; Supplemental Fig. S1). The Δ58 variant was expressed in E. coli as inclusion bodies, suggesting possible defects in the folding of this GDOsp variant and/or decreased solubility; no further study was pursued. The Δ51 variant was expressed and purified from E. coli as a soluble protein. It maintained tetramerization in an AUC analysis (data not shown), but showed low enzyme activity toward the gentisate substrate (Fig. 4B). The Δ19 variant was also expressed and purified from E. coli as a soluble protein; in contrast to Δ51, Δ19 maintained about two-thirds of the WT activity (Fig. 4B). Taken together, these mutagenesis data suggest that the mobile N-terminal 20-residue peptide has a positive yet limited role in the enzyme activity. Meanwhile, the structurally observable N-terminal peptide (residues 17−51) plays a significant role in the enzyme activity. However, the N-terminal peptide did not appear essential for the tetramerization, although a significant portion of the buried SAS during tetramerization was contributed by this peptide.
Figure 4.
Mutagenesis analysis. (A) Stereoview of the distribution of mutations in the 3D structure of the GDOsp subunit. The backbone traces of N and C domains are colored gray and yellow, respectively. Positions of the two Fe2+ ions are marked with green spheres. The interdomain disulfide bond, Cys100−Cys323, is colored in orange. The positions of three N-terminal truncation variants (Δ19, Δ51, and Δ200, i.e., C domain) and a C-domain deletion (Δ253–254) are marked with magenta triangles, mutants that did not produce 330 nm signal but yellow products are marked with red spheres, and positions of other point mutations are marked with blue spheres. In addition, mutants of WT-like activity are highlighted with a cyan ring. (B) The gentisate dioxygenase activity of GDOsp variants relative to that of the WT. The error bars represent standard deviation calculated from multiple measurements. Mutants that did not produce 330 nm signal, but yellow products, are labeled red.
Ferrous binding is important for GDOsp activity but not for oligomerization
Using an inductively coupled plasma-mass spectroscopy (ICP-MS) based metal-element analysis, we showed that the molar ratio of protein subunit to Fe in the recombinant protein sample directly purified from E. coli was 1.00:1.44 ([GDOsp subunit]:[Fe]). Furthermore, Fe2+ dependency of the enzyme activity was studied in solution. Consistent with the previous report (Liu et al. 2007), the initial velocity of the recombinant holo-GDOsp increased two- to 10-fold with additional Fe2+ in the concentration range of 0.1−1.0 mM (final). Contrarily, additional Fe3+ as low as 10 μM precipitated the GDOsp protein from solution. To determine the role of metal binding in oligomerization, we compared the biophysical properties of GDOsp both in the presence and in the absence of metal ions. Apo-GDOsp was prepared first by incubation of the holo-GDOsp with 100 mM EDTA (ethylene-diamine-tetra-acetic acid) for 2 d. It was followed by 1:2000 dialysis against a buffer of 20 mM Tris-HCl (pH 7.5) and 120 mM NaCl for overnight. The enzyme activity dropped to below 8% of the holo-enzyme but could be rescued partially by additional Fe2+ ion, confirming that most of the Fe2+ ion had been chelated by ETDA. When this apo-GDOsp sample was analyzed by gel-filtration chromatography, most of the protein eluted at the same position as the ferrous-bound enzyme corresponding to a tetramer (data not shown). AUC analysis further confirmed the tetramerization state of the apo-GDOsp (Fig. 1B). In addition, CD spectra analysis suggested that the apo-protein had secondary structures similar to those of the holo-enzyme (Fig. 1A), albeit it denatured at a lower temperature than the holo-enzyme (data not shown). In light of previous data (Adams et al. 2006; Liu et al. 2007) about bound Mn, we analyzed the effects of Mn2+ on the enzyme activity. Addition of Mn2+ (0.01−1.0 mM) to the apo-GDOsp protein failed to rescue the lost enzyme activity. Moreover, the enzyme activity of the holo-GDOsp was inhibited by Mn2+ at a concentration of 1 mM. Therefore, we concluded that Fe2+ ions are required for the GDOsp enzyme activity, but not essential for the protein folding.
The crystal structure of GDOsp showed that the enzyme contains two mononuclear metal-binding centers per subunit, in which the metal ions were refined as iron. Each ferrous ion was mainly coordinated by three histidine residues. Specifically, the N-domain metal was coordinated by the Nε2 atoms of His119, His121, and His160, and the C-domain ferrous ion was coordinated by the Nε2 atoms of His290, His292, and His330 (Fig. 2B; Supplemental Table S1). In addition, the orientation of most histidine ligands was stabilized by a hydrogen bond between the Nδ1 atom of the imidazole ring and a neighboring residue in the 3D structure. For example, His119 was fixed by a 2.8 Å hydrogen bond with the Tyr190 side-chain hydroxyl group, and His290, 2.9 Å with the Glu360 side-chain carboxyl group. Moreover, point mutations, Y190F and E360Q, reduced the enzyme activity, supporting the role of these residues in stabilizing the metal-binding sites in both domains. Taken together, we observed in the crystal structure that both metal-binding sites have well-defined, nearly identical geometry for the Fe2+ coordination.
