Abstract
In response to inflammatory stimuli, monocytes/macrophages secrete greater quantities of the proinflammatory cytokines tumour necrosis factor-α (TNF-α), interleukin-1β (IL-1β) and IL-6. The inflammatory process and the innate immune response are related to the activation of several transcription factors, such as nuclear factor κB (NF-κB) and activator protein 1 (AP-1). The proteasome is a multimeric protease complex, which plays a vital role in several cellular functions, including the regulation of transcription factors like NF-κB. In this study, we used the human monocyte cell line U937 stimulated with lipopolysaccharide (LPS) and phorbol 12-myristate 13-acetate (PMA) as a model to investigate the in vitro effects of MG132, a proteasome inhibitor, on the release of TNF-α, IL-1β and IL-6 and on the expression of their membrane and soluble receptors TNF-R1, IL-1R1 and IL-6R. We also analysed the effects of MG132 on the activation of NF-κB and AP-1 and on the IκB molecule. MG132 significantly inhibited the secretion of those proinflammatory cytokines. MG132 increased the release of the soluble receptors TNF-R1 and IL-1R1 from U937 cells and decreased their cell-surface expression. MG132 also increased IL-6R cell-surface expression and decreased its release. Proteasome inhibition also led to an increase in LPS+PMA-induced AP-1 activation and the attenuation of LPS+PMA-induced IκB degradation, resulting in the abolition of NF-κB activation. Our experiments strongly suggest that the proteasome is an important factor in the regulation of proinflammatory cytokines and their receptors.
Keywords: activator protein 1, cytokine receptors, nuclear factor κB, proinflammatory cytokines, proteasome
Introduction
Monocyte/macrophage cells play a central role in the innate immune response and inflammatory processes.1 These cells perform many functions: natural antitumour activity, antigen processing and secretion of biological products, such as the proinflammatory cytokines tumour necrosis factor-α (TNF-α), interleukin-1β (IL-1β) and IL-6, which act on different cells of the immune system, activating and producing a system of feedback that limits damage and restores homeostasis.1,2 However, the cytokines do not act alone. Their respective membrane and soluble receptors form a very important network that regulates the biological activity and bioavailability of these proinflammatory cytokines.3,4 The proteasome is a large multimeric protease complex, localized in the cellular cytoplasm, and consists of the 20S proteasome with proteolytic activity and the 19S regulatory complex.5 This complex is essential for several cellular processes, including protein degradation, cellular differentiation and antigen presentation.5,6 Recent studies have also increasingly implicated the proteasome in the regulation of some cell-surface cytokine receptors (IL-2R, IL-9R).7,8 Another very important activity of the proteasome is the regulation of transcription factors, including nuclear factor κB (NF-κB).9,10 This transcription factor induces the expression of a large number of genes related to the innate immune response and the inflammatory process, such as the TNF-α, IL-1β and IL-6 cytokines, cytokine receptors and other molecules.10 Other transcription factors are involved in the regulation of these inflammatory molecules, such as activator protein 1 (AP-1). This transcription factor is composed of Jun family members (c-Jun, JunB and JunD).11,12
In this study, we investigated the in vitro effects of the proteasome inhibitor MG132 on the release of TNF-α, IL-1β and IL-6 and their receptors, as well as on the activation of NF-κB and AP-1 and the inhibitor of the NF-κB (IκB) degradation, in the monocyte/macrophage-derived cell line U937.
Materials and methods
Chemicals and reagents
Lipopolysaccharide (LPS) from Escherichia coli 055:B5 (Sigma, St Louis, MO) 1 mg/ml was dissolved in phosphate-buffered saline (PBS), the proteasome inhibitor MG132 (Sigma) and phorbol 12-myristate 13-acetate (PMA; Sigma) were dissolved in sterile dimethyl sulphoxide (DMSO; Sigma) at a concentration of 42 mm and 60 ng/ml respectively. Solutions were kept frozen in aliquots at −20° for up to 80 days until use.
