Abstract
The expression of MICB, a member of the major histocompatibility complex class I chain-related gene B family, is induced in response to cellular stress. It is one of the ligands to the NKG2D receptor. MICB is polymorphic, but the distribution of MICB polymorphism in north-eastern Thais and their potential associations with cancer have not yet been elucidated. In this study, polymerase chain reaction–sequence-specific primers were developed to identify 15 MICB alleles and one group of alleles. We performed MICB typing in 100 healthy north-eastern Thai females (NETF) and 99 cervical cancer patients to evaluate the association of MICB polymorphisms and the risk of developing cervical cancer. Eight and nine alleles were detected in the NETF and cervical cancer respectively. MICB*00502 was associated negatively with a corrected P-value of 0·0009, suggesting the existence of a protective allele in cervical cancer. Amino acid substitutions carried by this allele were investigated for their potential involvement in natural killer (NK) cell activation. Although lysine at amino acid position 80 (Lys80) and aspartic acid at position 136 (Asp136) were associated negatively with cervical cancer, only MICB carrying Asp136 could induce NK cell killing more efficiently than MICB-Lys80 when the NK cells were blocked by anti-NKG2D. This result suggested that aspartic acid at position 136 may affect NKG2D binding, leading to different degrees of immune cell activation.
Keywords: activation, cancer, genetic, non-classical MHC
Introduction
Cervical cancer (CXCA) is currently the third most common type of cancer among women worldwide [1] and the second most common cancer among women in the developing world [2]. Even though screening programmes based on the Papanicolaou (Pap) smear and pelvic examination have led to a steep decline in incidence and deaths from CXCA, several hundred thousands of new cases are diagnosed each year, predominantly in both developing and industrialized countries. In Thailand, the incidence of CXCA is about 23·4 cases per 100 000 women [3]; however, the diagnosis is made usually in advanced stages. Infection with human papilloma virus (HPV) types 16 or 18 is the major cause of cervical neoplasia [4]. Although a high proportion of CXCA harbour HPV genomes, only a small number of women infected with high-risk papilloma viruses develop cervical tumours, suggesting that other environmental, genetic factors and/or immune surveillance contribute to cervical carcinogenesis.
Down-regulation of major histocompatibility complex (MHC) class I and induction of MHC class I chain-related molecules, MIC, have been observed in various tumour types. MIC proteins, mainly MHC class I chain-related gene A (MICA) and MHC class I chain-related gene B (MICB), are induced only in response to cellular stress on limited cell types, restricted essentially to epithelial-derived tumours [5,6]. MIC can activate killing activity of natural killer (NK) cells against cancerous cells via the NKG2D and DAP10 or DAP12 complex [7,8] expressed on the cell surface of many immune cells, e.g. NK cells, some cytotoxic CD8 αβT cells, γδT cells, NK T cells, macrophages and a small subset of CD4 αβT cells [7,9–11]. Thus, NKG2D-ligand recognition plays an important role in both innate and adaptive immune systems.
MICA and MICB are polymorphic [12,13]; however, MICB polymorphism seems to be lower than that of MICA. To date, at least 57 alleles of MICA were identified whereas 18 MICB alleles were reported [14]. The extent of MICB polymorphism and its distribution in populations remain under investigation. Consequently, the potential association of MICB to diseases has not been evaluated. In this study, we have described the development of a polymerase chain reaction (PCR) amplification method using sequence-specific primers (SSP) discriminating MICB alleles. This method was employed to type MICB of healthy north-eastern Thai females (NETF) as well as CXCA patients in order to evaluate the association of the MICB alleles with this disease. In addition, potentially functional MICB motifs identified by this association study were tested for their differential binding to NKG2D and NK cell-killing activities.
Materials and methods
Study subjects
The subjects were divided into two groups, CXCA and healthy control subjects. Peripheral blood mononuclear cells collected from citrate phosphate dextrose or ethylenediamine tetraacetic acid blood were used to prepare genomic DNA. The project was approved by the Khon Kaen University Ethical Committee (HE430205) and informed consents were obtained from all subjects participating in the project. Ninety-nine CXCA cases were recruited from the tumour clinic, Srinagarind Hospital, Faculty of Medicine, Khon Kaen University by gynaecological clinicians. All patients were north-eastern Thais and diagnosed histologically. The CXCA samples were classified into different stages following the International Federation of Gynecology and Obstetrics (FIGO system). These samples were categorized into two groups − low grade (stages I and II = 43 cases) and high grade (stages III and IV = 57 cases). In addition, 87% of these cases were positive with HPV (high-risk types: 16, 18 and 33 were 91·2% and low-risk types were 8·8%). Healthy NETF were screened by interview with negative Pap smear. One hundred unrelated female subjects whose families are Thai and have lived in the north-eastern part of Thailand for at least two generations were included as a healthy control group.
Oligonucleotide primers
In this study, we have developed a PCR-based typing system using SSP to identify MICB alleles. PCR reaction mixtures composed of 29 primer pairs were used for amplification of 15 MICB alleles and one group of alleles (Fig. 1) which have been identified previously [15–21]. The first set of 14 SSP pairs (reaction mixes numbers 1–14) were designed for specific amplifications of the 15 MICB alleles and one group of alleles, whereas the remaining primer set was included to identify homozygous alleles. Primers recognizing conserved homologous sequences in the human growth hormone (hGH) gene were used as internal control primers.
Fig. 1.