To identify the location(s) of the GDO active site(s), we constructed point mutations at both the N- and the C-domain metal-binding sites individually. The N-domain variant was the double-mutation H119A/H121A, where two of the three Fe2+-binding His residues were substituted by alanine; similarly, the C-domain variant was H290A/H292A. While the H290A/H292A mutant could not be expressed in a soluble form, the H119A/H121A mutant was expressed and purified in a process similar to that used for WT. This N-terminal Fe2+-binding-deficient mutant showed a WT-like CD wavelength-scan spectrum (Supplemental Fig. S2). In addition, this mutant was also crystallized under the same condition of the WT, indicating an overall structure similar to the WT (data not shown). Despite its WT-like structural properties, the H119A/H121A variant showed extremely low GDO activity. Even at 1 mg/mL protein concentration and 50 μM−1.0 mM gentisate concentration, the activity was barely detectable. These results suggest that the N-terminal Fe2+-binding site is essential for the enzyme activity but dispensable for the protein overall tertiary structure.
The enzyme activity of GDOsp depends on both cupin domains
To further define the roles of each cupin domain in GDOsp function, we carried out three more types of mutagenic analysis.
First, we constructed truncation variants of GDOsp N domain (residues 1−200, 1−236, and 51−236) and C domain (i.e., residues 201−374). Whereas all three N-domain constructs failed to express in E. coli as a soluble protein, the C-domain construct was expressed and purified in a process similar to that used for the full-length GDOsp. CD spectroscopy analysis on this C-domain recombinant protein showed a significant amount of secondary structures (Supplemental Fig. S2) while its enzyme activity was extremely low (Fig. 4B). This observation confirmed that the cupin domain is an independent folding unit.
Second, we constructed a series of point mutations inside each of the two cupin domain cavities and around the Fe2+-binding site. In the N-terminal domain, we had L39T, E47Q, Q108E, R127E, T154V, H162A, D175N, and Y190F variants, and a double-mutation variant Q108E/D175N; in the C-terminal domain, we had D255N, Q279A, Y298F, and E360Q variants as well as a double-deletion variant Δ253–254 (i.e., deletion of Pro253 and Tyr254) (Fig. 4A; Supplemental Fig. S1). These mutations were chosen to test the roles in enzymatic activity played by oligomerization (e.g., Leu39 and Glu47 from a neighboring subunit) hydrogen-bond networks inside the cavity (e.g., Gln108, Arg127, Thr154, His162, Asp175, and Asp255), and stability of the Fe2+ ligands (e.g., Tyr190). Most of the substitutions were introduced to minimize geometry disruption. All of these mutants were expressed in E. coli as soluble proteins, and their folding was confirmed by CD analysis (Supplemental Fig. S2). Using a standard activity assay with or without supplementary Fe2+ (50 μM), the mutagenic effects on catalysis were analyzed (Fig. 4B); in both cases, the overall patterns of mutational effects were the same. Almost all of these variants with point mutations around the Fe2+-binding sites showed decreased activity, ranging from virtually zero to <20% of the WT activity level. L39T, E47Q, and D225N were the exceptions, which had comparable activity to WT. In addition, the two-residue deletion (Δ253–254), which intended to increase the size of the C-domain cavity by shortening the loop around Asp255 (see Fig. 2B) such that a substrate molecule might be easier to access the C-domain metal center, actually reduced the activity to 30%. Among them, Gln108 and Gln279, Arg127 and Tyr298, and Tyr190 and Glu360 are three pairs of equivalent residues in the homologous N and C domains (Fig. 2B,D), and each pair of equivalent point mutations appeared to have similar effects on the enzyme activity. Inside the N-terminal domain cavity, Gln108 and Asp175 formed a hydrogen bond in the crystal structure, and both their mutations, Q108E and D175N, were inactive. To test whether a charge-conserved double mutation at this site could rescue the detrimental effect of the single mutations, we constructed the variant Q108E/D175N. The result showed that its activity was also near zero, indicating that a precise local hydrogen-bond network, instead of its overall charge, plays a critical role in the catalysis. These results demonstrate that residues around both Fe2+-binding sites contribute to the overall enzyme activity.
Third, we introduced mutations inside the domain interface by breaking the only interdomain disulfide bond between Cys100 and Cys323 (Fig. 4A). This disulfide bond is a unique structural feature present in GDOsp, but absent in other known members of the GDO family (Supplemental Fig. S1). It is completely buried in the domain interface of the subunit and inaccessible to solvent. It is distant from both potential substrate-binding pockets, thus it is unlikely to be directly involved in enzymatic catalysis. Single-point mutations C100A and C323A were constructed in an attempt to reduce potential coupling between the two homologous cupin domains. Both single-point mutations were further combined with other point mutations from the above second group, resulting in double mutations of Q108E/C323A and T154V/C100A. This latter group of double mutations was intended to invest the effects of active-site mutations in the background of a domain-decoupling mutant. All of these single- and double-mutant variants showed low activity (0%−16% WT level) (Fig. 4B), while their secondary structure (verified with CD) remained similar to WT (Supplemental Fig. S2). The observed decreases in catalytic activities suggest a possibility that an intimate communication between the two domains is necessary for the activity of GDOsp, albeit the mechanism is yet unclear.
