Abstract
Infection with Salmonella enterica serovar Typhimurium (S. Typhimurium) causes a severe and lethal systemic disease in mice, characterized by poor activation of the adaptive immune response against Salmonella-derived antigens. Recently, we and others have reported that this feature relies on the ability of S. Typhimurium to survive within murine dendritic cells (DCs) and avoid the presentation of bacteria-derived antigens to T cells. In contrast, here we show that infection of murine DCs with either S. Typhi or S. Enteritidis, two serovars adapted to different hosts, leads to an efficient T-cell activation both in vitro and in vivo. Accordingly, S. Typhi and S. Enteritidis failed to replicate within murine DCs and were quickly degraded, allowing T-cell activation. In contrast, human DCs were found to be permissive for survival and proliferation of S. Typhi, but not for S. Typhimurium or S. Enteritidis. Our data suggest that Salmonella host restriction is characterized by the ability of these bacteria to survive within DCs and avoid activation of the adaptive immune response in their specific hosts.
Keywords: dendritic cells, host restriction, Salmonella, T cell
Introduction
Dendritic cells (DCs) are a key link between innate and adaptive immunity. With their unique ability to prime antigen-specific naïve CD4+ and CD8+ T cells, DCs are critical for initiating adaptive immunity in response to infection by pathogenic bacteria.1,2 As a result of the detection of pathogen-associated molecular patterns (PAMPs) expressed by bacteria, DCs residing in peripheral tissues (i.e. intestinal and respiratory mucosa) become activated and experience functional changes, as part of a process known as DC maturation.3–6 Along with maturation, DCs acquire an enhanced capacity to process and present antigens on major histocompatibility complex (MHC) molecules7,8 and up-regulate the surface expression of costimulatory molecules, such as CD80, CD86 and CD40. Increased surface expression of peptide–MHC (pMHC) complexes and costimulation molecules empower DCs for priming pMHC-specific naïve T cells.1,9–11 Furthermore, maturation also increases the capacity of DCs to migrate from peripheral sites of infection to regional lymph nodes where naïve T cells normally reside.12,13
The DCs are critical components for the initiation of an effective immune response against infection so several microbial pathogens have evolved molecular mechanisms aimed at interfering with DC function.2,14 Recent studies have shown that virulent strains of Salmonella enterica serovar Typhimurium (herein S. Typhimurium) can interfere with the capacity of DCs to prime adaptive immunity against bacteria.15–20 This pathogen can survive within DCs in specialized vacuoles by delivering virulence effector proteins to the cytoplasm through a type 3 secretion system encoded in the Salmonella pathogenicity island 2 (SPI-2).15,17,20,21 SPI-2-encoded effector proteins delivered to the cytoplasm have been shown to interfere with normal vesicular trafficking in host cells.22 Consistent with this notion, recent studies indicate that vacuoles containing virulent S. Typhimurium deviate from the normal endocytic pathways and fail to colocalize with lysosomal markers in DCs.15,23 As a result, bacterial degradation and subsequent presentation to T cells of the bacteria-derived antigens on MHC molecules is prevented.15,16 These observations suggest that the capacity of S. Typhimurium to interfere with DC function could prevent the activation of T-cell-mediated immunity against antigens derived from this pathogen.24 Accordingly, infection with attenuated strains of S. Typhimurium that fail to interfere with DC function, induce strong T-cell responses in mice.12,15,16,25,26
S. Typhimurium is a member of the genus Salmonella enterica subspecies I whose members cause diseases in several warm-blooded animals.27,28 Some Salmonella enterica serovars are considered to be host generalists because they cause either systemic disease or gastrointestinal infection in several hosts.29S. Typhimurium and S. Enteritidis are examples of generalist serovars that cause disease in mice, chicken, calves and humans.30,31 In contrast, other S. enterica serovars cause systemic disease in only a few particular hosts and are rarely associated with diseases in other species. An example of this latter type is S. Typhi, the etiological agent of typhoid fever in humans. S. Typhi is a host-adapted serovar that only causes disease in humans and higher primates.32 Several studies have suggested that the host’s immune response could be a key component in Salmonella host restriction.29,33,34 For instance, S. Typhimurium can avoid adaptive immunity and cause a severe systemic disease in mice, but it seems unable to do this in other hosts.35,36 Supporting this notion is the observation that S. Typhimurium induces a strong immune response in other hosts, such as chickens and humans,37–39 and can only cause a systemic disease when cellular adaptive immunity is deficient in these hosts.40–42
Dendritic cells play a crucial role in initiating adaptive immunity in the host, capturing Salmonella at an early stage of infection, so interference with the ability of DCs to process and present bacterial antigens could be advantageous for Salmonella dissemination within its specific host. Accordingly, it is likely that Salmonella serovars that fail to cause disease within a specific host could also be unable to interfere with DC function, which would then result in the activation of the adaptive immune response and bacterial clearance.
In this study we evaluated the ability of three different Salmonella enterica serovars to interfere with the function of DCs and their capacity to prevent activation of adaptive immunity in the mouse. We observed that S. Typhi and S. Enteritidis failed to avoid antigen presentation by murine DCs and consequently specific T cells were activated both in vitro and in vivo in response to bacterial challenge. Accordingly, these two Salmonella serovars were unable to survive within murine DCs and vacuoles containing them colocalized with lysosomal markers. In contrast, in human DCs only S. typhi was shown to replicate, while S. Typhimurium and S. Enteritidis were readily degraded by these DCs. Our data suggest that the host specificity of Salmonella enterica serovars may be determined by the ability of these bacteria to interfere with DC function and avoid host adaptive immunity. We provide new evidence supporting a role for DCs as cellular components influencing pathogen–host specificity, a function not described before.
Experimental procedures
Mice
C57BL/6 mice were purchased and maintained at the Pontificia Universidad Católica de Chile animal facility (Santiago, Chile). OT-I and OT-II transgenic mice were a generous gift from Dr Ralph Steinman (The Rockefeller University, New York, NY). OT-I and OT-II are transgenic mice with specific T-cell receptors for H-2Kb/OVA257–264 and I-Ab/OVA323–339, respectively. All animal work was performed according to institutional guidelines.
Bacterial strains and DC cultures
Salmonella enterica serovars used in this study were: S. Typhimurium 14028s, S. Typhi STH2370 and S. Enteritidis phage type (PT) 1. The S. typhimurium was obtained from the American Type Culture Collection (ATCC, Manassas, VA). The S. Typhi and S. Enteritidis were clinical strains, isolated from infected Chilean patients: S. Typhi was obtained from the Lucio Córdova Hospital for Infectious Disease in Santiago, Chile, and S. Enteritidis was provided by the Instituto de Salud Pública, Chile. Recombinant Salmonella serovars for ovalbumin (OVA) were obtained by electrotransformation of S. Typhimurium, S. Typhi and S. Enteritidis with the plasmid pKK-OVA, which was provided by Jean-Claude Sirard (Pasteur Institute, Lille, France), and OVA expression for each serovar was detected by Western blot assay. As a loading control, OmpC protein was detected in each Western blot using a rabbit anti-OmpC serum, provided by Dr Alejandro Venegas (Pontificia Universidad Católica de Chile, Santiago, Chile). Bacteria were grown on Luria–Bertani broth and recombinant bacteria were selected on carbenicillin (50 μg/ml; Sigma-Aldrich, St Louis, MO).