Culture medium
RPMI-1640 culture medium (Sigma) was supplemented with 10% fetal calf serum (Gibco, Carlsbad, CA), 2 mm l-glutamine (Gibco) and antibiotics (Sigma); this medium is referred to as RPMI-S.
Cell culture and in vitro treatment
U937 cells13 were cultured in RPMI-S at 37° in a humidified atmosphere containing 5% CO2 and 95% air until they reached the exponential phase (2–3 weeks), then the cells were washed and resuspended in RPMI-S and seeded in a 12-well flat-bottom tissue culture plate (Corning-Costar, Lowell, MA) at a density of 1 × 106 cells in 2 ml of RPMI-S per well. The cells were either treated or not treated with the proteasome inhibitor MG132 (final concentration 10 μm), and incubated for 2 hr at 37°. At the end of the incubation period, the cells were washed three times with RPMI-1640 tissue medium culture. The control and experimental cultures were resuspended in either RPMI-S with or without LPS (1 μg/ml) + PMA (30 ng/ml) (LPS+PMA) in a final volume of 2 ml and cultured for 24 hr. The same amount of DMSO was added to the control samples as was added to the experimental cultures.
Assessment of apoptosis by flow cytometry
Cells (1 × 106) were incubated for 10 min with Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide, according to the kit instructions (Annexin-V-Fluos; Roche, Mannheim, Germany) and analysed by flow cytometry with a Beckman Coulter model EPICS-XL cytometer (Beckman Coulter, Fullerton, CA). The data were processed using the System II software package (Beckman Coulter). At least 20 000 events were analysed for each treated sample. Viability was confirmed before and after each experiment (Viability > 95%).
ELISA for TNF-α, IL-1β, IL-6 and soluble cytokine receptors
The levels of TNF-α, IL-1β, IL-6, soluble TNF receptor 1 (sTNF-R1), sIL-1R1 and sIL-6R receptors were determined in the supernatants of U937 cell cultures using a sandwich enzyme-linked immunosorbent assay (ELISA) technique. The TNF-α, IL-1β and IL-6 were measured using kits from Amersham Biosciences (Little Chalfont, UK). The sTNF-R1 and sIL-6R were quantified using kits from Biosource Europe SA (Nivelles, Belgium); and the concentration of sIL-1R1 was measured with a kit from R&D Systems (Minneapolis, MN).The assays were performed according to the manufacturers’ instructions. Absorbance was read on an Opsys MR™ 96-well microplate reader (DYNEX Technologies Inc., Chantilly, VA).
Analysis of TNF-R1, IL-1R1 and IL-6R membrane expression by flow cytometry
The expression of TNF-R1, IL-1R1 and IL-6R was assessed by flow cytometry. Independent assays were performed for each receptor. Briefly, U937 cells (1 × 106) were washed twice with ice-cold PBS (0·1% bovine serum albumin, 0·1% NaN3) and resuspended in PBS. Then, the cells were incubated with hamster monoclonal anti-human TNF-R1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), rat monoclonal anti-human IL-1R1 antibody (Serotec, Oxford, UK), or mouse monoclonal anti-human IL-6R antibody (Biosource, Camarillo, CA) for 30 min on ice. The cells were washed three times with ice-cold PBS, and incubated with phycoerythrin-conjugated goat F(ab′)2 anti-hamster immunoglobulin G (IgG; Serotec), FITC-conjugated goat F(ab′)2 anti-rat IgG (Biosource) or FITC-conjugated goat F(ab′)2 anti-mouse IgG (Sigma) secondary antibody for TNF-R1, IL-1R1 and IL-6R, respectively, for 20 min at 4°. The cells were then washed and fixed in 1% paraformaldehyde, and assessed for fluorescence by flow cytometry using a Beckman Coulter model EPICS XL MCL cytometer as described earlier. An appropriate isotype control was used in each test to adjust for background fluorescence (Serotec) and the results are reported as mean fluorescence intensity (MFI).