In addition, we observed that all variants that contained mutations Q108E, D175N, and H119A/H121A, as well as the C-domain and Δ51 construct discussed earlier, did not catalyze gentisate cleavage to maleylpyruvic acid as monitored by an ultraviolet absorbance signal at 330 nm. Instead, after 4−12 h incubation, all of them produced a yellow color in the reaction mixture, suggesting an alternative, slow reaction with the substrate. A similar result was not observed for the other variants listed above, in which case the reaction mixture remained colorless. In a control experiment, overnight storage of the gentisate substrate in the same buffer did not turn yellow. It seems that at least two conditions must be met for the reaction to produce a yellow product. First, the normal GDO activity must be abolished so that it does not compete with the slow, yellow color-producing reaction (Harpel and Lipscomb 1990b). Second, the mutation permitted some substrate-binding alternative to the major, active-site binding. Interestingly, the recombinant protein of C domain alone (i.e., residues 200−374) can produce a yellow reaction mixture, suggesting a possibility that an intact C domain is responsible for the yellow-producing reaction in this group of variants.
Discussion
GDOsp is a novel member of the cupin family
Based on its amino acid sequence and 3D structure, GDOsp belongs to the cupin superfamily (Mills et al. 2002; Dunwell et al. 2004; Gopal et al. 2005; Straganz and Nidetzky 2006; Liu et al. 2007). This superfamily includes functionally diverse proteins from Archaea, Eubacteria, and Eukaryota. A cupin protein may contain single- (Woo et al. 2000) or double (Adams et al. 2006)-cupin domains. Bicupin proteins often form homodimers (Suarez et al. 1996; Werwath et al. 1998), or homotetramers (Crawford et al. 1975; Harpel and Lipscomb 1990a; Suarez et al. 1996; Fu and Oriel 1998; Werwath et al. 1998; Feng et al. 1999). In contrast, single cupin proteins have been observed to form homohexamers (Woo et al. 2000). GDOsp belongs to the bicupin subfamily and forms a homotetramer both in solution and in crystal (Figs. 1B, 3B). Its folding is similar to a previously reported E. coli GDO structure (PDB ID 2D40) (Adams et al. 2006) at both subunit and tertiary structure levels. For example, the RMSD between their subunits is 1.2 Å for 277 Cα atoms (using a 3 Å cutoff). On the other hand, our observation on the GDOsp oligomerization is in sharp contrast to the conclusion based on size-exclusion chromatography from a previous report (Liu et al. 2007).
Bicupin proteins are believed to result from a gene duplication event followed by divergent evolution. For example, in GDOsp, the N and C domains show a recognizable sequence homology and an outstanding 3D structural similarity (Fig. 2). In fact, all known GDO proteins are bicupin proteins (Supplemental Fig. S1). Although both of their N and C domains share similar cupin-motif sequences, usually only one domain binds metal and maintains the active site. Little is known about the potential functions of the domain that loses the metal-binding ability. GDOsp appears to be a rare case in which both the N and C domain maintain the histidine triad and bind with Fe2+ ions. The two conserved HXH-containing motifs are located in strands β4 and β9 of the smaller β-sheet in the N domain, and are similarly in β16 and β21 of the C domain (Fig. 2). The GDO from Haloferax sp. (GDOhs) may provide another example of double metal binding in bicupin GDO proteins, although the enzyme activity in each individual cupin domain from such a protein remains to be demonstrated (Dunwell et al. 2004). GDOsp provides the first example that a bicupin protein may contain two intact, functionally related Fe2+-binding sites, both of which contribute to the enzymatic activity in an interdependent manner (Fig. 4).
Our mutagenesis data show that knocking out such a metal-binding site (e.g., by H119A/H121A mutation) does not affect the protein overall tertiary structure, at least in the case of the GDOsp N domain (Supplemental Fig. S2), but it indeed abrogates the enzymatic activity (Fig. 4). Meanwhile, indirect disruption of either Fe2+-binding site by mutating a ligand (His)-stabilizing residue (e.g., Y190F or E360Q) resulted in a decrease of enzymatic activity but had no significant effect on protein second structure and oligomerization. Similarly, Fe depletion of a GDOsp sample decreases specific activity without changing the WT-like secondary structure (Fig. 1A) or its oligomerization state (Fig. 1B). Together with the results of the H119A/H121A mutation, these observations are in sharp contrast to results from another bacterial GDO, GDOxp (Hirano et al. 2007). In the GDOxp case tetramerization, which is shown to be essential for its enzyme activity, can be disrupted by either taking off ferrous ions or truncation of its N-terminal peptide (residues 1−30) (Hirano et al. 2007). Thus, ferrous ions are considered to be cooperative effectors to form an active GDOxp tetramer. Conversely, an inactive monomer GDOxp protein could reconstitute a tetramer structure and restore enzyme activity in a cooperative manner upon the addition of ferrous ions. In the GDOsp case, however, we could not dissociate the tetramer by chelating ferrous ions. Our result is consistent with the structural observations that tetramerization interface is extensive and independent of the metal-binding site.