Murine bone marrow-derived DCs were prepared as previously described15,20 Briefly, DCs were grown from bone marrow progenitors in RPMI-1640 containing 5% fetal calf serum, 1 mm pyruvate, 2 mm glutamine, 1 mm non-essential amino acids and 10 ng/ml of recombinant murine granulocyte–macrophage colony-stimulating factor (Peprotech, Rocky Hill, NJ). Cultures of DCs were routinely analysed by flow cytometry for expression of surface markers CD11c, I-Ab, H-2Kb, CD80, CD86 and CD40; revealing 60–70% of CD11c-positive cells with an immature phenotype, as previously described.15 Human DCs were prepared from buffy coats obtained from healthy blood donors from Hospital Clínico de la Universidad de Chile. Leucocytes were isolated by density gradient separation with Ficoll–Hypaque (Axis-Shield, Oslo, Norway). Cells (4 × 107/well) were incubated in serum-free AIM-V therapeutic medium (Gibco BRL, Div. of Invitrogen, Carlsbad, CA) at 37° in 5% CO2 for 2 hr in a six-well plate (Falcon Becton Dickinson, Hershey, PA). Non-adherent cells were removed and the remaining cells were incubated for 5 days in the presence of recombinant human interleukin-4 (rhIL-4) (500 U/ml) (US Biological, Swampscott, MA) and 800 U/ml granulocyte–macrophage colony-stimulating factor (Leucomax; Shering Plough, Brinny Co, Ireland). Cultures were maintained for 5 days, replacing the medium every 2 days. On day 5, cells were analysed by fluorescence-activated cell sorting (FACS) for quantification of CD11c-positive cells and used for the experiments. Between 80 and 90% of immature CD11c-positive cells were obtained in each preparation.
To evaluate the survival for each Salmonella serovar inside DCs, overnight bacteria cultures were subcultured starting at a 1/100 dilution until reaching exponential phase (OD600 = 0·6), then were washed twice with ice cold phosphate-buffered saline (PBS) and resuspended on RPMI-1640 medium supplemented with 5% fetal calf serum, without antibiotics. Day 5 DCs were infected with Salmonella serovars at a multiplicity of infection (MOI) of 50 for 1 hr, treated with 100 μg/ml gentamicin for 1hr to kill extracellular bacteria, washed and incubated for the indicated times with 50 μg/ml gentamicin. To recover intracellular bacteria, DCs were washed twice with ice-cold PBS, permeabilized for 30 min with 0·1% Triton X-100 in PBS and cellular lysates were plated on Luria–Bertani agar.
To evaluate the percentage of Salmonella-infected DCs at different time-points, infected DCs were permeabilized with PBS–bovine serum albumin (BSA) 3%–Saponin for 30 min and incubated with rabbit anti-Salmonella antibodies (Becton Dickinson, Francis Lakes, NJ) for 2 hr on ice. Then, cells were washed and incubated with phycoerythrin (PE)-conjugated anti-CD11c (Becton Dickinson Pharmingen) and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (FITC-IgG; Pierce, Rockford, IL) antibodies for 1 hr. Cells were washed and fixed on PBS–1% paraformaldehyde and analysed by flow cytometry. All acquisitions were performed on a FACScalibur cytometer (BD Biosciences, San José, CA) and analysed using WinMDI 2.8 software (The Scripps Research Institute, La Jolla, CA; http://facs.scripps.edu/help/html/read1ptl.htm).
Detection of bacteria-derived antigen presentation on cultured DCs
As previously described,15 day 5 DCs were pulsed for 1 hr with either wild-type or OVA-expressing Salmonella serovars at an MOI equal to 50. After the pulse, DCs were washed and treated for 1 hr with gentamicin (100 μg/ml) to eliminate extracellular bacteria. After 16 hr of culture in the presence of 50 μg/ml gentamicin, DC viability was determined by exclusion of trypan blue and different numbers of live DCs were cocultured with either 1 × 105 OT-I or 1 × 105 OT-II T cells, obtained from lymph nodes of transgenic mice.43,44 After 20 hr of DC–T-cell coculture, IL-2 release was measured by cytokine enzyme-linked immunosorbent assay (ELISA). Briefly, ELISA plates (NuncTM, Rochester, NY) were activated overnight at 4° with 65 ng/well of purified anti-mouse IL-2 (clone JES6-1A12, BD Biosciences Pharmingen) in 1 × PBS, blocked for 1 hr with 3% BSA in 1 × PBS at room temperature and incubated with 200 μl of cell-free supernatant from DC–T-cell cocultures, for 24 hr at 4°. Then, plates were incubated with 35 ng/well of biotin anti-mouse IL-2 (clone JES6-5H4, BD Biosciences Pharmingen) for 1 hr at room temperature, washed and incubated with streptavidin–horseradish peroxidase (BD Biosciences Pharmingen) for 1 hr at room temperature. 3-3′-5-5′-Tetramethyl-benzidine, at a final concentration of 100 μg/ml (Sigma-Aldrich), was used as a colorimetric substrate. The enzymatic reaction was stopped with 2 m H2SO4, and absorbance was recorded at 450 nm. For detection of H-2Kb complexes on the cellular surface, DCs were infected with non-recombinant or OVA-expressing Salmonella serovars at MOI equal to 50 for 1 hr. After this time, DCs were treated with 100 μg/ml gentamicin for 1 hr to kill extracellular bacteria, washed at 4° with ice-cold PBS, and incubated for 16 hr at 37° with 50 μg/ml gentamicin. Then, DCs were stained with anti-CD11c-PE and supernatant from 25-D1.16 hybridoma cells, which produce a monoclonal antibody that is specific for the H-2Kb/SIINFEKL complex (provided by Dr Ronald N. Germain).45 After washing, cells were stained with goat anti-mouse IgG-FITC (Pharmingen) and analysed by FACS. As a positive control, DCs were pulsed for 16 hr either with 10 μg/ml of purified OVA or 10 ng/ml of SIINFEKL peptide and treated as described above. These concentrations were obtained from titration curves performed on DCs (data not shown). All acquisitions were performed on a FACScalibur cytometer (BD Biosciences) and analysed using WinMDI 2.8 software (The Scripps Research Institute, La Jolla, CA. http://facs.scripps.edu/help/html/read1ptl.htm). The percentage of CD11c+ cells showing positive 25-D1.16 staining was derived from histograms for FITC fluorescence, in which a marker region was set based on the background staining observed for unpulsed DCs. Cells contained within the green fluorescence marker, were considered positive for 25-D1.16 staining.
Detection of bacterial antigen presentation in vivo
C57BL/6 mice that were 6–8 weeks old were infected in the footpad with 105 colony-forming units (CFUs) of OVA-expressing strains of Salmonella serovars. After 36 hr, popliteal lymph nodes were harvested and 1 × 106 cells were cocultured with either OT-I or OT-II transgenic T cells. After 20 hr of coculture, IL-2 release was measured as described above. For detection of H-2Kb/SIINFEKL complexes, cells derived from popliteal lymph nodes were washed twice with ice-cold PBS and incubated with anti-CD11c-PE and supernatant from 25-D1.16 hybridoma cells, then processed as described above and analysed by FACS.
In vivo T-cell proliferation
Single cell suspensions obtained from spleens derived either from OT-I or OT-II mice were stained with carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes, Invitrogen, Carlsbad, CA) and injected intravenously to syngeneic wild-type recipient mice. A total of 1 × 106 Vα2+ CD8+ (for OT-I) or Vα2+ CD4+ (for OT-II) CFSE+ cells were injected into recipient animals. After 24 hr, 1 × 105 CFU of OVA-expressing Salmonella serovars were injected intravenously into recipient animals. Spleens and lymph nodes were extracted from recipient mice 3 days after bacterial injection and OT-I or OT-II cell proliferation was evaluated by detecting the dilution of CFSE staining in CD8+ and CD4+ populations, respectively.