Preparation of cell extracts
Nuclear and cytoplasmic extracts were prepared using a modification of a previously published method.14 Briefly, after the experiments, the U937 cells were washed in PBS, and then 100 μl buffer A [10 mm HEPES (pH 7·9), 1·5 mm MgCl2, 10 mm KCl, 0·5 mm dithiothreitol, 0·2% Nonidet P-40, 100 μl protease inhibitor cocktail (Sigma) per millilitre of buffer A] was added for extraction of cytoplasmic extracts. For nuclear extracts we used 50 μl buffer B [20 mm HEPES (pH 7·9), 25% glycerol, 0·42 mm NaCl, 1·5 mm MgCl2, 0·5 mm dithiothreitol, 0·2 mm ethyldiaminetetraacetic acid (EDTA) and 100 μl protease inhibitor cocktail per millilitre of buffer B]. We used 5 μl of all the extract fractions to quantify their proteins by the Bradford method.15 The nuclear and cytoplasmic extracts were frozen and stored at −80° until use.
Western blot analysis of IκB-α
For Western blot analysis, 50 μg of the cytoplasmic proteins from each sample was fractionated on a sodium dodecyl sulphate (Gibco) –polyacrylamide gel (12%) and electrophoretically transferred to a nitrocellulose membrane (Amersham, Arlington Heights, IL). The membrane was washed twice with Tris-buffered saline [TBS; 50 mm Tris–HCl, 150 mm NaCl (pH 7·5)] and the non-specific protein-binding sites on the membrane were blocked with 1% blocking solution (Roche) with shaking for 1 hr. The membrane was incubated at 4° overnight, without shaking, with a primary antibody directed against IκB-α (Oncogene, San Diego, CA). Excess antibody was removed by washing the membrane twice with TBST (TBS with 0·1% Tween 20) for 10 min each. The membrane was then probed with horseradish peroxidase-labelled secondary antibody (anti-mouse IgG–POD/anti-rabbit IgG–POD; Roche). The immobilized protein, bound to the antibody of interest, was then detected with the BM Chemiluminescence Western Blotting Kit (mouse/rabbit) (Roche).
NF-κB and AP-1 electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSAs) were performed using a LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, IL). The oligonucleotide sequences were: for NF-κB: 5′-AGTTGAGGGGACTTTCCCAGGC-3′ and 5′-GCCTGGGAAAGTCCCCTCAACT-3′; for AP-1: 5′-CGCTTGATGACTCAGCCGGAA-3′ and 5′-TTCCGGCTGAGTCATCAAGCG-3′. Oligonucleotides for NF-κB and AP-1 were annealed, and end-labelled with the Biotin 3′ End DNA Labeling Kit (Pierce). Binding reactions were performed as follows: nuclear extract (5 μg) was preincubated at 4° for 5 min in binding buffer [10 mm Tris–HCl (pH 7·5), 50 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, 5% glycerol and 1 μg/μl poly(dI·dC)]. The binding reaction was initiated by the addition of 20 fmol of labelled oligonucleotide. After 20 min incubation at room temperature, DNA–protein complexes were resolved on non-denaturing 6% polyacrylamide gels in low-ionic-strength buffer (0·5 × TBE) at 200 V for 2 hr. After gel electrophoresis, the DNA–protein complexes were transferred to a nylon membrane (Biodyne B Positively Charged Nylon Membrane; Pierce). After transfer, the membrane was immediately cross-linked for 15 min on an ultraviolet transilluminator equipped with 312-nm bulbs. A chemiluminescent detection method using a luminol/enhanced solution and a stable peroxide solution (Pierce) was used as described by the manufacturer, and the membranes were exposed to radiographic film for 2–5 min before development. Relative intensities were analysed using the ImageJ package. Transcriptional activity is expressed in fold change, considering 1 as the basal activity (untreated cells).
Statistical analysis
The results represent the means ± standard deviations (SD) of the values obtained from at least four independent experiments, carried out in triplicate. Comparisons between groups were made with anova, the Kruskal–Wallis test and the Mann–Whitney U-test. For correlations, we used Pearson’s test. A value of P< 0·05 was considered significant.