Substrate specificity in the N domain is determined by a group of conserved residues in GDO proteins
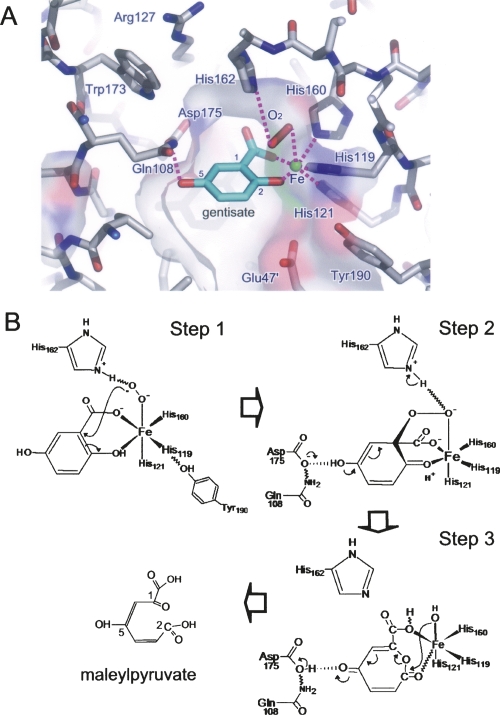
Since the N domain is significantly more conserved than the C domain among GDO proteins (Supplemental Fig. S1), we speculated that the corresponding N-domain Fe2+-binding site is one of the substrate (gentisate and O2) binding and catalytic sites, if not the only one. Toward understanding the catalytic mechanism and substrate specificity of GDOsp and other GDO proteins in general, we manually modeled both substrate gentisate and oxygen (O2) into the N-domain active site of GDOsp (Fig. 5A) following the hints from the crystal structure of 2,3-HPCD (homo-protocatechuate 2,3-dioxygenase) complexed with substrates. 2,3-HPCD is a Fe2+-containing, extradiol, noncupin dioxygenase. Its complex crystal structure with a slow substrate has been recently reported, showing reaction intermediates at three different stages (Kovaleva and Lipscomb 2007). The enzyme forms a pseudosymmetric homotetramer, where three different intermediates are observed in the four subunits. The reason for the structural difference at the individual active sites is attributed to the distinct crystal packing of each subunit. The 2,3-HPCD protein does not have a cupin folding, and there is no detectable sequence homology between 2,3-HPCD and GDOsp. The 2,3-HPCD protein uses a His-His-Glu triad of metal-binding motif, and the relative positions of these metal-binding residues in the primary sequence have no correlation with those in bicupin GDO proteins. In particular, the metal-binding site triad residues in 2,3-HPCD come from three β-strands, while those in GDOsp come from two β-strands. Nevertheless, the active site of 2,3-HPCD is located inside the cavity of a β-barrel (but not a cupin). The similarity between the dioxygenase functions of 2,3-HPCD and GDOsp are likely to be the result of convergent evolution. According to the crystal structure of 2,3-HPCD−substrate complex, some mechanistic insight from the 2,3-HPCD structure may be useful in understanding the dioxygenases of the cupin family, in particular GDOsp.
Figure 5.
Putative substrate-binding mode and catalytic mechanism of GDOsp. (A) Molecular modeling of gentisate and O2 binding to the vicinity of the Fe2+ ion in the N-domain cupin cavity. Glu47 from a neighboring subunit is marked with a prime. Oxygen, nitrogen, and Fe atoms are colored red, blue, and green, respectively. Carbon atoms are colored gray and cyan in the protein and substrate, respectively. Hydrogen bonds between substrates and GDOsp as well as the iron ion are shown as magenta dashed lines. (B) Schematic diagram of a putative catalytic mechanism by GDOsp.
Based on the 2,3-HPCD complex structure and our structural and mutagenesis data, we performed a molecular modeling to identify possible substrate-binding modes in the N-terminal domain of GDOsp (also see the Supplemental material). The result is illustrated in Figure 5. Such a two-ends binding mode for gentisate is supported by previous spectroscopic data and has been proposed by Harpel and Lipscomb before (Harpel and Lipscomb 1990a). Other residues involved in this substrate-binding mode include Arg127, Thr137, Thr154, His162, and Trp173, which are conserved among GDO proteins (Supplemental Fig. S1). Our model of substrate binding provides structural explanations for observations from our mutagenesis and functional studies and allows us to formulate a putative catalytic mechanism.
The current GDOsp model supports an extradiol mechanism
Based on available structural information, including the above described molecular modeling and our functional studies on a collection of GDOsp mutations, as well as previously reported studies on similar enzymes (Harpel and Lipscomb 1990b; Mendel et al. 2004; Kovaleva and Lipscomb 2007), we propose a putative catalytic mechanism for the N domain of GDOsp, which is depicted in Figure 5B (Steps 1−3). First, gentisate diffuses to the active site and displaces two water molecules to chelate with the Fe2+ ion by its deprotonated carboxylate at C1 and phenolic hydroxyl at C2. Hydroxyl at C5 forms a hydrogen bond with the carboxylic side chain of Asp175, which, in turn, is stabilized by the side chain of Gln108. The chelation and hydrogen-bond network precisely position the substrate gentisate at the active site. The substrate dioxygen then coordinates as the last ligand to the Fe2+ ion forming a Fe–superoxide complex as shown in the mechanisms of other extradiol dioxygenases (Kovaleva and Lipscomb 2007). This complex is stabilized by the imidazole side chain of conserved His162 through electron transfer and hydrogen-bond rearrangement (Step 1), and His162 forms a hydrogen bond with conserved Arg127. In addition, Thr154 appears required for gentisate binding. The attack of the Fe-bound superoxide at C1 yields an alkylperoxo intermediate. A subsequent Criegee rearrangement of this intermediate results in O-O bond cleavage and insertion of the first oxygen atom into the aromatic ring to form an anhydride intermediate. This rearrangement is accelerated by a proton transfer from the hydroxyl group at C5 to the Asp175–Gln108 hydrogen bond network and by electron transfer to the ring associated with ketone formation (Step 2). The subsequent transfer of the hydroxyl group (i.e., the second oxygen) from Fe2+ ion to C2 and resonance rearrangement would yield the product maleylpyruvic acid (Step 3). Results from our mutagenesis analysis on the key residues Gln108, His162, Asp175, and their microenvironments (e.g., Arg127) support such a catalytic mechanism.