Confocal and electron microscopy
To evaluate colocalization analyses between Salmonella and lysosomal markers, DCs from 5-day cultures were seeded on round coverslips (400 000 cells/ml) in complete RPMI medium and infected with S. Typhi wild-type or S. Typhimurium or S. Enteritidis transformed with the plasmid pGFP (MOI equal to 50). After 1 hr of infection, DCs were washed twice with cold PBS and then incubated for an additional hour with complete RPMI medium supplemented with 100 μg/ml gentamicin to kill extracellular bacteria. Then, the culture medium was replaced with complete RPMI medium supplemented with 50 μg/ml gentamicin and the DCs were incubated for a further 14 hr. Afterwards, the DCs were fixed and permeabilized for 10 min at − 20° with methanol (100%) and blocked for 3 hr with PBS–BSA 3%. Salmonella typhi-specific mouse polyclonal antiserum was produced to detect S.typhi inside DCs and was used at dilution 1/300 to 1/750 in PBS–BSA 1%. Then, DCs were incubated for 2 hr with 1/500 goat anti-mouse IgG-FITC (Pierce). For the detection of LAMP-2, covers were incubated for 12 hr with 1/50 rabbit anti-LAMP-2 (Zymed, Invitrogen, Carlsbad, CA) on PBS–BSA 1% at 4°, washed three times with PBS–BSA 1% and incubated with 1/50 goat anti-rabbit IgG-rhodamine (Santa Cruz Biotechnology, Santa Cruz, CA) for 2 hr at 4°. After incubation, covers were washed three times, mounted on microscope slices and analysed on Fluoview FV100 Olympus confocal microscope. Fluorescence extension analyses were performed using the image-pro plus® software. Quantitative analyses were performed counting the number of intracellular bacteria that showed colocalization with LAMP-2 in several random fields (at least 50 cells were analysed per experiment). Colocalization was considered positive when red and green fluorescence histograms overlapped by more than 50%.
For electron microscopy analyses, DCs were infected for 24 hr with Salmonella serovars, fixed overnight in PLP (4% Paraformaldehyde, 0·01 m periodate and 0·2 m l-lysine on 0·1 m phosphate buffer, pH 7·4). Samples were rinsed in distilled water and post-fixed for 30 min at 4° in 1% osmium tetroxide, dehydrated in ethanol and acetone, and embedded in Epon. Thin sections were cut with an OmU2 Reichert Ultramicrotome and were observed under a Phillips Tecnai 21 electron microscope. Quantification of intact bacteria residing inside DCs was performed by counting at least 100 cells per experiment (in two independent experiments).
Results
S. Typhi and S. Enteritidis fail to evade antigen presentation in murine DCs
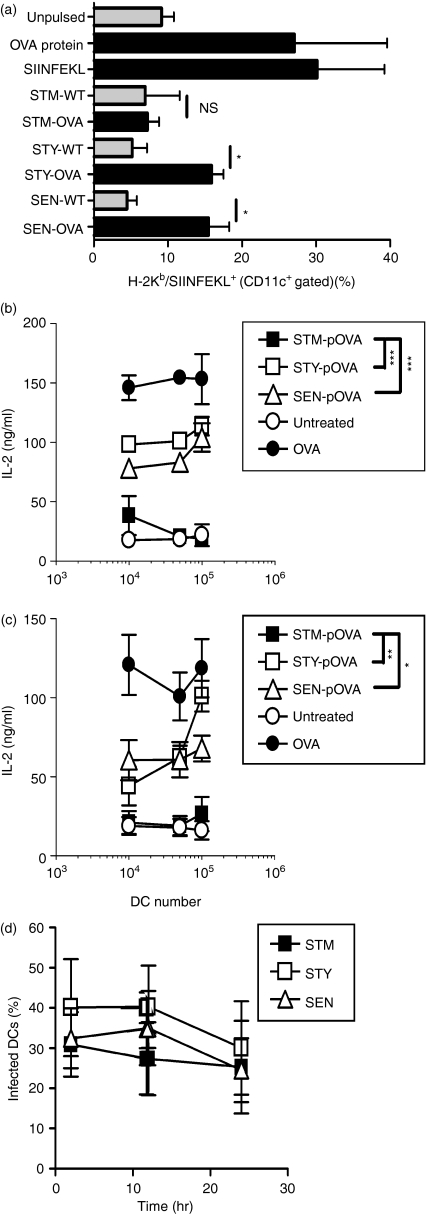
We evaluated whether S. Typhi and S. Enteritidis were able to avoid antigen presentation by murine DCs, as previously described for S. Typhimurium.16,19,20 To track OT-I and OT-II T-cell activation, we generated S. Typhimurium, S. Typhi and S. Enteritidis strains recombinant for the OVA antigen. Western blot assays demonstrated that each serovar expressed similar amounts of the OVA antigen (Fig. 1). Murine DCs were infected either with OVA-expressing S. Typhimurium, S. Typhi or S. Enteritidis and after 16 hr the density of H-2Kb/SIINFEKL complexes was measured on the surface of murine DC using a monoclonal antibody that binds specifically to this pMHC complex.45 As a positive control we used DCs pulsed either with 10 μg/ml purified OVA or 10 ng/ml SIINFEKL peptide. At these antigen concentrations, approximately 40% of DCs were positive for 25-D1.16 staining. As shown in Fig. 2a, H-2Kb/SIINFEKL complexes were detected on the surface of DCs infected either with OVA-expressing S. Typhi or S. Enteritidis. In contrast, H-2Kb/SIINFEKL complexes could not be detected on the surface of murine DCs infected with OVA-expressing S. Typhimurium, which showed similar levels to uninfected cells (Fig. 2a). These data suggest that although murine DCs fail to process and present antigens expressed by S. Typhimurium on MHC-I molecules, they can efficiently do so for antigens expressed by S. Typhi and S. Enteritidis. Consistent with these data was the observation that murine DCs infected with OVA-expressing S. Typhi and S. Enteritidis were able to activate H-2Kb/OVA-specific and I-Ab/OVA-specific transgenic T cells (OT-I and OT-II T cells, respectively, Fig. 2b and c). In contrast, neither OT-I nor OT-II T cells were activated by DCs infected with OVA-expressing S. Typhimurium (Fig. 2b and c). The differences observed in the antigen presentation assays shown in Fig. 2 for each Salmonella serovar were not the result of differential entrance by bacteria to DCs. As shown in Fig. 2d, equivalent numbers of DCs were infected by each serovar at a given time-point.
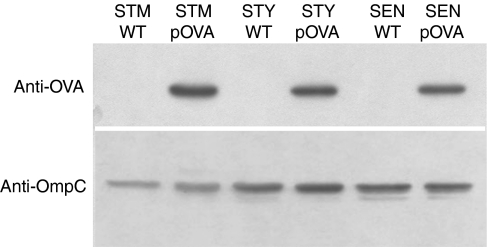
Figure 1.
Salmonella serovars express similar amount of recombinant ovalbumin (OVA). S. Typhimurium (STM), S. Typhi (STY) and S. Enteritidis (SEN) were transformed with the plasmid pKK-OVA and the expression of recombinant OVA was tested by Western blot. As a loading control, the expression of the OmpC protein (porin) was also tested in all strains. As a negative control, wild-type strains for each serovar were included.
Figure 2.