Results
MG132 reduces the LPS+PMA-induced production of TNF-α, IL-1β and IL-6
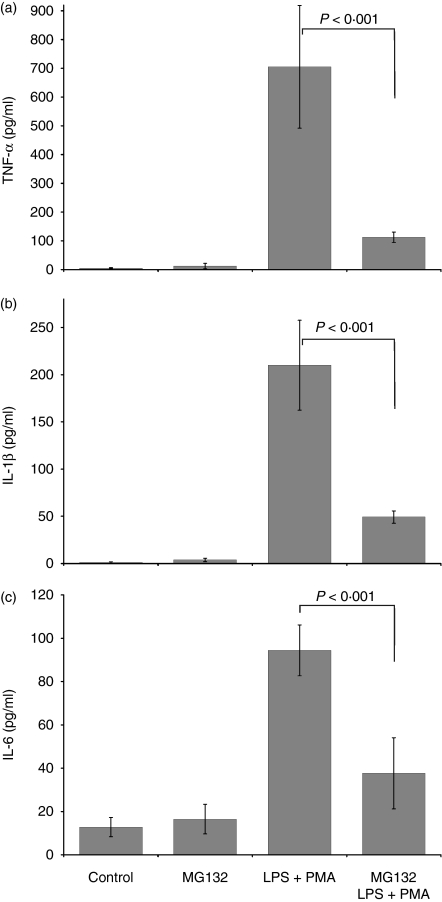
We first investigated whether the proteasome inhibitor MG132 reduces the production of TNF-α, IL-1β and IL-6 in U937 monocytic cells. Figure 1 shows the basal levels of TNF-α, IL-1β and IL-6 in control (untreated cells) (5·20 ± 1·80, 1·18 ± 0·8 and 12·79 ± 4·50 pg/ml, respectively). The spontaneous liberation of these cytokines from U937 cells was similar in cultures treated only with MG132. In contrast, a significant increase in the cytokine concentrations (P< 0·001) was observed in the supernatants from untreated cells stimulated with LPS + PMA (TNF-α, 705·12 ± 213·26 pg/ml; IL-1β, 210·03 ± 47·56 pg/ml; and IL-6, 94·40 ± 11·76 pg/ml). When the cells were treated with MG132 and stimulated with LPS+PMA, the proinflammatory cytokine concentrations decreased dramatically, with the supernatant concentrations 6·3-, 4·3- and 2·5-fold lower for TNF-α, IL-1β and IL-6, respectively, than those of untreated cells stimulated with LPS+PMA (P< 0·001). These results indicate that the proteasome inhibitor MG132 decreases the production of TNF-α, IL-1β and IL-6 in cells stimulated with LPS+PMA but apparently does not modify their expression in unstimulated cells.
Figure 1.
Proteasome inhibition induced a decrease in TNF-α, IL-1β, and IL-6. 1 × 106 U937 cells were cultured in RPMI-S medium in the presence or absence of the proteasome inhibitor MG132 (10 μm final concentration) for 2 hr. The control and experimental cultures were washed and stimulated or not with LPS (1 μg/ml) + PMA (30 ng/ml) for 24 hr. The culture supernatants were then recollected and the TNF-α, IL-1β, and IL-6 concentrations determined by ELISA. The results represent the means ± SD, Mann–Whitney U test. P < 0·001.