Interpretation of major histocompatibility complex class I chain-related gene B (MICB) alleles based on polymerase chain reaction (PCR)–sequence-specific primers (PCR) of 29 PCR reaction mixes. MICB alleles are listed on the left. The numbers of PCR reaction mixes are shown on the top. Black boxes represent positive PCR reactions with the primer mixes, as indicated at the top of each column. Primer mixes numbers 15–29 were designed for substitutions at allele-specific sites in order to identify a sample carrying homozygous MICB allele.
Primer sequences and locations relative to the MICB gene are given in Table 1. DNA samples extracted from International Histocompatibility Workshop cell lines with known MICB alleles, kindly provided by Dr Campbell Witt, Royal Perth Hospital, Western Australia, were used as positive controls to validate specific amplifications of all designed primers.
Table 1.
The locations and sequences of oligonucleotide primers used for major histocompatibility complex class I chain-related gene B (MICB) typing by polymerase chain reaction (PCR)–sequence-specific primers (PCR).
| Mix no. | Names and primer sequence | Location | Specificity |
|---|---|---|---|
| 1 | 7473A*: GGT GCT GTC CCA GGA TGA | 7456–7473 | MICB * 001 |
| 8224RG*: ACC CCG GAT TTC AGA TAT CG | 8224–8205 | ||
| 2 | MICB3F: CAC TTG GGT GGA AAG GTG ATG | 7895–7915 | MICB * 002, 007, 008, 014 |
| P14R*: GAG GAA GAG CTC CCC ATT | 8051–8034 | ||
| 3 | MICB4F: CAG CCC TGT TCC CTG CAT | 8765–8782 | MICB * 003, 009N |
| P18R: CTG AGA CCT CGC TGC AGA | 8871–8854 | ||
| 4 | MICB2F: GAG GTC GGG ACA GCA GAC | 7341–7358 | MICB * 004 |
| A104R: CTC AGC TCC CAG GAC ATT | 7597–7580 | ||
| 5 | C9089T: GGA ACA CAG CGG GAA TCA T | 9071–9089 | MICB * 00501 |
| T9236T: CAT ATG GAA AGT CTG TCC GT | 9255–9236 | ||
| 6 | A022*: TCC AGC TTC TAT CCC CGA | 8901–8918 | MICB * 00503, 013 |
| MICB4R: ACA GCC GTC CCT GCT GTT | 9171–9154 | ||
| 7 | A023C*: AAT GGA ACC TAC CAG ACG | 9000–9017 | MICB * 00504 |
| MICB4R: ACA GCC GTC CCT GCT GTT | 9171–9154 | ||
| 8 | A006K*: TGA ATG TCA CCT GCA GCA | 8845–8862 | MICB * 006, 015 |
| MICB4R: ACA GCC GTC CCT GCT GTT | 9171–9154 | ||
| 9 | A106: ATT AGG GTC TGT GAG ATG | 7974–7991 | MICB * 008 |
| MICB3R: GAA TTG CGG GAA CAG TAG AGC | 8281–8261 | ||
| 10 | MICB3F: CAC TTG GGT GGA AAG GTG ATG | 7895–7915 | MICB * 009N |
| A107NR: CCC GGA TTT CAG ATA TCA | 8222–8205 | ||
| 11 | MICB2F: GAG GTC GGG ACA GCA GAC | 7341–7358 | MICB * 011 |
| A-005R: CTG CCC ACT GTC CCT GGT | 7577–7560 | ||
| 12 | A7671G*: ACC CTG ACT CAT ATC AAG GG | 7652–7671 | MICB * 012 |
| MICB3R: GAA TTG CGG GAA CAG TAG AGC | 8281–8261 | ||
| 13 | A022*: TCC AGC TTC TAT CCC CGA | 8901–8918 | MICB * 013 |
| A022_301VR: GGG CAC AGG GTG AGT GCT | 9107–9090 | ||
| 14 | 533G‡: CAC ACT ATC GCG CTA TGC AG | 8161–8180 | MICB * 014, 013, 015 |
| A022_301VR: GGG CAC AGG GTG AGT GCT | 9107–9090 | ||
| 15 | 7473G*: GGT GCT GTC CCA GGA TGG | 7456–7473 | All but not MICB * 001 |
| 8224RG*: ACC CCG GAT TTC AGA TAT CG | 8224–8205 | ||
| 16 | MICB3Fn: AGG AAT AGG GTC AGG GAG G | 7790–7808 | All but not MICB * 002, 007, 008, 014 |
| PB3R*: GAG GAA GAG CTC CCC ATC | 8051–8034 | ||
| 17 | P17†: CCC CAT GGT GAA TGT CAC | 8837–8854 | All but not MICB * 003, 009N |
| MICB4R: ACA GCC GTC CCT GCT GTT | 9171–9154 | ||
| 18 | MICB2Fn: TTT CCT GCC TCC TCA GGG AG | 7324–7343 | All but not MICB * 004 |
| 7597RC: CTC AGC TCC CAG GAC ATC | 7597–7580 | ||
| 19 | MICB4Fn: ACA GAA GGT CTG GGA TCT GT | 8709–8728 | All but not MICB * 00501 |
| 9105RG: GGC ACA GGG TGA GTG CCG | 9106–9089 | ||
| 20 | 8918G*: TCC AGC TTC TAT CCC CGG | 8901–8918 | All but not MICB * 00503, 013 |
| MICB4R: ACA GCC GTC CCT GCT GTT | 9171–9154 | ||
| 21 | 9017C*: AAT GGA ACC TAC CAG ACC | 9000–9017 | All but not MICB * 00504 |
| MICB4R: ACA GCC GTC CCT GCT GTT | 9171–9154 | ||
| 22 | 8862G*: TGA ATG TCA CCT GCA GCG | 8845–8862 | All but not MICB * 006, 015 |