Antigens derived from S. Typhi (STY) and S. Enteritidis (SEN) are presented on major histocompatibility complex class I (MHC-I) and MHC-II molecules by murine dendritic cells (DCs) to specific T cells. (a) Murine DCs were infected with ovalbumin (OVA)-recombinant-Salmonella serovars (multiplicity of infection 50) for 1 hr, washed and treated with gentamicin (50 μg/ml) to kill extracellular bacteria. After 16 hr of gentamicin treatment, DCs were incubated with a monoclonal antibody against H-2Kb/SIINFEKL complex, washed, stained with a secondary fluorescein isothiocyanate (FITC) -conjugated anti-mouse immunoglobulin G (IgG) antibody and analysed by fluorescence-activated cell sorting. The graph shows the percentage of murine CD11c+ cells positive for H-2Kb/SIINFEKL, after infection with OVA-expressing S. Typhimurium (STM), S. Typhi or S. Enteritidis. As a positive control, DCs were pulsed with 10 μg/ml OVA or 10 ng/ml SIINFEKL peptide. As a negative control, uninfected DCs or DCs infected with parental Salmonella strains (non-recombinant for OVA) were included. All data acquisition was compensated for background using unpulsed cells stained only with secondary FITC-labelled anti-mouse IgG antibody. Data shown are means of five independent experiments and error bars represent SE (Student’s t-test: *P = 0·02). (b,c) Murine DCs were pulsed for 1 hr with OVA-expressing S. Typhimurim, S. Typhi, S. Enteritidis or purified OVA (100 μg) and treated with gentamicin for 16 hr to kill extracellular bacteria. Then, DCs were cocultured with OT-I T cells, specific for H2-Kb/SIINFEKL (b) or with OT-II T cells, specific for I-Ab/ ISQAVHAAHAEINEAGR (c). After 20 hr of coculture, interleukin-2 (IL-2) release was measured on culture media by enzyme-linked immunosorbent assay. Data shown are means of three independent experiments and error bars represent SE (Student’s t-test: ***P < 0·0001; **P = 0·002, *P = 0·02). (d) Percentage of DCs infected by each Salmonella serovar at 2, 12 and 24 hr after bacterial challenge. Percentage of Salmonella-infected DCs was determined by flow cytometry as described in Experimental procedures. Data shown in the graph are means (± SE) for the fractions of CD11c/FITC double-positive cells obtained for each time-point in three independent experiments.
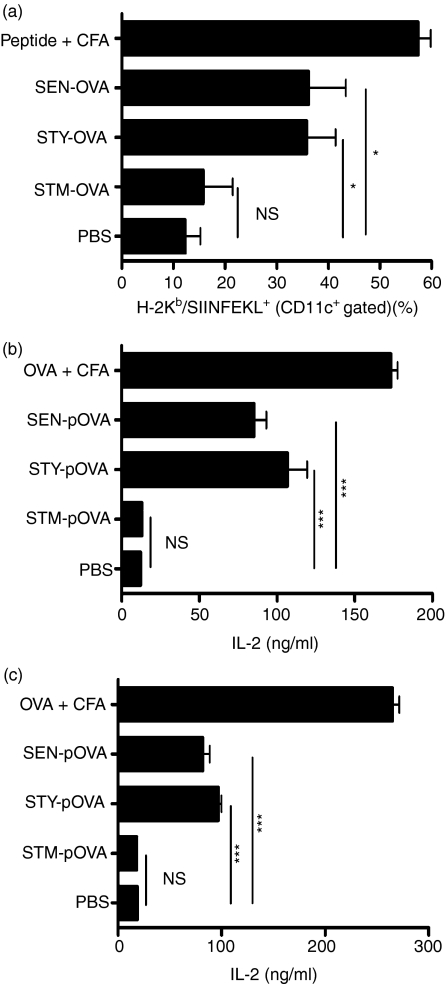
Additionally, we evaluated the antigen presentation capacity of antigen-presenting cells (APCs) obtained from draining lymph nodes of mice challenged either with OVA-expressing S. Typhi, S. Typhimurium or S. Enteritidis measuring the presence of H-2Kb/SIINFEKL complexes on the surface of CD11c+ APCs. As shown in Fig. 3a, measurable amounts of H-2Kb/SIINFEKL complexes could be observed only for CD11c+ APCs obtained from lymph nodes derived from mice infected with OVA-expressing S. Typhi and S. Enteritidis. Furthermore, only APCs obtained from mice infected with OVA-expressing S. Typhi or S. Enteritidis could activate OT-I and OT-II transgenic T cells (Fig. 3b and c). Neither H-2Kb/SIINFEKL complexes (Fig. 3a) nor activation of OT-I and OT-II transgenic T cells (Fig. 3b and c) could be detected for APCs obtained from lymph nodes derived from mice infected with OVA-expressing S. Typhimurium. These data suggest that S. Typhi and S. Enteritidis fail to avoid antigen presentation by murine DCs and this leads to T-cell activation.
Figure 3.
S. Typhi-derived and S. Enteritidis-derived antigens are presented on major histocompatibility complex class I (MHC-I) and MHC-II molecules in vivo by antigen-presenting cells (APCs) to specific T cells. Mice were infected in each footpad with 1 × 105 colony-forming units (CFU) and after 36 hr regional lymph nodes were harvested. (a) Lymph node-derived APCs were incubated with phycoerythrin-conjugated anti-CD11c antibody and monoclonal antibody against H-2Kb/SIINFEKL complex, washed and stained with a secondary fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G (IgG) antibody. The graphics shows the percentage of CD11c+ cells positive for H-2Kb/SIINFEKL (Student’s t-test: **P = 0·008; *P = 0·02). (b,c) Lymph node APCs (100 000) derived from mice infected with S. Typhimurim (STM), S. Typhi (STY) and S. Enteritidis (SEN) or injected with purified OVA (50 μg) in Freund’s complete adjuvant were cocultured with OT-I T cells, specific for H-2Kb/SIINFEKL (b) or with OT-II T cells, specific for I-Ab/ISQAVHAAHAEINEAGR (c). After 20 hr of coculture, interleukin-2 (IL-2) release was measured on culture media by enzyme-linked immunosorbent assay. Data shown are means of three independent experiments and error bars represent SE (Student’s t-test: ***P < 0·0001).
In vivo T-cell activation in response to infection with S.Typhi or S. Enteritidis
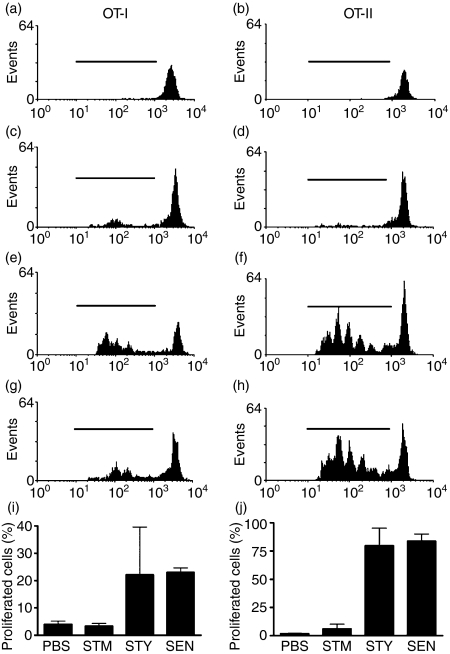
To determine whether the capacity of murine DCs to present antigens derived from S. Typhi and S. Enteritidis translated into an efficient activation of T cells in vivo, OVA-specific T-cell proliferation was evaluated upon infection with OVA-expressing Salmonella serovars. CFSE-labelled OT-I or OT-II transgenic T cells were adoptively transferred to C57BL/6 mice and proliferation was determined in response to bacterial challenge by dilution of CFSE fluorescence, as previously described.20,46–48 Twenty-four hours after adoptive transfer of transgenic T cells, mice were infected intravenously with 105 CFU of OVA-expressing S. Typhimurium, S. Typhi or S. Enteritidis. To determine the proliferation of transferred T cells, cell suspensions derived from the spleens of recipient mice were analysed by flow cytometry 3 days after the bacterial challenge. As shown in Fig. 4, OT-I and OT-II T-cell populations obtained from recipient mice infected with OVA-expressing S. Typhi and S. Enteritidis showed significant dilution of CFSE-derived fluorescence, which is indicative of T-cell proliferation (Fig. 4e–j). In contrast, no measurable T-cell proliferation was observed for recipient mice challenged with OVA-expressing S. Typhimurium or for naïve mice (Fig. 4a–d, i and j). These results suggest that, in contrast to S. Typhimurium, S. Typhi and S. Enteritidis lead to an efficient antigen-specific T-cell response in the mouse.