It is worth noting that a few of the above-mentioned critical residues in this N-domain substrate-binding model (e.g., Arg127, Thr154, and Asp175) are not conserved in the C-domain of GDOsp (Fig. 2D; Supplemental Fig. S1). In addition, the cavity around the C-domain Fe-binding site appears smaller than that of the N domain (Fig. 2B). Particularly, the loop around Asp255 between α7 and β14 plunged into the C-domain cavity, and side chains of Asn255, Gln279, and Tyr298 protrude toward the metal binding, preventing a gentisate molecule from occupying a similar position as in the N-domain. It is interesting to ask whether the C-domain site is also functional as a gentisate dioxygenase active site. Indeed, the recombinant protein of the GDOsp C domain alone could convert gentisate into a yellow product, suggesting a direct interaction of the C domain with the substrate gentisate. Furthermore, a group of point mutations in the C domain showed WT-like folding, but significantly lower GDO activity (e.g., Y298F), suggesting that residues in the C domain are also critical for the activity. Thus, the C domain may not explore the same substrate-binding mode should it also function as a fully active gentisate dioxygenase catalytic site. It remains for the future substrate complex structure to illustrate the exact roles of each domain and of the associated metal-binding site.
Since most of our point mutations, including those of the C domain, were carefully selected to disrupt neither the oligomerization interface nor the domain folding, we can rule out the possibility that mutations interfere with proper oligomerization, thus resulting in activity loss. Moreover, point mutations in either domain can abolish the overall activity, indicating that the two domains do not function independently during catalysis. This argument prompted us to test the role of domain interaction within a GDOsp subunit. A group of point mutations to break the interdomain disulfide bond in each subunit was constructed in an attempt to decouple a putative domain–domain interaction. This disulfide bond is buried in the domain interface, thus it is likely to be intact in vivo even in a reducing environment. Such a disulfide-breaking mutation (e.g., C100A) typically behaves like most other inactive point mutations within either the N or the C domain, suggesting that an intact domain–domain communication is as important as metal-binding sites in both domains. Although long-distance communication within a protein molecule or complex has been observed before (Wei et al. 2003; Clarkson and Lee 2004), this is the first time, to the best of our knowledge, that it is reported from a bicupin protein. As an alternative explanation, such a disulfide bond may be essential for the overall stability of the bicupin folding, thus affecting the activity indirectly. Given the fact that this interdomain disulfide bond is unique for GDOsp, we favor a more direct role of the disulfide bond in the GDO activity.
Our current study, including crystallography analysis and extensive structure-based mutagenic and functional analyses on the gentisate 1,2-dioxygenase from S. pomeroyi, allows us to identify key residues in the GDO catalysis. A phenomenon of domain interaction in the GDOsp subunit is observed and attributed to the double metal-binding sites in this unusual bicupin protein. One explanation for the interdomain interaction could be an allosteric regulation to the N-domain active site by a C-domain substrate-binding. This and other possibilities remain to be further explored in GDOsp and verified in other GDO proteins.
Materials and Methods
Protein expression, purification, and mutagenesis
The cDNA of GDOsp (GenBank ID YP_164947) (Liu et al. 2007) was cloned into the pET28a vector (Novagen/Merck). Recombinant GDOsp protein was expressed in E. coli BL21 (DE3) cells, and it had a calculated molecular weight of 44.3 kDa in the presence of an amino (N)-terminal 20-residue His-tag adapted from the expression vector. Point mutations were introduced into the pET28a-GDOsp parental construct using overlap PCR site-directed mutagenesis methods (Sambrook and Russell 2001). All mutations were confirmed by DNA sequencing over the full-length coding region. The protein sample was first purified with nickel-NTA resin (Qiagen). It was followed by resource-Q ion-exchange chromatography (Amshambioscience) and was eluted with a NaCl gradient (10−300 mM); the GDOsp protein peaked at about 160 mM salt. It was concentrated and subsequently purified by size-exclusion chromatography on a Superdex-200 column (Amshambioscience) eluted with a store buffer of 20 mM Tris-HCl (pH 8.0) and 120 mM NaCl supplemented with 10 mM β-mercaptoethanol. The purified GDOsp enzyme was concentrated to 30 mg/mL and stored at −80°C. Protein concentrations were determined using 280 nm absorption and extinction coefficients calculated by the program ProtParam (http://www.expasy.org/tools/protparam.html), e.g., 99.2 AU/mM (i.e., 0.446 mg/mL per OD280 nm) for the His-tagged, full-length WT GDOsp.