| MICB4R: ACA GCC GTC CCT GCT GTT | 9171–9154 | ||
| 23 | 9055G: CCA AGG AGA GGA GCA GAG | 9038–9055 | All but not MICB * 007 |
| MICB4R: ACA GCC GTC CCT GCT GTT | 9171–9154 | ||
| 24 | 7991C: ATT AGG GTC TGT GAG ATC | 7974–7991 | All but not MICB * 008 |
| MICB3R: GAA TTG CGG GAA CAG TAG AGC | 8281–8261 | ||
| 25 | MICB3F: CAC TTG GGT GGA AAG GTG ATG | 7895–7915 | All but not MICB * 009N |
| 8224RG: ACC CCG GAT TTC AGA TAT CG | 8224–8205 | ||
| 26 | 7560C: GAA ACG CAG GGC AAA GCC | 7543–7560 | All but not MICB * 011 |
| 8224RG: ACC CCG GAT TTC AGA TAT CG | 8224–8205 | ||
| 27 | MICB2F: GAG GTC GGG ACA GCA GAC | 7341–7358 | All but not MICB * 012 |
| 7671RT: TCT CAC CTC CTT TCT GGT | 7688–7671 | ||
| 28 | 8918G*: TCC AGC TTC TAT CCC CGG | 8901–8918 | All but not MICB * 013 |
| 9107RC: GGG CAC AGG GTG AGT GCC | 9107–9090 | ||
| 29 | 533G‡: CAC ACT ATC GCG CTA TGC AG | 8161–8180 | All but not MICB * 013, 014, 015 |
| 9107RC: GGG CAC AGG GTG AGT GCC | 9107–9090 |
Amplification of the MICB gene using PCR–SSP
PCR conditions for all designed primers were optimized with their corresponding reference DNA samples. To confirm DNA amplification, each primer mix contained internal control primers which amplified the 439 base pairs (bp) product of the hGH gene. The condition initially followed the standard condition procedure of the 12th International Workshop for human leucocyte antigen typing based on PCR–SSP, with modifications. The final volume of 13 μl of PCR containing 100–200 ng of DNA, 1× PCR buffer, 2·0 mM MgCl2, 0·2 mM 2′-deoxynucleosides 5′-triphosphate mix, 0·5–1·0 μM of specific primers, 0·1 μM of internal control primers and 0·5 units Taq DNA polymerase [25]. PCR amplification was carried out using the PTC 200 thermocycler (MJ Research, Oldendorf, Germany). The cycling condition was performed with one cycle of denaturation at 96°C for 2 min and followed subsequently with three different cycling steps: five cycles of 96°C 30 s, 65°C 60 s, 72°C 40 s; 21 cycles of 96°C 30 s, 58°C 60 s, 72°C 40 s; four cycles of 96°C 30 s, 55°C 75 s, 72°C 120 s and finally with one cycle of extension step at 72°C for 10 min. MICB allele-specific PCR products were finally investigated by agarose gel electrophoresis. Successful PCR amplification must be found with a 439 bp product of the hGH internal control primers.
Molecular genetic techniques and construction of plasmids
Genomic DNA of MICB*014 was obtained by PCR. Forward PCR primer was synthesized corresponding to the MICB gene (exon 2–exon 6, 4·2 Kb) by designing the 5′ end of the primers containing a four-nucleotide sequence (CACC) and the signal sequence of MICB (PB112-TOPO: CACCATGGGGCTGGGCCGGGTCCTGCTGTTTCTGGCCGTCGCCCTCCCTTTTGCACCCCCGGCAGCCGCCGCTGAGCCCCACAGTCTTCGT) and the 3′ end of primer containing stop codon (PB11-CYTB-TOPO: GGCGCCCTCAGTGGA (A/G)CCAGTGGAC). The 4·2 Kb PCR product was inserted into the pcDNA 3·1 TOPO vector using the pcDNA 3·1 Directional TOPO Expression Kit (Invitrogen, Carlsbad, CA, USA). The plasmid sequence was validated by sequencing. Then, the codon for Lys80 (AAG > GAG) in α1 and Asp136 (GAT > AAT) in α2 of MICB*014 (Table 2) were mutated by the site-directed mutagenesis technique. The primers used for mutagenesis were:
Table 2.
Allelic amino acids of major histocompatibility complex class I chain-related gene B (MICB).
| Amino acid position | ||||||
|---|---|---|---|---|---|---|
| MICB | 75 | 80 | 121 | 136 | 212 | 291 |
| 002 | D | E | I | N | T | G |
| 003 | D | K | I | D | I | G |
| 004 | N | K | I | D | T | G |
| 00501/00502/00503 | D | K | I | D | T | G |
| 008 | D | E | M | N | T | G |
| 013 | D | K | I | D | T | S |
| 014 | D | E | I | N | T | S |
E80K forward (5′-CCTGGGAGCTAAGACCTGGGAC-3′),
reverse (5′-GTCCCAGGTCTTAGCTCCCAGG-3′); and
N136D forward (5′-CATTTCTACTACGATGGGGAGCTC-3′), and reverse (5′-GAGCTCCCCATCGTAGAAATG-3′).