Figure 4.
Infection with S. Typhi and S. Enteritidis leads to activation of naïve T cells in vivo. Carboxyfluorescein succinimidyl ester (CFSE)-labelled OT-I or OT-II transgenic T cells were adoptively transferred to C57BL/6 mice and after 24 hr were intravenously infected with 105 colony-forming units (CFU) of ovalbumin (OVA)-expressing S. Typhimurium, S. Typhi and S. Enteritidis. Three days after infection, splenic cell suspensions were evaluated for in vivo activation of OT-I or OT-II transgenic T cells using fluorescence-activated cell sorting (FACS). (a,b) Uninfected mice; (c,d) S. Typhimurium-infected mice; (e,f), S. Typhi-infected mice; (g,h), S. Enteritidis-infected mice. Histograms show representative FACS profiles of CFSE-derived fluorescence for CD8+ T cells (OT-I) or CD4+ T cells (OT-II). (i,j) Percentage of CD4+ and CD8+ cells that proliferated (cells within the marker in histograms) from two independent experiments.
Salmonella intracellular survival within murine or human DCs is serovar specific
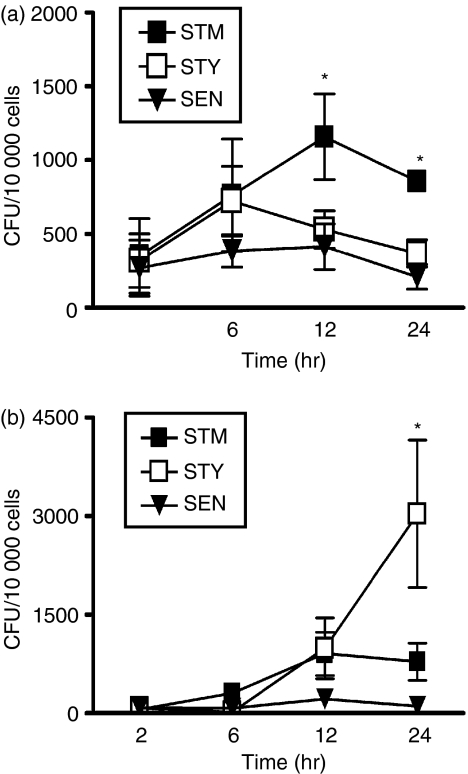
To determine whether S. Typhi and S. Enteritidis can survive inside murine DCs, as has been observed for S. Typhimurium,15,17,18,20 murine DCs were infected with each of the Salmonella serovars. Intracellular bacterial load was quantified at different time-points using a gentamicin protection assay that has been previously described.15,20 Although all three Salmonella serovars were able to survive within murine DCs up to 24 hr post-challenge, the number of intracellular S. Typhi and S. Enteritidis recovered from murine DCs was significantly reduced at 12 and 24 hr, compared to S. Typhimurium (Fig. 5a). These results suggest that although all three serovars can infect murine DCs, only S. Typhimurium can survive inside murine DCs for extended periods of time and proliferate. In contrast, S. Typhi and S. Enteritidis had reduced survival within murine DCs and faster degradation rates.
Figure 5.
Survival of Salmonella serovars within murine or human dendritic cells (DCs) is host specific. Number of Salmonella serovars as colony-forming units (CFU) within murine DCs (a) or human DCs (b) after 2, 6, 12 and 24 hr of infection. DCs were infected with Salmonella serovars [S. Typhimurium, S. Typhi and S. Enteritidis; multiplicity of infection 50] and lysed with Triton X-100 at the indicated times to release intracellular Salmonella. Intracellular bacteria were plated on Luria–Bertani agar plates. After 12 hr of incubation at 37°, bacterial colonies were quantified. Graphics show means of four independent experiments; bars represent SE (Student’s t-test: *P < 0·04).
To determine whether the capacity of Salmonella to interfere with DC function is host-specific, human DCs were infected with S. Typhimurium, S. Typhi or S. Enteritidis and the ability of these bacterial serovars to survive within these cells was evaluated using a gentamicin protection assay. As shown in Fig. 5b, all evaluated Salmonella serovars could invade human DCs and survive within them for up to 24 hr. However, after 24 hr S. Typhimurium and S. Enteritidis showed significantly reduced growth rates compared to S. Typhi.
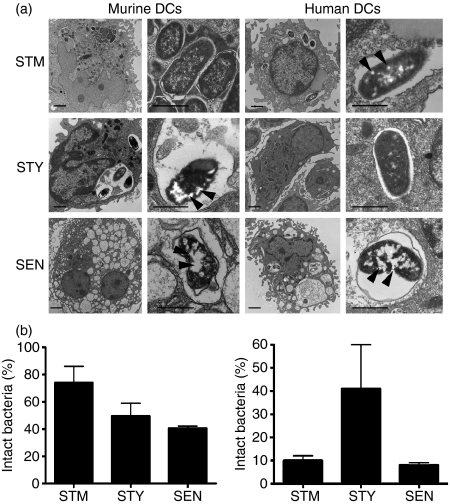
Consistent with the findings described above, a high frequency of bacterial degradation signs, appearing as electron-light structures, were observed for S. Typhi and S. Enteritidis residing within murine DC endosomes (Fig. 6a and b). In contrast, the majority of S. Typhimurium residing inside murine DC endosomes showed intact ultrastructure and no signs of degradation (Fig. 6a and b). However, a strikingly different scenario was observed when electron microscopy analyses were performed on human DCs. While the majority of intracellular S. Typhi appeared to be intact inside DC vacuoles, S. Enteritidis and S. Typhimurium showed a high prevalence of degradation signs (Fig. 6a and b).
Figure 6.
Transmission electron microscopy of dendritic cells (DCs) infected with Salmonella serovars. Murine or human DCs were infected with either of the Salmonella serovars (multiplicity of infection 50) and 24 hr later were processed for transmission electron microscopy (experimental procedures). (a) Representative micrographs showing S. Typhimurium (STM), S. Typhi (STY) and S. Enteritidis (SEN) inside murine or human DCs. Bacteria on the left panels (6800 ×, bars = 1 μm) are magnified in the right panels (16 500 ×, bars = 5 μm). Arrowheads show degradation patterns within bacteria. (b) Percentage of intact bacteria inside DCs from murine host (left graph) or human host (right graph) after 24 hr of infection. Graphs show means of two independent experiments and bar represent SE.