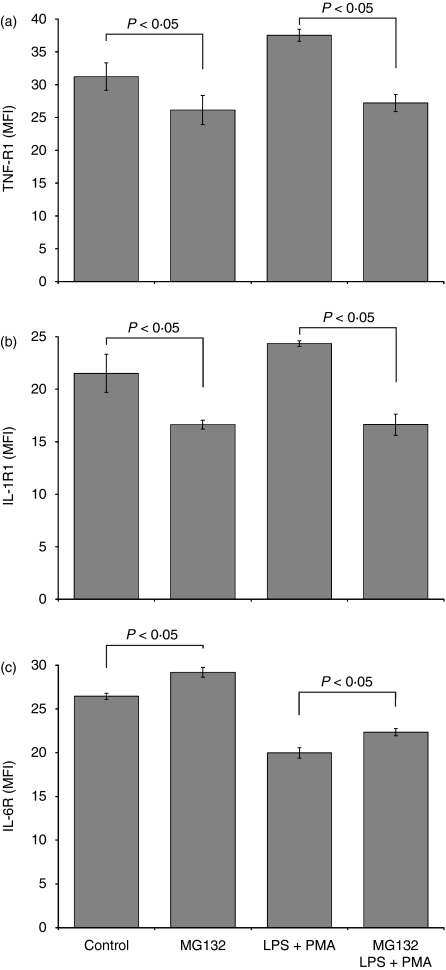
MG132 decreases TNF-R1, IL-1R1 expression and increases IL-6R expression on U937 cells
To investigate the effects of MG132 on membrane TNF-R1, IL-1R1 and IL-6R expression in U937 cells, we analysed the expression of these receptors by flow cytometry on U937 cells stimulated or not with LPS+PMA. Figure 2 illustrates that MG132 decreased the expression of TNF-R1 (26·15 ± 2·20 MFI) and IL-1R1 (16·62 ± 0·42 MFI) compared with the levels on untreated cells (31·23 ± 2·10 MFI for TNF-R1 and 26·15 ± 2·20 MFI for IL-1R1, P< 0·05). In contrast, the addition of the MG132 proteasome inhibitor induced an increase on the membrane expression of IL-6R (MG132-treated cells, 29·19 ± 0·57 MFI versus untreated cells, 26·44 ± 0·30 MFI; P< 0·05). Similarly, when MG132-treated cultures were stimulated for 24 hr with LPS+PMA, we observed a reduction in TNF-R1 and IL-1R1 expression and an increase in IL-6R expression relative to that in the MG132-untreated cells stimulated with LPS+PMA. Thus, proteasome inhibition decreased the expression of TNF-R1 and IL1-R1 and increased the expression of IL-6R.
Figure 2.
Regulation of TNF-R1, IL-1R1, and IL-6R expression in U937 cells treated with MG132. 1 × 106 U937 cells were cultured in the presence or absence of the proteasome inhibitor MG132 (10 μm final concentration) for 2 hr. The control and experimental cultures were washed and stimulated or not with LPS (1 μg/ml) + PMA (30 ng/ml) for 24 hr. We then measured by flow cytometry the expression of these receptors. The results represent the means ± SD, Kruskall–Wallis test. P< 0·05.
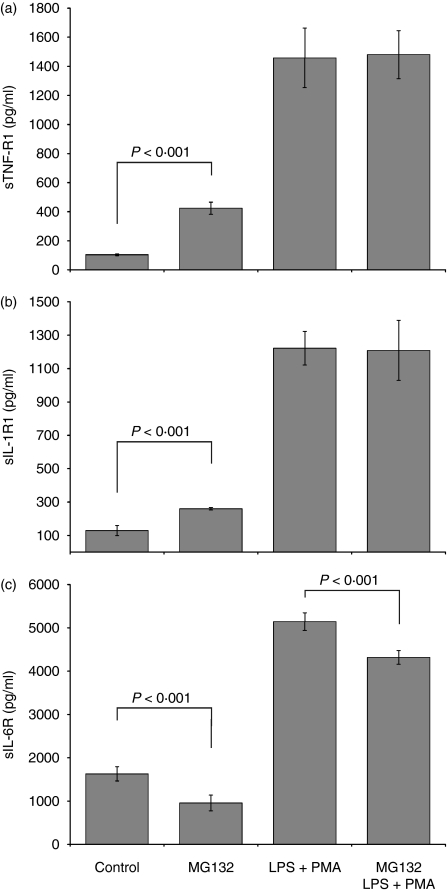
Effects of MG132 on sTNF-R1, sIL-1R1 and sIL-6R in U937 cells
Our next goal was to investigate whether proteasome inhibition altered sTNF-R1, sIL-1R1 and sIL-6R release in the U937 monocyte cell line. In Fig. 3(a) and 3(b), it is shown that the supernatants from U937 cells treated exclusively with MG132 show high concentrations of sTNF-R1 (424·34 ± 41·37 pg/ml) and sIL-1R1 (259·80 ±7·70 pg/ml), which are significantly greater than those observed in the untreated control cells (104·20 ± 5·72 pg/ml for sTNF-R1 and 129·03 ± 30·03 pg/ml for sIL-1R1, P< 0·001). There were no differences in the liberation of sTNF-R1 and sIL-1R1 between the cell group stimulated with LPS+PMA and the group treated with MG132 and later stimulated with LPS+PMA. Finally, we determined the MG132 effect on sIL-6R (Fig. 3c). The inhibitor diminished the liberation of sIL-6R (956·68 ± 180·06 pg/ml) compared with untreated cells (1628·50 ± 165·97 pg/ml; P< 0·001). Cells treated with MG132 and later stimulated with LPS+PMA liberated less sIL-6R (4315·04 ± 155·31 pg/ml) relative to that liberated by cells stimulated with only LPS+PMA (5143·13 ± 203·44 pg/ml; P< 0·001).