The PCR control reaction with no primer and experiment reaction were performed at the same time. After PCR products were amplified, PCR reactions (both experiment and control reactions) were treated with DpnI (Promega, Madison, WI, USA). Then, both of the treated PCR reactions were transformed into competent cells (TOP10 Esherichia coli). Colonies were selected by amiplicilin (Sigma-Aldrich, St. Louis, MO, USA). The experimental reaction was accepted when the control reaction had no grown colony on the plate. The resulting plasmid DNAs carrying Lys80 and Asp136 of MICB were validated by sequencing to confirm that no inappropriate mutations had occurred.
Major histocompatibility complex class I chain-related molecules stably transfected cells
The C1R cells were transfected with MICB*014, mutated MICB*014-Lys80 and MICB*014-Asp136 to produce stable transfectants by the electroporation method (250 volt and 950 μF) using Genepulser (Biorad, Foster City, CA, USA). Populations of cells expressing MICB were fluorescence activated cell sorter (FACS) sorted and maintained in complete RPMI-1640 with 1 mg/ml G418.
Flow cytometry analysis of transfectants
For flow cytometry analysis, 105 cells were preincubated in PBS containing 1% bovine serum albumin, 0·1% sodium azide (Sigma-Aldrich). Cells were stained with different concentrations (20–50 μg/ml) of NKG2D-immunoglobulin (Ig) fusion protein [26] (kindly provided by Dr Hugh T. Reyburn, University of Cambridge, Cambridge, UK) followed, after washing, by phycoerythrin-labelled F(ab′)2 fragments of goat anti-human Ig (Beckman Coulter, Fullerton, CA, USA). Staining with mouse anti-MICA/B monoclonal antibodies (mAbs) (1D10, a kind gift of Dr Andrew Brooks, University of Melbourne, Melbourne, Australia) was visualized with phycoerythrin-labelled F(ab′)2 fragments of goat anti-mouse Ig (Dako, Carpinteria, CA, USA). Samples were analysed using a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA).
Natural killer cell isolation and propagation
Primary polyclonal NK cells were prepared from healthy adult donors, as described previously [27]. Pure NK cell populations (> 95% CD3-, CD56+) were obtained from negative selection (Miltenyi Biotec, Gladbach, Germany) and were monitored periodically for the presence of T cells. All the NK cells expressed the activating receptors, NKG2D; mAbs to CD3 and CD56 were purchased from BD Pharmingen (San Diego, CA, USA) and mAb to NKG2D was purchased from R&D Systems (Minneapolis, MN, USA).
Cytotoxic assay
The cytolytic activity of NK cells against transfected C1R cells expressing MICB*014, mutated MICB*014-Lys80, mutated MICB*014-Asp136 and untransfected target cell lines was assessed in the 4-h 51Cr release assay [28]. Target cells were labelled with sodium chromate (Na51Cr2O4; Invitrogen) and incubated for 4 h with NK cells in triplicate at various effector-to-target ratios. In experiments with antibody blocking, NK cells were preincubated with different amounts of the anti-NKG2D mAbs (0·3, 1, 3, 10 μg) for 1 h before addition to the target cells. Specific lysis was determined using the formula: % lysis = 100 × [(mean experimental cpm − mean spontaneous cpm)/(mean maximum cpm − mean spontaneous cpm)]. The spontaneous release of 51Cr was < 25% (on average ∼ 10%) of the maximal release. The maximum release value was determined from target cells treated with 5% (v/v) Triton X-100 (Sigma-Aldrich).
Statistical analysis
Phenotype frequencies of MICB were determined by number of alleles in samples/number of samples. Alleles and amino acids in case–control associations were evaluated on phenotype frequencies using the χ2 or Fisher's exact tests by spss version 9 (SPSS, Inc., Chicago, IL, USA). A statistical correction in phenotype frequencies was applied for multiple comparison according to the number of alleles tested at each locus (correction factor = 9). Significance of the differences in the distributions of MICB alleles among low and high grades of the CXCA and control groups and HPV typing was tested by χ2 as indicated by P-value.
Results
Major histocompatibility complex class I chain-related gene B genotyping in healthy NETF and CXCA patients
Typing of 15 MICB alleles and one group of alleles could be validated using our described PCR–SSP method with 29 primer mixes. The interpretation diagram for typing is shown in Fig. 1. Comparison of MICB allele distributions between 99 CXCA patients and 100 NETF is listed in Table 3. For NETF, seven MICB alleles and one group of alleles were found, of which the four most common MICB alleles were MICB*00502 (31·0%), MICB*002/007 (29·5%), MICB*004 (11·5%) and MICB*008 (8·5%) respectively.
Table 3.
The distributions of major histocompatibility complex class I chain-related gene B (MICB) alleles in north-eastern Thai females (NETF) and cervical cancer (CXCA).
| NETF (n = 100) | CXCA (n = 99) | ||||||
|---|---|---|---|---|---|---|---|
| MICB | n | % PF | n | % PF | P-value | Pc | Odds ratio |
| 00502 | 62 | 62·0 | 34 | 34·3 | 0·0001 | 0·0009 | 0·32 |
| 002/007 | 59 | 59·0 | 53 | 53·5 | 0·44 | ||
| 004 | 23 | 23·0 | 26 | 26·3 | 0·59 | ||
| 008 | 17 | 17·0 | 19 | 19·2 | 0·69 | ||
| 014 | 7 | 7·0 | 13 | 13·1 | 0·15 | ||
| 00503 | 6 | 6·0 | 6 | 6·1 | 0·99 | ||
| 013 | 4 | 4·0 | 8 | 8·1 | 0·23 | ||
| 003 | 1 | 1·0 | 3 | 3·0 | 0·31 | ||
| 00501 | 0 | 0·0 | 6 | 6·1 | 0·0124 | 0·1119 | |
| Blank† | 21 | 21·0 | 30 | 30·3 | 0·13 | ||
Blank demonstrates the percentage of homozygous alleles in the populations.