To evaluate whether the observations obtained from gentamicin protection assays and electron microscopy could be explained by differential targeting to lysosomal degradation, the recruitment of lysosomal markers to Salmonella-containing vacuoles was determined both in murine and human DCs infected with each of the Salmonella serovars. Quantitative confocal microscopy analyses showed that a minority of vacuoles containing S. Typhimurium in murine DCs colocalized with the lysosomal marker LAMP-2 (Fig. 7a and g). In contrast, higher levels of colocalization between LAMP-2 and vacuoles containing S. Typhi were observed in murine DCs (Fig. 7b and g). Higher amounts of colocalization between S. Enteritidis and LAMP-2 were also observed within murine DCs (Fig. 7c and g). As shown by fluorescence extension analyses, colocalization between fluorescence derived from LAMP-2 and bacteria inside murine DCs was observed for S. Typhi and S. Enteritidis but not for S. Typhimurium (Fig. 7, insets). In contrast, when equivalent experiments were performed using human DCs, we observed a higher frequency of colocalization between vacuoles containing S. Typhimurium and the lysosomal marker LAMP-2, as compared with human DCs infected with S. Typhi (Fig. 7d, e and g). Surprisingly, there was less colocalization of LAMP-2 with vacuoles containing S. Enteritidis within human DCs (Fig. 7f and g), indicating that the growth restriction of S. Enteritidis inside human DCs cannot be explained by colocalization with lysosomal markers, as seems to be the case for S. Typhimurium and S. Typhi.
Figure 7.
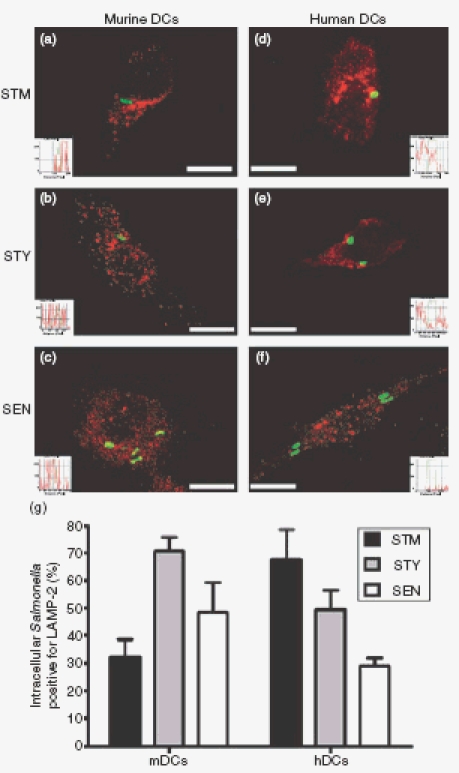
Colocalization of Salmonella serovars with LAMP-2 within murine and human dendritic cells (DCs). Murine or human DCs were infected with either S. Typhimurium, S. Typhi or S. Enteritidis (MOI equal 50) and, after 16 hr of infection, DCs were stained for Lamp-2 (rhodamine). (a–f) Representative merged images of S. Typhimuirum, S. Typhi or S. Enteritidis (green) and LAMP-2 (red) within murine or human DCs. Fluorescence intensity histograms show the degree of overlay between green and red fluorescence (insets). Bars represent 5 μm. (g) Quantitative analyses of Salmonella serovars and LAMP-2 colocalization within murine and human DCs. The graphs show means of three independent experiments and bar are SE.
Discussion
Host restriction remains an important yet unresolved issue of Salmonella biology and pathogenicity. Previous studies have contributed to the identification of bacterial factors that can restrict the capacity of different Salmonella serovars to invade host cells, survive inside them and cause diseases in different hosts by the transfer of genes from one serovar to another.33,49–52 In this study we provide evidence that Salmonella serovars differ in their ability to avoid adaptive immunity in mice, because of interference with DC function. According to our data, this interference feature of Salmonella is host-restricted.
Our results show that the enhanced capacity of murine DCs to degrade S. Typhi and S. Enteritidis can lead to efficient presentation of bacteria-expressed antigens on MHC class I and class II molecules, both in vitro and in vivo. With this attribute, murine DCs were able to prime in vitro antigen-specific MHC-I- and MHC-II-restricted T cells only when infected with S. Typhi or S. Enteritidis. Furthermore, murine DCs were also very efficient at activating bacteria-specific T cells in vivo in response to infection with S. Typhi or S. Enteritidis. Consistent with these findings, the impaired capacity of S. Typhi or S. Enteritidis to survive within murine DCs for extended times could partially explain why adaptive immunity in the mouse can be activated. Our data also suggest that the ability of Salmonella serovars to replicate inside murine DCs is conditioned to the escape from lysosomal degradation. This notion is supported by the observation that Salmonella serovars unable to replicate within murine DCs were targeted for phagosome–lysosome fusion. Our results are consistent with previous studies, which showed that host-restricted serovars are degraded at higher rates than S. Typhimurium by other murine cells, such as macrophages.49,50 The inability of S. Typhi and S. Enteritidis to impair murine DC function was consistent with the observation that these Salmonella serovars were more immunogenic in mice than was S. Typhimurium.
We also showed that the different ability of S. Typhimurium and S. Typhi to survive inside murine DCs was inverted when DCs were derived from human hosts. Only S. Typhi could replicate within human DCs, while S. Typhimurium and S. Enteritidis failed to replicate within these APCs. Additionally, our electron microscopy analyses revealed that only S. Typhi maintains an intact structure inside human DCs after 24 hr of infection, in contrast to S. Typhimurium and S. Enteritidis, which showed signs of degradation within these cells at similar infection times. However, our confocal microscopy results suggest that the recruitment of the lysosomal marker LAMP-2 to the Salmonella-containing vacuoles in human DCs is not necessarily related to the ability of the different serovars to replicate inside human DCs. It is probable that different human and mouse degradation pathways account for this apparent discrepancy. Although equivalent amounts of CD11c-positive cells with an immature phenotype were obtained from both mouse bone marrow and human blood samples, we cannot rule out that the differences observed in our experiments could be influenced by differences in DC preparation.
S. Typhi is one of the most host-adapted serovars within the Salmonella enterica genus.32 In C57BL/6 mice, S. Typhi is unable to colonize internal organs after oral or intraperitoneal infection (our unpublished results); however, it causes a systemic disease in human after oral ingestion. Here we show that S. Typhi cannot interfere with murine DC function nor avoid the activation of the adaptive immune response in the mouse. However, our results show that this bacterium can survive a greater period within human DCs, where they avoid degradation. Since S. Typhi causes diseases only in humans, these results suggest that the capacity to interfere with DC function could be characteristic of host adaptation. S. Enteritidis, on the other hand, is a host generalist serovar responsible for infections in a large number of animal and bird hosts. S. Enteritidis is unable to impede antigen presentation by murine DCs and cannot replicate inside murine or human DCs. When considered together, these observations suggest that S. Enteritidis is less adapted to mice and humans than S. Typhimurium and S. Typhi. In part, the reduced adaptation to mice of S. Enteritidis could be the result of an impaired ability to evade the host’s immune response.
Despite the differential abilities shown by Salmonella enterica subspecies I serovars to evade antigen presentation by DCs, these bacteria share more than 90% of their genomes and are therefore genetically very similar.28,53 Differences between these three serovars are primarily deletions and/or insertions of gene blocks,54 which could harbour genes encoding proteins that act as virulence factors specific for certain hosts. The genetic differences between the Salmonella serovars studied here, which could account for the variation in antigen presentation avoidance by DCs, remain to be elucidated. It has been recently shown that a functional SPI-2-coded type 3 secretion system is required for diminished colocalization of S. Typhimurium with lysosomal markers and for reduced antigen presentation.15,16,20 Although S. Typhi and S. Enteritidis possess the SPI-2,55,56 comparative analyses between Salmonella serovar genomes, using microarray technology, show that some genes present in S. Typhimurium SPI-2 are missing from the SPI-2 of S. Typhi and S. Enteritidis.28 Moreover, S. Typhimurium harbours a virulence genetic locus, known as spv, which resides in a virulence plasmid.57 Genes within spv are required for the onset of systemic infection in mice and deletion of this locus renders S. Typhimurium less virulent in mice.58 Importantly, the virulence plasmid and the spv locus are missing from both the S. Typhi and S. Enteritidis genomes.28 However, S. Typhi possesses a considerable amount of genomic DNA that is absent in other Salmonella serovars.54,56,59,60 This additional DNA could constitute pathogenicity islands that would account for disease in humans and probably interfere with the function of human DCs.