Enzyme activity measurement and kinetic characterizations
Gentisate 1,2-dioxygenase activity of the GDOsp variant was determined by monitoring the increase in absorbance at 330 nm that accompanies the formation of maleylpyruvate (Lack 1959). In a standard assay, each 500-μL mixture contained 1.5 μM enzyme, 200 μM substrate gentisate (unless otherwise mentioned), 20 mM Tris-HCl (pH 8.0), and 120 mM NaCl; for a low-activity mutant, the assay was repeated with higher (up to 100 μM) protein concentrations. For the kinetic assay, varied concentrations (0.5−200 μM) of substrate were used, and the assay was initiated by addition of a fixed quantity of GDOsp enzyme (13 μM final). Increase in absorbance at 330 nm was recorded for 60 s at room temperature using a U-2010 spectrophotometer (Hitachi). To calculate specific activities, a molar extinction coefficient of 10,200 M−1cm was used for the product maleylpyruvate (Wheelis et al. 1967). Assuming a single binding site per subunit, apparent V max and K M values were calculated from the Michaelis-Menten equation fitted by using a linear fit program Origin7.0. All reported kinetic values were the means of multiple separate measurements.
Crystallization, data collection, and processing
Initial crystallization condition was identified by using crystallization screening kits from Hampton Research. The recombinant GDOsp sample in the store buffer was mixed with an equal volume (1 μL) of the reservoir solution to perform the hanging-drop vapor diffusion crystallization at 16°C. The optimal reservoir solution contained 20% (w/v) polyethylene glycol (PEG) 3350 and 200 mM sodium iodide. These crystals were rectangular cuneiforms and required ∼5−7 d to attain their maximum size of 0.1 × 0.1 × 0.05 mm.
The crystal was soaked in a cryoprotectant solution containing 20% (w/v) PEG 3350, 200 mM sodium iodide, and 15% (v/v) glycerol immediately before flash-cooling at cryogenic temperature for data collection. X-ray diffraction data were collected on an in-house MarResearch image plate equipped with a Rigaku copper rotating anode X-ray generator. The data were processed, scaled, and merged using the HKL2000 program package (Otwinowski and Minor 1997).
Structure determination and refinement
Phases of the GDOsp enzyme crystal structure were determined by the molecular replacement method using the program Molrep (Vagin and Isupov 2001) and the E. coli GDO crystal structure (eGDO, PDB file 2D40) (Adams et al. 2006) as the search model that shares 37% sequence identity with GDOsp. Multiple cycles of rebuilding and refinement with the programs Refmac5 (Murshudov et al. 1997) and Coot (Emsley and Cowtan 2004) were then carried out. In the final refined structure, three residues (Arg83 of molecule A and Asp196 of both molecules A and B) were found on the border of a disallowed region of the Ramachandran plot. The Arg83 residue was involved in crystal packing, and the two Asp196 residues were located in a Pro-containing surface loop. All of them were distant from the active site.
Analytical ultracentrifugation
Boundary sedimentation velocity (SV) measurements were performed with an Optima XL-A/I analytical ultracentrifuge (Beckman-Coulter Instruments) and a four-cell An-60 Ti rotor. Freshly prepared GDOsp was used for the AUC experiment. The SV analysis was carried out at 20°C at a rotor speed of 38,000 rpm. Absorbance scans were performed at 280 nm with a radial step size of 30 μm. Sedimentation coefficient distribution analysis, c (M), was deduced using the computer program SEDFIT (www.analyticalultracentrifugation.com/ download.htm) with a 95% confidence limit setting.
Circular dichroism spectrometry
Far-UV circular dichroism (CD) spectra, which detect the secondary structure content of the protein, were obtained on a π*-180 spectropolarimeter (Applied Photophysics). The protein concentration was adjusted to 0.3−0.5 mg/mL in 20 mM Tris-HCl (pH 8.0) and 120 mM NaCl. The optical path of the quartz cell was 1 mm. A wavelength scan was performed at 20°C between 200 and 260 nm. Multiple measurements were carried out for each protein sample.
Inductively coupled plasma–MS measurements
To identify the metal elements that bind to the GDOsp recombinant protein sample, the metal content was determined and quantified with the inductively coupled plasma–mass spectrometry (ICP–MS) technique using a Bruker instrument by the Analysis Center of Tsinghua University. The experiment was performed using a 25-μM protein sample in a buffer of 20 mM Tris-HCl (pH 8.0) and 120 mM NaCl without supplementation of iron.
Data deposition
The atomic coordinates and structure factors of GDOsp have been deposited in the RCSB Protein Data Bank (www.pdb.org) with PDB accession code 3BU7.
Acknowledgments
We are grateful to staff members of the Structural Biology Core Facility at the Institute of Biophysics, Chinese Academy of Sciences (CAS), for their excellent technical assistance, especially to Xiaoxia Yu, Zhongnian Zhou, Meirong Zhang, and Yi Han. We thank Dr. Walter Baase of the University of Oregon for helpful discussions on CD experiments. This work was supported by “863” Project grant 2006AA02A322 from the Chinese NFS and CAS grant 2006CB911002 to X.L., grant 30221033 from NSFC and Ministry of Science and Technology International Cooperation Project 2006DFB32420, and CAS grant KSCX2-YW-05 to Z.R.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Xuejun C. Zhang, National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, Beijing 100101, People's Republic of China; e-mail: zhangc@omrf.org; fax: (405) 271-7953; or Zihe Rao, Laboratory of Structural Biology, Tsinghua University, Beijing 100084, People's Republic of China; e-mail: raozh@xtal.tsinghua.edu.cn; fax: 86-10-64872026.