Association of MICB and CXCA
According to the allele distribution in CXCA patients, eight MICB alleles and one group of alleles were identified. Among these alleles, MICB*00501 was found only in CXCA patients with a frequency of 6·1% (P-value = 0·012). However, when the corrected P-value was calculated, no distinct allele was associated significantly positively with CXCA. Interestingly, MICB*00502 was associated negatively with CXCA, suggesting a protective allele with a corrected P-value of 0·0009 and an odds ratio of 0·32, as shown in Table 3. These data were re-analysed and it was found that statistically significant differences of the MICB allele distributions among low and high grades of CXCA and NETF were observed in MICB*00502. There were statistically significant differences in the distributions of this allele between NETF and low-grade (P-value < 0·001), and between NETF and high-grade cancer (P-value = 0·013). However, the differences of percentage PF between low and high grades of CXCA were not significant (P-value = 0·064). The data reconfirmed that MICB*00502 was associated negatively with the disease, but not stages, of CXCA. In addition, the association between the MICB alleles and HPV typing status of patients, i.e. positive or negative HPV PCR, was not significant (P-value = 0·960) (data not shown).
Association of amino acid substitutions of MICB and CXCA
According to the reported alleles, there were 12 amino acid substitutions in the α1, α2 and α3 domains but only six substitutions were identified in the north-eastern Thai population containing D75N, E80K, I121M, N136D, T212I and G291S (Table 4). The MICB*00501, 00502 and 00503 alleles carry synonymous amino acid substitutions. Lys80 and Asp136 were associated negatively with CXCA (P-value = 0·0021, odds ratio = 0·3134; Table 4). It should be noted that MICB*00502 carries Lys80 and Asp136. Thus, this result is in agreement with the allele association study, suggesting that these amino acid substitutions may bind strongly to NKG2D, leading to stronger immune activation.
Table 4.
The amino acid substitutions of major histocompatibility complex B class I chain-related gene B (MICB) alleles in north-eastern Thai females (NETF) and cervical cancer (CXCA).
| NETF (n = 100) | CXCA (n = 99) | |||||
|---|---|---|---|---|---|---|
| Position | n | % PF | n | % PF | P-value | Odds ratio |
| Aspartic75 | 96 | 96·0 | 95 | 96·0 | 0·99 | |
| Asparagine75 | 22 | 22·0 | 26 | 26·3 | 0·48 | |
| Glutamic80 | 80 | 80·0 | 70 | 70·7 | 0·08 | |
| Lysine80 | 89 | 89·0 | 71 | 71·7 | 0·0021 | 0·31 |
| Isoleucine121 | 97 | 97·0 | 99 | 100·0 | 0·08 | |
| Methionine121 | 17 | 17·0 | 19 | 19·2 | 0·69 | |
| Aspartic136 | 89 | 89·0 | 71 | 71·7 | 0·0021 | 0·31 |
| Asparagine136 | 80 | 80·0 | 70 | 70·7 | 0·13 | |
| Isoleucine212 | 1 | 1·0 | 2 | 2·0 | 0·55 | |
| Glycine291 | 99 | 99·0 | 94 | 94·9 | 0·09 | |
| Serine291 | 10 | 10·0 | 20 | 20·2 | 0·05 | |
Amino acid substitutions of MICB affecting NKG2D induced NK cell activation
To investigate which of these positions affected NKG2D binding and activation, plasmid DNAs encoding the MICB*014 gene was constructed by the pcDNA 3·1 Directional TOPO Expression kit. Site-directional mutagenesis was then performed to mutate amino acid position 80 from glutamic acid to lysine, called MICB*014-Lys80, and position 136 from asparagine to aspartic acid, called MICB*014-Asp136 (Table 2). These plasmid DNAs were transfected into the C1R cells to produce stable transfectants expressing the three MICB proteins. By staining with anti-MICA/B mAb (1D10), all transfectants expressed MICB on the cell surface but at different levels (Fig. 2a). In addition, the NKG2D-Ig fusion protein bindings varied substantially, with MICB*014-Asp136 transfectants representing strong binding, whereas MICB*014 and MICB*014-Lys80 were weaker (Fig. 2b). When transfected cells were cultured with NK cells and measured for the cytolytic activity using the chromium release assay, both MICB*014 and mutants could comparably induce NK cells to kill target cells (Fig. 3a). To determine whether the different binding capacity of MICB and mutated MICB to NKG2D would affect NK cell activity the MICB transfectants were co-cultured with NK cells pretreated by anti-NKG2D at different concentrations, from 0·3 to 10 μg of anti-NKG2D, to block the killing activity caused by NKG2D. Transfected cells carrying MICB*014-Asp136 was blocked by anti-NKG2D less efficiently than those carrying MICB*014-Lys80 and MICB*014 (Fig. 3b). It should be noted that MICB*014-Lys80 and MICB*014 were expressed at a comparable level on transfected cells (Fig. 2), with MICB*014-Lys80 transfectants showing a slightly weaker staining pattern. However, MICB*014 transfectants could not induce killing activity when the NK cells were pretreated with anti-NKG2D (Fig. 3). In contrast, MICB*014-Asp136 induced elevated killing activities at all concentrations of anti-NKG2D tested. This result suggested that although MICB*014 wild-type and mutants could induce NK cell killing comparably, aspartic acid at position 136, affecting NK cell-killing activity, could be revealed only by anti-NKG2D blocking experiments. This motif was carried by MICB*00502, which was associated negatively with CXCA.