Figure 3.
Quantification of soluble TNF-R1, IL-1R1, and IL-6R in U937 cells treated with MG132 and stimulated with LPS + PMA. 1 × 106 U937 cells were cultured in the presence or absence of the proteasome inhibitor MG132 (10 μm final concentration) for 2 hr. The control and experimental cultures were washed and stimulated or not with LPS (1 μg/ml) + PMA (30 ng/ml) for 24 hr. Then sTNF-R1, sIL-1R1, and sIL-6R soluble receptor release was determined by ELISA. The results represent the average concentrations expressed in pg/ml. Statistical analysis was performed with anova. P < 0·001.
Pearson’s correlation test was used to investigate a possible relationship between the membrane expression and liberation of soluble forms of the TNF-R1, IL-1R1 and IL-6R receptors induced by proteasome inhibition. We found a significant positive correlation for TNF-R1 (P< 0·02), even though the r-value was low (0·285) and negative correlation, with r= −0·954 (P< 0·001) for IL-6R. No correlation was found for IL-1R1. These results together strongly suggest that the MG132 plays an important role in the control of membrane and soluble receptors.
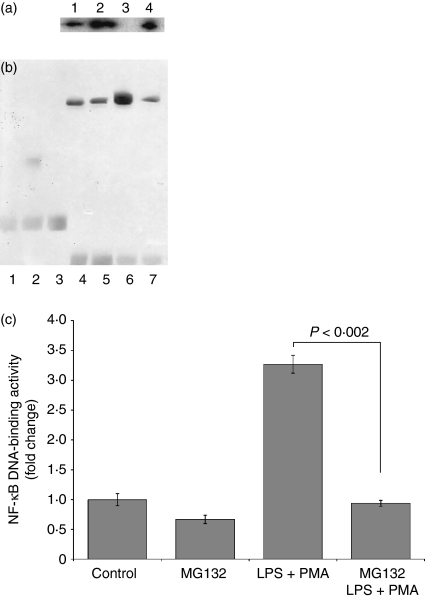
Proteasome inhibitor MG132 reverses the effects of LPS+PMA on IκB degradation
The degradation of IκB constitutes the first step in NF-κB activation, we performed a set of experiments to determine whether MG132 blocks the effects of LPS+PMA on IκB degradation in U397 cells. As illustrated in Fig. 4(a), the addition of LPS+PMA for 2 hr resulted in a rapid loss of IκB from the cytoplasmic extracts (LPS+PMA; lane 3). However, pretreatment with MG132 reversed the effects of LPS+PMA on IκB degradation (lane 4). Similarly, a slight increase in IκB was seen in cells treated with MG312 alone (lane 2), compared with untreated cells (lane 1).
Figure 4.