Abbreviations: AUC, analytical ultracentrifugation; CD, circular dichroism spectroscopy; EDTA, ethylene-diamine-tetra- acetic acid; GDOsp, gentisate 1,2-dioxygenase from S. pomeroyi; ICP-MS, inductively coupled plasma-mass spectroscopy; RMSD, root-mean-square deviation; SAS, solvent accessible surface; WT, wild type.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035881.108.
References
- Adams, M.A., Singh, V.K., Keller, B.O., Jia, Z. Structural and biochemical characterization of gentisate 1,2-dioxygenase from Escherichia coli O157:H7. Mol. Microbiol. 2006;61:1469–1484. doi: 10.1111/j.1365-2958.2006.05334.x. [DOI] [PubMed] [Google Scholar]
- Arciero, D.M., Lipscomb, J.D., Huynh, B.H., Kent, T.A., Munck, E. EPR and Mossbauer studies of protocatechuate 4,5-dioxygenase. Characterization of a new Fe2+ environment. J. Biol. Chem. 1983;258:14981–14991. [PubMed] [Google Scholar]
- Bayly, R.C., Barbour, M.G. Microbial degradation of aromatic compounds. In: Gibson D.T., editor. Marcel Dekker; New York: 1984. pp. 253–294. [Google Scholar]
- Boldt, Y.R., Sadowsky, M.J., Ellis, L.B., Que L., Jr, Wackett, L.P. A manganese-dependent dioxygenase from Arthrobacter globiformis CM-2 belongs to the major extradiol dioxygenase family. J. Bacteriol. 1995;177:1225–1232. doi: 10.1128/jb.177.5.1225-1232.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, P.J. National Academy of Sciences; Washington, D.C: 1972. Degradation of synthetic organic molecules in the biosphere; pp. 15–55. [Google Scholar]
- Clarkson, M.W., Lee, A.L. Long-range dynamic effects of point mutations propagate through side chains in the serine protease inhibitor eglin c. Biochemistry. 2004;43:12448–12458. doi: 10.1021/bi0494424. [DOI] [PubMed] [Google Scholar]
- Crawford, R.L., Hutton, S.W., Chapman, P.J. Purification and properties of gentisate 1,2-dioxygenase from Moraxella osloensis . J. Bacteriol. 1975;121:794–799. doi: 10.1128/jb.121.3.794-799.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagley, S., Evans, W.C., Ribbons, D.W. New pathways in the oxidative metabolism of aromatic compounds by microorganisms. Nature. 1960;188:560–566. doi: 10.1038/188560a0. [DOI] [PubMed] [Google Scholar]
- Dunwell, J.M., Culham, A., Carter, C.E., Sosa-Aguirre, C.R., Goodenough, P.W. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 2001;26:740–746. doi: 10.1016/s0968-0004(01)01981-8. [DOI] [PubMed] [Google Scholar]
- Dunwell, J.M., Purvis, A., Khuri, S. Cupins: The most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Feng, Y., Khoo, H.E., Poh, C.L. Purification and characterization of gentisate 1,2-dioxygenases from Pseudomonas alcaligenes NCIB 9867 and Pseudomonas putida NCIB 9869. Appl. Environ. Microbiol. 1999;65:946–950. doi: 10.1128/aem.65.3.946-950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W., Oriel, P. Gentisate 1,2-dioxygenase from Haloferax sp. D1227. Extremophiles. 1998;2:439–446. doi: 10.1007/s007920050090. [DOI] [PubMed] [Google Scholar]
- Fujisawa, H., Uyeda, M., Kojima, Y., Nozaki, M., Hayaishi, O. Protocatechuate 3,4-dioxygenase. II. Electron spin resonance and spectral studies on interaction of substrates and enzyme. J. Biol. Chem. 1972;247:4414–4421. [PubMed] [Google Scholar]
- Fusetti, F., Schroter, K.H., Steiner, R.A., van Noort, P.I., Pijning, T., Rozeboom, H.J., Kalk, K.H., Egmond, M.R., Dijkstra, B.W. Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus . Structure. 2002;10:259–268. doi: 10.1016/s0969-2126(02)00704-9. [DOI] [PubMed] [Google Scholar]
- Gibello, A., Ferrer, E., Martin, M., Garrido-Pertierra, A. 3,4-Dihydroxyphenylacetate 2,3-dioxygenase from Klebsiella pneumoniae, a Mg2+-containing dioxygenase involved in aromatic catabolism. Biochem. J. 1994;301:145–150. doi: 10.1042/bj3010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal, B., Madan, L.L., Betz, S.F., Kossiakoff, A.A. The crystal structure of a quercetin 2,3-dioxygenase from Bacillus subtilis suggests modulation of enzyme activity by a change in the metal ion at the active site(s) Biochemistry. 2005;44:193–201. doi: 10.1021/bi0484421. [DOI] [PubMed] [Google Scholar]
- Harpel, M.R., Lipscomb, J.D. Gentisate 1,2-dioxygenase from pseudomonas. Purification, characterization, and comparison of the enzymes from Pseudomonas testosteroni and Pseudomonas acidovorans . J. Biol. Chem. 1990a;265:6301–6311. [PubMed] [Google Scholar]