Fig. 2.
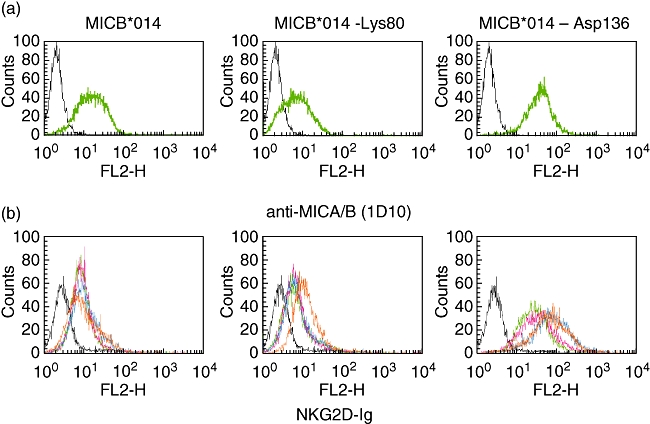
Differential expression and binding of NKG2D by allelic variants of major histocompatibility complex class I chain-related gene B (MICB). Transfected C1R cells were stained with anti-MICA/B (a) and different concentrations of NKG2D-immunoglobulin (Ig) fusion protein (green, pink, blue and orange lines represent 20, 30, 40 and 50 μg/ml of NKG2D-Ig respectively) (see pdf online for colour) (b) to detect the MICB expression on the cell surface and NKG2D binding capacity. MICB*014-Asp136 could be expressed on the cell surface higher than MICB*014 or MICB*014-Lys80, and thus could bind to NKG2D-Ig more than other clones respectively.
Fig. 3.
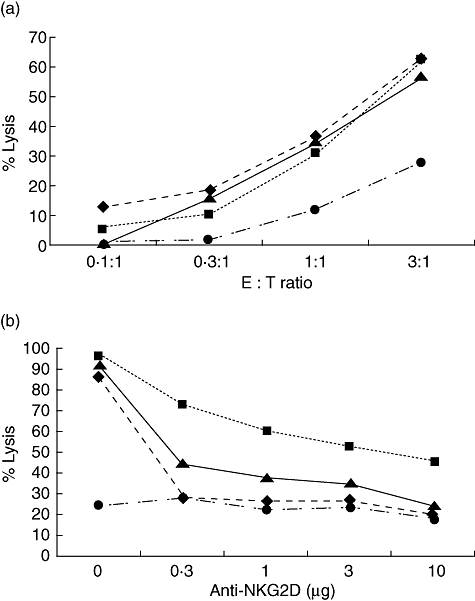
Anti-NKG2D could inhibit MICB*014-Asp136 less efficiently in NKG2D-mediated lysis. (a) Major histocompatibility complex class I chain-related gene B (MICB) transfected cells were cultured with natural killer (NK) cells. Both MICB*014 wild-type and mutants could induce NK cells to kill target cells. (b) MICB transfected C1R cells were cultured with NK cells treated with different concentrations of anti-NKG2D at an effector : target ratio of 3 : 1. Although MICB*014 (▴), MICB*014-Lys80 (♦) and MICB*014-Asp136 (▪) could induce NK cells to kill target cells compared with untransfected cells (•), MICB*014-Asp136 needed a high concentration of anti-NKG2D to block the activity of NK cells.
Discussions
In this study, we have described the distribution of MICB alleles in NETF using PCR–SSP modified from Ahmad et al. [22], Gonzalez et al. [23] and Collins et al. [24] for typing of 15 MICB alleles and one group of alleles. However, this set could not discriminate MICB*002 and MICB*007 and could not identify MICB*010, which carries polymorphism in exon 6. Because PCR–SSP is recognized generally to be a highly robust and reproducible technique, the PCR–SSP MICB typing system described here will allow amplification of all MICB alleles and their possible combination as heterozygous alleles under identical conditions. Control DNA samples were obtained from homozygous typed cell lines (HTCLs) for method validation. The specificity of all MICB amplifications was verified using a set of oligoprimers combined in 29 mixtures of PCR reactions. During the development of a strategy for MICB typing by PCR–SSP, the similar result of inability to amplify MICB*001 and MICB*00504 using reference HTCLs (HELA and RML cell lines) confirmed previous observations by Gonzalez et al. [23] and Schroeder et al. [29] that the sequences of these alleles were artefacts. Thus, only 14 from 15 MICB alleles and one group of alleles were typed successfully. It is essential that every primer mix should be validated with standard DNA to ensure the absence of multiple amplifications and specificity. This is due to the multi-copy nature of the MIC gene family [12,13]. To avoid this situation, in some cases a first PCR was performed to amplify specifically a MICB fragment carrying exons 2–4 (Wongsena et al. in preparation). Then, the MICB typing was performed using this PCR product as a template.