Overall, our observation supports a new component for Salmonella host specificity: the capacity to interfere with DC function. This information may contribute to the identification of new molecular factors determining host-range specificity of Salmonella and help with the design of new and improved vaccines against this intracellular pathogen.
Acknowledgments
We would like to thank Drs R. Steinman (The Rockefeller University) for the OT-I and OT-II mice, J. C. Sirard (Pasteur Institute, Lille) for the pKK-OVA plasmid, R. Germain (National Institutes of Health) for the 25-D1.16 hybridoma and A. Maldonado (Instituto de Salud Pública) for the S. Enteritidis strain. We are grateful to Drs J. R. Schwebach and A. Nguyen (Albert Einstein College of Medicine) for their critical reading of this manuscript. The authors are supported by grants FONDECYT #1070352, #11075060, #1050979 and #3060041, DIPUC #2002/11E, IFS #A/3639-1 and #B/3764-1, SavinMuco-Path-INCO-CT-2006-032296 and Millennium Nucleus on Immunology and Immunotherapy (P04/030-F). P.A.G. and L.J.C. are CONICYT fellows.
References
- 1.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Rescigno M. Dendritic cells and the complexity of microbial infection. Trends Microbiol. 2002;10:425–61. doi: 10.1016/s0966-842x(02)02425-3. [DOI] [PubMed] [Google Scholar]
- 3.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 4.Lopez CB, Moltedo B, Alexopoulou L, Bonifaz L, Flavell RA, Moran TM. TLR-independent induction of dendritic cell maturation and adaptive immunity by negative-strand RNA viruses. J Immunol. 2004;173:6882–9. doi: 10.4049/jimmunol.173.11.6882. [DOI] [PubMed] [Google Scholar]
- 5.Means TK, Hayashi F, Smith KD, Aderem A, Luster AD. The Toll-like receptor 5 stimulus bacterial flagellin induces maturation and chemokine production in human dendritic cells. J Immunol. 2003;170:5165–75. doi: 10.4049/jimmunol.170.10.5165. [DOI] [PubMed] [Google Scholar]
- 6.Michelsen KS, Aicher A, Mohaupt M, Hartung T, Dimmeler S, Kirschning CJ, Schumann RR. The role of toll-like receptors (TLRs) in bacteria-induced maturation of murine dendritic cells (DCS). Peptidoglycan and lipoteichoic acid are inducers of DC maturation and require TLR2. J Biol Chem. 2001;276:25680–6. doi: 10.1074/jbc.M011615200. [DOI] [PubMed] [Google Scholar]
- 7.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–3. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 8.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 9.Turley SJ, Inaba K, Garrett WS, Ebersold M, Unternaehrer J, Steinman RM, Mellman I. Transport of peptide–MHC class II complexes in developing dendritic cells. Science. 2000;288:522–7. doi: 10.1126/science.288.5465.522. [DOI] [PubMed] [Google Scholar]
- 10.Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, Lander ES, Hacohen N. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–5. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 11.Dustin ML, Chan AC. Signaling takes shape in the immune system. Cell. 2000;103:283–94. doi: 10.1016/s0092-8674(00)00120-3. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins SA, Niedergang F, Corthesy-Theulaz IE, Kraehenbuhl JP. A recombinant Salmonella typhimurium vaccine strain is taken up and survives within murine Peyer’s patch dendritic cells. Cell Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-5822.2000.00035.x. [DOI] [PubMed] [Google Scholar]
- 13.Rescigno M, Rotta G, Valzasina B, Ricciardi-Castagnoli P. Dendritic cells shuttle microbes across gut epithelial monolayers. Immunobiology. 2001;204:572–81. doi: 10.1078/0171-2985-00094. [DOI] [PubMed] [Google Scholar]
- 14.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–82. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 15.Tobar JA, Gonzalez PA, Kalergis AM. Salmonella escape from antigen presentation can be overcome by targeting bacteria to Fc gamma receptors on dendritic cells. J Immunol. 2004;173:4058–65. doi: 10.4049/jimmunol.173.6.4058. [DOI] [PubMed] [Google Scholar]
- 16.Cheminay C, Mohlenbrink A, Hensel M. Intracellular Salmonella inhibit antigen presentation by dendritic cells. J Immunol. 2005;174:2892–9. doi: 10.4049/jimmunol.174.5.2892. [DOI] [PubMed] [Google Scholar]
- 17.Jantsch J, Cheminay C, Chakravortty D, Lindig T, Hein J, Hensel M. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell Microbiol. 2003;5:933–45. doi: 10.1046/j.1462-5822.2003.00334.x. [DOI] [PubMed] [Google Scholar]
- 18.Petrovska L, Aspinall RJ, Barber L, et al. Salmonella enterica serovar typhimurium interaction with dendritic cells: impact of the sifA gene. Cell Microbiol. 2004;6:1071–84. doi: 10.1111/j.1462-5822.2004.00419.x. [DOI] [PubMed] [Google Scholar]
- 19.Alaniz RC, Cummings LA, Bergman MA, Rassoulian-Barrett SL, Cookson BT. Salmonella typhimurium coordinately regulates FliC location and reduces dendritic cell activation and antigen presentation to CD4+ T cells. J Immunol. 2006;177:3983–93. doi: 10.4049/jimmunol.177.6.3983. [DOI] [PubMed] [Google Scholar]
- 20.Tobar JA, Carreno LJ, Bueno SM, Gonzalez PA, Mora JE, Quezada SA, Kalergis AM. Virulent Salmonella enterica serovar typhimurium evades adaptive immunity by preventing dendritic cells from activating T cells. Infect Immun. 2006;74:6438–48. doi: 10.1128/IAI.00063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hensel M, Shea JE, Waterman SR, et al. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–74. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 22.Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 2003;5:501–11. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Del Portillo F, Jungnitz H, Rohde M, Guzman CA. Interaction of Salmonella enterica serotype typhimurium with dendritic cells is defined by targeting to compartments lacking lysosomal membrane glycoproteins. Infect Immun. 2000;68:2985–91. doi: 10.1128/iai.68.5.2985-2991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bueno SM, Tobar JA, Iruretagoyena MI, Kalergis AM. Molecular interactions between dendritic cells and Salmonella: escape from adaptive immunity and implications on pathogenesis. Crit Rev Immunol. 2005;25:389–403. doi: 10.1615/critrevimmunol.v25.i5.40. [DOI] [PubMed] [Google Scholar]
- 25.O’Callaghan D, Maskell D, Tite J, Dougan G. Immune responses in BALB/c mice following immunization with aromatic compound or purine-dependent Salmonella typhimurium strains. Immunology. 1990;69:184–9. [PMC free article] [PubMed] [Google Scholar]
- 26.Valentine PJ, Devore BP, Heffron F. Identification of three highly attenuated Salmonella typhimurium mutants that are more immunogenic and protective in mice than a prototypical aroA mutant. Infect Immun. 1998;66:3378–83. doi: 10.1128/iai.66.7.3378-3383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Minor L. Typing of Salmonella species. Eur J Clin Microbiol Infect Dis. 1988;7:214–8. doi: 10.1007/BF01963091. [DOI] [PubMed] [Google Scholar]