- Harpel, M.R., Lipscomb, J.D. Gentisate 1,2-dioxygenase from Pseudomonas. Substrate coordination to active site Fe2+ and mechanism of turnover. J. Biol. Chem. 1990b;265:22187–22196. [PubMed] [Google Scholar]
- Hirano, S., Morikawa, M., Takano, K., Imanaka, T., Kanaya, S. Gentisate 1,2-dioxygenase from Xanthobacter polyaromaticivorans 127W. Biosci. Biotechnol. Biochem. 2007;71:192–199. doi: 10.1271/bbb.60444. [DOI] [PubMed] [Google Scholar]
- Jin, S., Zhu, T., Xu, X., Xu, Y. Biodegradation of dibenzofuran by Janibacter terrae strain XJ-1. Curr. Microbiol. 2006;53:30–36. doi: 10.1007/s00284-005-0180-1. [DOI] [PubMed] [Google Scholar]
- Kovaleva, E.G., Lipscomb, J.D. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science. 2007;316:453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack, L. The enzymic oxidation of gentisic acid. Biochim. Biophys. Acta. 1959;34:117–123. doi: 10.1016/0006-3002(59)90239-2. [DOI] [PubMed] [Google Scholar]
- Liu, D., Zhu, T., Fan, L., Quan, J., Guo, H., Ni, J. Identification of a novel gentisate 1,2-dioxygenase from Silicibacter pomeroyi . Biotechnol. Lett. 2007;29:1529–1535. doi: 10.1007/s10529-007-9421-7. [DOI] [PubMed] [Google Scholar]
- Mendel, S., Arndt, A., Bugg, T.D. Acid-base catalysis in the extradiol catechol dioxygenase reaction mechanism: Site-directed mutagenesis of His-115 and His-179 in Escherichia coli 2,3-dihydroxyphenylpropionate 1,2-dioxygenase (MhpB) Biochemistry. 2004;43:13390–13396. doi: 10.1021/bi048518t. [DOI] [PubMed] [Google Scholar]
- Mills, E.N., Jenkins, J., Marigheto, N., Belton, P.S., Gunning, A.P., Morris, V.J. Allergens of the cupin superfamily. Biochem. Soc. Trans. 2002;30:925–929. doi: 10.1042/bst0300925. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymo. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Que L., Jr, Lipscomb, J.D., Zimmermann, R., Munck, E., Orme-Johnson, N.R., Orme-Johnson, W.H. Mossbauer and EPR spectroscopy of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa . Biochim. Biophys. Acta. 1976;452:320–334. doi: 10.1016/0005-2744(76)90182-0. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Russell, D. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. Molecular cloning: A laboratory manual. [Google Scholar]
- Straganz, G.D., Nidetzky, B. Variations of the 2-His-1-carboxylate theme in mononuclear non-heme FeII oxygenases. ChemBioChem. 2006;7:1536–1548. doi: 10.1002/cbic.200600152. [DOI] [PubMed] [Google Scholar]
- Suarez, M., Ferrer, E., Martin, M. Purification and biochemical characterization of gentisate 1,2-dioxygenase from Klebsiella pneumoniae M5a1. FEMS Microbiol. Lett. 1996;143:89–95. doi: 10.1111/j.1574-6968.1996.tb08466.x. [DOI] [PubMed] [Google Scholar]
- Vagin, A.A., Isupov, M.N. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1451–1456. doi: 10.1107/s0907444901012409. [DOI] [PubMed] [Google Scholar]
- Wei, C.L., Yang, Y.B., Wang, W.C., Liu, W.C., Hsu, J.S., Tsai, Y.C. Engineering Streptomyces clavuligerus deacetoxycephalosporin C synthase for optimal ring expansion activity toward penicillin G. Appl. Environ. Microbiol. 2003;69:2306–2312. doi: 10.1128/AEM.69.4.2306-2312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werwath, J., Arfmann, H.A., Pieper, D.H., Timmis, K.N., Wittich, R.M. Biochemical and genetic characterization of a gentisate 1, 2-dioxygenase from Sphingomonas sp. strain RW5. J. Bacteriol. 1998;180:4171–4176. doi: 10.1128/jb.180.16.4171-4176.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelis, M.L., Palleroni, N.J., Stanier, R.Y. The metabolism of aromatic acids by Pseudomonas testosteroni and P. acidovorans . Arch. Mikrobiol. 1967;59:302–314. doi: 10.1007/BF00406344. [DOI] [PubMed] [Google Scholar]
- Whiting, A.K., Boldt, Y.R., Hendrich, M.P., Wackett, L.P., Que L., Jr Manganese(II)-dependent extradiol-cleaving catechol dioxygenase from Arthrobacter globiformis CM-2. Biochemistry. 1996;35:160–170. doi: 10.1021/bi951979h. [DOI] [PubMed] [Google Scholar]
- Whittaker, J.W., Lipscomb, J.D., Kent, T.A., Munck, E. Brevibacterium fuscum protocatechuate 3,4-dioxygenase. Purification, crystallization, and characterization. J. Biol. Chem. 1984;259:4466–4475. [PubMed] [Google Scholar]
- Woo, E.J., Dunwell, J.M., Goodenough, P.W., Marvier, A.C., Pickersgill, R.W. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 2000;7:1036–1040. doi: 10.1038/80954. [DOI] [PubMed] [Google Scholar]