The data of MICB distribution in our population are in agreement with a previous report on a Spanish population [23], that alleles MICB*00502 (31·0%), MICB*002/007 (29·5%), MICB*004 (11·5%) and MICB*008 (8·5%) are the most frequent alleles. Four alleles, MICB*006, MICB*009N, MICB*011 and MICB*012, have not been detected in both the CXCA group and our control population group. Even though MICB*009N can be found in Japanese and east Asians [30], the absence of this and other alleles suggests that these alleles are also rare in our population, as reported previously in Spain [23]. Interestingly, MICB*00502 represented the most common allele found in the control population and was associated negatively with CXCA (Pc = 0·0009), suggesting a protective MICB allele.
Major histocompatibility complex class I chain-related gene B is represented polymorphically by 18 alleles that variably include 12 bi-allelic amino acid substitutions with fairly random distribution in the extracellular domains [15–21]. However, there has been no evidence for functional significance of this allelic variation. Our study showed that two amino acid substitutions of MICB found in MICB*00502 (E80K and N136D) were associated negatively with CXCA. MICB*00501 and MICB*00503 also shared these substitutions, but they were not found to be associated with CXCA. This may be because they were rare in our population. Although some MICB motifs have been predicted to be involved in NKG2D binding [31], these two non-synonymous amino acid substitutions were not included. It is conceivable that these two amino acid changes could affect protein structure and/or function, as they are non-conservative. We have tested these two substitutions functionally regarding NK cell activation. Apparently, MICB*014 with aspartic acid at 136 was expressed at a higher level when compared with MICB*014 wild-type or MICB*014-Lys80. This could be the effect of stable transfectant selection or the gene sequence affecting expression level [32]. The NKG2D binding capability of these MICB proteins was in accordance with protein expression detected by antibody. Interestingly, these MICB proteins could, comparably, induce cytolytic activities of NK cells but differently in the anti-NKG2D blocking experiments. NK cells were treated with anti-NKG2D and washed before incubation with target cells. Thus, the NKG2D receptors on NK cells should be blocked at the same level regardless of the different expressions of MICB on transfectants. Consequently, the different cytolytic activities should be resulted from the kinetic differences reflected by binding affinity between NKG2D and MICB or MICB mutants, suggesting that aspartic acid at position 136 could probably induce NK cell-killing more efficiently. It is also possible that the level of MICB expression may contribute to this phenomenon and that the polymorphic residue at 136 has no effect on NK cell activation. A more sensitive method, such as the use of the Biacore system, may be needed to evaluate these differences. However, our study is the first report on the functional polymorphism of MIC proteins on NK cell activation by not only NKG2D binding [33].
In conclusion, our typing method of MICB alleles enabled us to show the overview picture of the MICB distribution in our population. The associations of MICA and haplotype analysis of MICA and MICB are presented elsewhere (Jumniansong et al. unpublished data). No distinct MICB allleles with strong positive associations indicating a risk factor for CXCA were identified, but MICB*00502 had a significantly negative association, indicating a protective allele. This allele carries an aspartic acid at position 136 that may affect expression and NKG2D binding, leading to a higher degree of NK cell-killing activities.
Acknowledgments
We are most grateful to Dr Hugh Reyburn and Dr Mar Vales-Gomez, Department of Pathology, University of Cambridge, Cambridge, UK who assisted and supported the functional experiments. The MICA/B antibody was kindly provided by Dr Andrew Brooks, University of Melbourne, Australia. We are grateful to Miss Chiraporn Chonanant, Master's candidate of the Medical Science Program, Graduate School, Khon Kaen University, the tumour clinic team, Srinagarind Hospital and department of pathology for technical assistance, clinical sample recruitment, diagnosis and clinical data. A. J. was a Royal Golden Jubilee (RGJ) scholarship (PHD/0035/2544) supported by Thailand Research Fund (TRF). C. L. was a Thailand Research Fund Scholar (RSA/5/2543) supported by the Thailand Research Fund (TRF). This work was supported by RGJ (BGJ47K0016) and the Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences, Khon Kaen University, Thailand.