Proteasome inhibition and its effects on IκB-α degradation and NF-κB activation. 1 × 106 U937 cells were cultured in the presence or absence of the proteasome inhibitor MG132 (10 μm final concentration) for 2 hr. The control and experimental cultures were washed and stimulated or not with LPS (1 μg/ml) + PMA (30 ng/ml). After 2 hr, the cytoplasmic extracts were harvested and analysed by western blot (a). Lane 1, control; lane 2, MG132; lane 3, LPS + PMA; lane 4, MG132 and LPS + PMA. The test was repeated four times with similar results. Nuclear extracts were collected following 4 hr of LPS + PMA stimulation and analysed by EMSA. Representative blot showing the NF-κB-specific complexes (b), and the means ± SD of binding activity of four experiments (c). Lane 1, biotin–EBNA control DNA; lane 2, biotin–EBNA control DNA + EBNA extracts; lane 3, biotin–EBNA control DNA + EBNA extracts + 200-fold molar excess of unlabeled EBNA DNA; lane 4, Control; lane 5, MG132; lane 6, LPS + PMA; lane 7, MG132 and LPS + PMA. Statistical analysis was performed with the Kruskal–Wallis test. P < 0·002.
Proteasome inhibition decreases LPS+PMA-mediated NF-κB activation
To examine whether the proteasome participates in the regulation of the transcription factor NF-κB in U937 cells, these cells were treated with MG132 before stimulation with LPS+PMA. As shown in Fig. 4(b) and 4(c), LPS+PMA increased NF-κB activation 3·27-fold relative to that in untreatead cells (P< 0·002). The increase in NF-κB activation induced by LPS+PMA diminished significantly from 3·27-fold to 0·94-fold in the group treated with MG132 and later stimulated with LPS+PMA (P< 0·002). The activation of NF-κB induced by LPS+PMA was blocked by MG132.
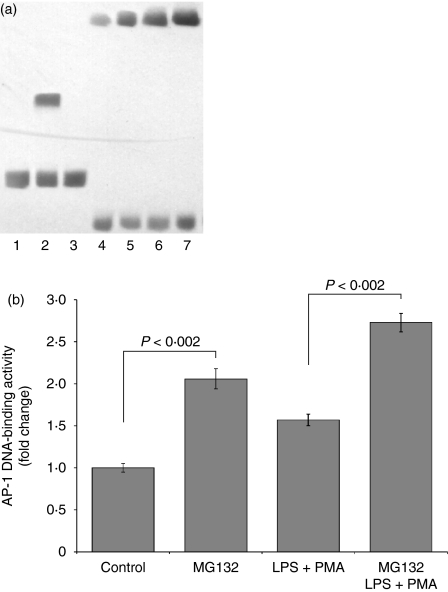
Proteasome inhibition enhances LPS+PMA-mediated AP-1 activation
We studied whether the proteasome regulates AP-1 activation in U937 cells. Addition of LPS+PMA increased AP-1 activation 1·57-fold relative to its activation in untreated cells (P< 0·002; Fig. 5a,b). MG132 increased the levels of AP-1 activation 2·06-fold compared with the levels in the untreated cells (P< 0·002). There was also a 2·46-fold increase in the activation of this transcription factor in cells treated with MG132 and later stimulated with LPS+PMA compared with the 1·57-fold increase in cells only stimulated with LPS+PMA (P< 0·002).
Figure 5.
Proteasome inhibition and its effects on AP-1 activation. 1 × 106 U937 cells were cultured in the presence or absence of the proteasome inhibitor MG132 (10 μm final concentration) for 2 hr. The control and experimental cultures were washed and stimulated or not with LPS (1 μg/ml) + PMA (30 ng/ml). After 4 hr, the nuclear extracts were collected and analyzed by EMSA. Representative blot showing the AP-1 specific complexes (a), and the means ± SD of binding activity of four experiments (b). Lane 1, biotin–EBNA control DNA; lane 2, biotin–EBNA control DNA + EBNA extracts; lane 3, biotin–EBNA control DNA + EBNA extracts + 200-fold molar excess of unlabeled EBNA DNA; lane 4, Control; lane 5, MG132; lane 6, LPS + PMA; lane 7, MG132 and LPS + PMA. Statistical analysis was performance with the Kruskal–Wallis test. P < 0·002.