- 28.Chan K, Baker S, Kim CC, Detweiler CS, Dougan G, Falkow S. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar typhimurium DNA microarray. J Bacteriol. 2003;185:553–63. doi: 10.1128/JB.185.2.553-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumler AJ, Tsolis RM, Ficht TA, Adams LG. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–87. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fierer J, Guiney DG. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J Clin Invest. 2001;107:775–80. doi: 10.1172/JCI12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Kingsley RA, Santos RL, et al. Molecular pathogenesis of Salmonella enterica serotype typhimurium-induced diarrhea. Infect Immun. 2003;71:1–12. doi: 10.1128/IAI.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edsall G, Gaines S, Landy M, Tigertt WD, Sprinz H, Trapani RJ, Mandel AD, Benenson AS. Studies on infection and immunity in experimental typhoid fever. I. Typhoid fever in chimpanzees orally infected with Salmonella typhosa. J Exp Med. 1960;112:143–66. doi: 10.1084/jem.112.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raffatellu M, Sun YH, Wilson RP, et al. Host restriction of Salmonella enterica serotype typhi is not caused by functional alteration of SipA, SopB, or SopD. Infect Immun. 2005;73:7817–26. doi: 10.1128/IAI.73.12.7817-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raffatellu M, Chessa D, Wilson RP, Tukel C, Akcelik M, Baumler AJ. Capsule-mediated immune evasion: a new hypothesis explaining aspects of typhoid fever pathogenesis. Infect Immun. 2006;74:19–27. doi: 10.1128/IAI.74.1.19-27.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beal RK, Wigley P, Powers C, Hulme SD, Barrow PA, Smith AL. Age at primary infection with Salmonella enterica serovar typhimurium in the chicken influences persistence of infection and subsequent immunity to re-challenge. Vet Immunol Immunopathol. 2004;100:151–64. doi: 10.1016/j.vetimm.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Robertsson JA, Fossum C, Svenson SB, Lindberg AA. Salmonella typhimurium infection in calves: specific immune reactivity against O-antigenic polysaccharide detectable in in vitro assays. Infect Immun. 1982;37:728–36. doi: 10.1128/iai.37.2.728-736.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berndt A, Methner U. B cell and macrophage response in chicks after oral administration of Salmonella typhimurium strains. Comp Immunol Microbiol Infect Dis. 2004;27:235–46. doi: 10.1016/j.cimid.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Berndt A, Methner U. Gamma/delta T cell response of chickens after oral administration of attenuated and non-attenuated Salmonella typhimurium strains. Vet Immunol Immunopathol. 2001;78:143–61. doi: 10.1016/s0165-2427(00)00264-6. [DOI] [PubMed] [Google Scholar]
- 39.La Brooy JT, Shearman DJ, Rowley D. Antibodies in serum and secretions 1 year after salmonella gastroenteritis. Clin Exp Immunol. 1982;48:551–4. [PMC free article] [PubMed] [Google Scholar]
- 40.Foley JE, Orgad U, Hirsh DC, Poland A, Pedersen NC. Outbreak of fatal salmonellosis in cats following use of a high-titer modified-live panleukopenia virus vaccine. J Am Vet Med Assoc. 1999;214:67–70. [PubMed] [Google Scholar]
- 41.van Diepen A, van de Gevel JS, Koudijs MM, Ossendorp F, Beekhuizen H, Janssen R, van Dissel JT. Gamma irradiation or CD4+-T-cell depletion causes reactivation of latent Salmonella enterica serovar typhimurium infection in C3H/HeN mice. Infect Immun. 2005;73:2857–62. doi: 10.1128/IAI.73.5.2857-2862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruenewald R, Blum S, Chan J. Relationship between human immunodeficiency virus infection and salmonellosis in 20- to 59-year-old residents of New York City. Clin Infect Dis. 1994;18:358–63. doi: 10.1093/clinids/18.3.358. [DOI] [PubMed] [Google Scholar]
- 43.Clarke SR, Barnden M, Kurts C, Carbone FR, Miller JF, Heath WR. Characterization of the ovalbumin-specific TCR transgenic line OT-I: MHC elements for positive and negative selection. Immunol Cell Biol. 2000;78:110–7. doi: 10.1046/j.1440-1711.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- 44.Robertson JM, Jensen PE, Evavold BD. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323–339 epitope. J Immunol. 2000;164:4706–12. doi: 10.4049/jimmunol.164.9.4706. [DOI] [PubMed] [Google Scholar]
- 45.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide–MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–26. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 46.Hasbold J, Gett AV, Rush JS, Deenick E, Avery D, Jun J, Hodgkin PD. Quantitative analysis of lymphocyte differentiation and proliferation in vitro using carboxyfluorescein diacetate succinimidyl ester. Immunol Cell Biol. 1999;77:516–22. doi: 10.1046/j.1440-1711.1999.00874.x. [DOI] [PubMed] [Google Scholar]
- 47.Bonifaz L, Bonnyay D, Mahnke K, Rivera M, Nussenzweig MC, Steinman RM. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J Exp Med. 2002;196:1627–38. doi: 10.1084/jem.20021598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carreno LJ, Gonzalez PA, Kalergis AM. Modulation of T cell function by TCR/pMHC binding kinetics. Immunobiology. 2006;211:47–64. doi: 10.1016/j.imbio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Pascopella L, Raupach B, Ghori N, Monack D, Falkow S, Small PL. Host restriction phenotypes of Salmonella typhi and Salmonella gallinarum. Infect Immun. 1995;63:4329–35. doi: 10.1128/iai.63.11.4329-4335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwan WR, Huang X-Z, Hu L, Kopecko DJ. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect Immun. 2000;68:1005–13. doi: 10.1128/iai.68.3.1005-1013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehmann J, Bellmann S, Werner C, Schroder R, Schutze N, Alber G. IL-12p40-dependent agonistic effects on the development of protective innate and adaptive immunity against Salmonella enteritidis. J Immunol. 2001;167:5304–15. doi: 10.4049/jimmunol.167.9.5304. [DOI] [PubMed] [Google Scholar]
- 52.Barrow PA, Huggins MB, Lovell MA. Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect Immun. 1994;62:4602–10. doi: 10.1128/iai.62.10.4602-4610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crosa JH, Brenner DJ, Ewing WH, Falkow S. Molecular relationships among the Salmonelleae. J Bacteriol. 1973;115:307–15. doi: 10.1128/jb.115.1.307-315.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards RA, Olsen GJ, Maloy SR. Comparative genomics of closely related salmonellae. Trends Microbiol. 2002;10:94–9. doi: 10.1016/s0966-842x(01)02293-4. [DOI] [PubMed] [Google Scholar]
- 55.Amavisit P, Lightfoot D, Browning GF, Markham PF. Variation between pathogenic serovars within Salmonella pathogenicity islands. J Bacteriol. 2003;185:3624–35. doi: 10.1128/JB.185.12.3624-3635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parkhill J, Dougan G, James KD, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar typhi CT18. Nature. 2001;413:848–52. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 57.Gulig PA, Danbara H, Guiney DG, Lax AJ, Norel F, Rhen M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993;7:825–30. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 58.Gulig PA, Doyle TJ. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect Immun. 1993;61:504–11. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bueno SM, Santiviago CA, Murillo AA, Fuentes JA, Trombert AN, Rodas PI, Youderian P, Mora GC. Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar typhi. J Bacteriol. 2004;186:3202–13. doi: 10.1128/JB.186.10.3202-3213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McClelland M, Sanderson KE, Spieth J, et al. Complete genome sequence of Salmonella enterica serovar typhimurium LT2. Nature. 2001;413:852–6. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]