References
- 1.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37:4–66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 2.Pisani P, Parkin DM, Bray F, Ferlay J. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer. 1999;83:18–29. doi: 10.1002/(sici)1097-0215(19990924)83:1<18::aid-ijc5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 3.Vatanasapt V, Sriamporn S, Vatanasapt P. Cancer control in Thailand. Jpn J Clin Oncol. 2002;32:S82–91. doi: 10.1093/jjco/hye134. [DOI] [PubMed] [Google Scholar]
- 4.Munoz N, Bosch FX, de Sanjose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 5.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–50. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci USA. 1999;96:6879–84. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer S, Groh V, Wu J, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Song Y, Bakker AB, et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–2. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Groh V, Spies T. T cell antigen receptor engagement and specificity in the recognition of stress-inducible MHC class I-related chains by human epithelial gamma delta T cells. J Immunol. 2002;169:1236–40. doi: 10.4049/jimmunol.169.3.1236. [DOI] [PubMed] [Google Scholar]
- 10.Groh V, Bruhl A, El-Gabalawy H, Nelson JL, Spies T. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc Natl Acad Sci USA. 2003;100:9452–7. doi: 10.1073/pnas.1632807100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda M, Shadeo A, MacFadyen AM, Takei F. CD1d-independent NKT cells in beta 2-microglobulin-deficient mice have hybrid phenotype and function of NK and T cells. J Immunol. 2004;172:6115–22. doi: 10.4049/jimmunol.172.10.6115. [DOI] [PubMed] [Google Scholar]
- 12.Leelayuwat C, Townend DC, Degli-Esposti MA, Abraham LJ, Dawkins RL. A new polymorphic and multicopy MHC gene family related to nonmammalian class I. Immunogenetics. 1994;40:339–51. doi: 10.1007/BF01246675. [DOI] [PubMed] [Google Scholar]
- 13.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1994;91:6259–63. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors HLA system, 2004. Hum Immunol. 2005;66:571–636. doi: 10.1016/j.humimm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Ando H, Mizuki N, Ota M, et al. Allelic variants of the human MHC class I chain-related B gene (MICB) Immunogenetics. 1997;46:499–508. doi: 10.1007/s002510050311. [DOI] [PubMed] [Google Scholar]
- 16.Bahram S, Shiina T, Oka A, Tamiya G, Inoko H. Genomic structure of the human MHC class I MICB gene. Immunogenetics. 1996;45:161–2. doi: 10.1007/s002510050184. [DOI] [PubMed] [Google Scholar]
- 17.Pellet P, Renaud M, Fodil N, et al. Allelic repertoire of the human MICB gene. Immunogenetics. 1997;46:434–6. doi: 10.1007/s002510050299. [DOI] [PubMed] [Google Scholar]
- 18.Visser CJ, Tilanus MG, Schaeffer V, et al. Sequencing-based typing reveals six novel MHC class I chain-related gene B (MICB) alleles. Tissue Antigens. 1998;51:649–52. doi: 10.1111/j.1399-0039.1998.tb03008.x. [DOI] [PubMed] [Google Scholar]
- 19.Fischer G, Arguello JR, Perez-Rodriguez M, et al. Novel intronic variants of MICB (MHC class I chain-related gene B) Eur J Immunogenet. 1999;26:399–404. doi: 10.1046/j.1365-2370.1999.00182.x. [DOI] [PubMed] [Google Scholar]
- 20.Fischer G, Arguello JR, Perez-Rodriguez M, et al. Sequence-specific oligonucleotide probing for MICB alleles reveals associations with MICA and HLA-B. Immunogenetics. 2000;51:591–9. doi: 10.1007/s002510000179. [DOI] [PubMed] [Google Scholar]
- 21.Fischer G, Perez-Rodriguez M, Arguello JR, et al. Three novel MICB alleles. Tissue Antigens. 2000;55:166–70. doi: 10.1034/j.1399-0039.2000.550210.x. [DOI] [PubMed] [Google Scholar]
- 22.Ahmad T, Marshall SE, Mulcahy-Hawes K, et al. High resolution MIC genotyping: design and application to the investigation of inflammatory bowel disease susceptibility. Tissue Antigens. 2002;60:164–79. doi: 10.1034/j.1399-0039.2002.600207.x. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez S, Rodriguez-Rodero S, Martinez-Borra J, Lopez-Vazquez A, Rodrigo L, Lopez-Larrea C. MICB typing by PCR amplification with sequence specific primers. Immunogenetics. 2003;54:850–5. doi: 10.1007/s00251-002-0533-x. [DOI] [PubMed] [Google Scholar]
- 24.Collins RW, Stephens HA, Clare MA, Vaughan RW. High resolution molecular phototyping of MICA and MICB alleles using sequence specific primers. Hum Immunol. 2002;63:783–94. doi: 10.1016/s0198-8859(02)00425-1. [DOI] [PubMed] [Google Scholar]
- 25.Leelayuwat C, Srisuk T, Paechaiyaphum R, Limpaiboon T, Romphruk A. Production and evaluation of Taq DNA polymerase. J Med Assoc Thai. 1997;80:S129–37. [PubMed] [Google Scholar]
- 26.Vales-Gomez M, Browne H, Reyburn HT. Expression of the UL16 glycoprotein of human cytomegalovirus protects the virus-infected cell from attack by natural killer cells. BMC Immunol. 2003;4:4–14. doi: 10.1186/1471-2172-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandelboim O, Reyburn HT, Vales-Gomez M, et al. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–22. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chisholm SE, Reyburn HT. Recognition of vaccinia virus-infected cells by human natural killer cells depends on natural cytotoxicity receptors. J Virol. 2006;80:2225–33. doi: 10.1128/JVI.80.5.2225-2233.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroeder MEH, Kim TD, Blasczyk R. Eight novel MICB alleles, including a null allele identified in gastric MALT lymphoma pateints. Tissue Antigens. 2004;64:276–80. doi: 10.1111/j.1399-0039.2004.00286.x. [DOI] [PubMed] [Google Scholar]
- 30.Komatsu-Wakui M, Tokunaga K, Ishikawa Y, et al. Wide distribution of the MICA-MICB null haplotype in East Asians. Tissue Antigens. 2001;57:1–8. doi: 10.1034/j.1399-0039.2001.057001001.x. [DOI] [PubMed] [Google Scholar]
- 31.Holmes MA, Li P, Petersdorf EW, Strong RK. Structural studies of allelic diversity of the MHC class I homolog MIC-B, a stress-inducible ligand for the activating immunoreceptor NKG2D. J Immunol. 2002;169:1395–400. doi: 10.4049/jimmunol.169.3.1395. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Groh V, Strong RK, Spies T. A single amino acid substitution causes loss of expression of a MICA allele. Immunogenetics. 2000;51:246–8. doi: 10.1007/s002510050039. [DOI] [PubMed] [Google Scholar]
- 33.Steinle A, Li P, Morris DL, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53:279–87. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]