Discussion
To study the effects of proteasomal inhibition on the interrelationship between cytokines and their receptors, we initially determined the effects of MG132 on proinflammatory cytokines (TNF-α, IL-1β and IL-6). Our results are consistent with previous observations of the proteasome inhibitor MG132, which indicated that MG132 significantly diminishes proinflammatory cytokine release.16,17
It has been reported that the proteasome regulates the levels of receptors on the cell surface in different ways. For example, inhibitors of the proteasome [(e.g. MG132, Z-Ile-Glu-(O-t-butyl)-Ala-leucinal (PSI) and N-acetyl-L-leucinyl-L-leucinyl-norleucinal (LLnL)] prevent the internalization of the interleukins, including IL-2 and IL-9, and affect the densities of the receptors IL-2R, IL-9R, the growth hormone receptor and the erythropoietin receptor.7,8,18
We chose to investigate TNF-R1, IL-1R1 and IL-6R because most biological events induced by TNF-α, IL-1β and IL-6 are mediated by these receptors.19,20 Interestingly, the proteasome inhibitor MG132 induced a decrease in the membrane receptors TNF-R1 and IL-1R1 and an increase in the soluble receptors sTNF-R1 and sIL-1R1. However, at the same time, the proteasome inhibitor increased the IL-6R and decreased sIL-6R. These results together indicate a complex and specific activity of MG132 on TNF-α, IL-1β and IL-6, and their receptors, which cannot be explained as toxic effects of MG132 because cellular viability was not affected. Moreover, these observations strongly suggest the existence of a novel mechanism by which the activity of the proteasome regulates the bioavailability of these inflammatory cytokines and consequently their bioactivity.
We found a statistically significant correlation between membrane and soluble receptor for TNF-R1 and IL-6R but not for IL-1R1. This could be explained by the fact that membrane receptors can be internalized into the cells and at the same time shed from the cell surface.21,22
Peiretti and Levine and their colleagues have elegantly demonstrated that proteasomal inhibition activates the shedding of TNF-α receptors from endothelial and epithelial cells, respectively. In those studies, they demonstrated that the proteasome plays an important role in the regulation of transmembrane components of the TNF-α–TNFR axis.23,24 In our work, we observed the same behaviour in monocytic cells. This work also presents the first data demonstrating proteasome participation in the regulation of membrane and soluble IL-6R and IL-1R1 in monocytic cells, which play principal roles in the inflammatory process.
The MG132 induces different effects at the molecular level. One involves the attenuation of NF-κB activation, after which NF-κB can neither translocate into the nuclei of cells nor activate the genes that encode proinflammatory cytokines. This may be because the most important target of MG132 is the proteasome.25 This is supported by the fact that, after treatment with MG132, there was no degradation of the IκB molecule, which is required for NF-κB translocation into the nucleus.10 MG132 also acts on AP-1, which activates genes related to the expression of proinflammatory cytokines and the induction of apoptosis.26,27 Our results for the AP-1 transcription factor suggest a defence mechanism for the maintenance of some capacity of response for the same or novel injury.
Another possible explanation of our results is that the proteasome regulates the basal levels of extracellular signal-regulated kinase 1/2 (ERK1/2), p38 and Jun N-terminal kinase (JNK) activation. Other studies have also shown that the proteasome regulates these kinases in THP-1 and RAW 264.7 cells.17,28 These kinases induce the activation of some enzyme members of the ADAM (a disintegrin and metalloproteinase) family of zinc metalloproteases, which participate in the cleavage and shedding of cytokine receptors, including ADAM17 for TNF-R1 and IL-1R1, and ADAM17 and ADAM10 for IL-6R. It has been reported that the proteasome is involved in the regulation of these enzymes and others, including protein phosphatase 2a, which induces the degradation of gp130 (a subunit of the IL-6 receptor).3,4,29,30 In this context, the observations made in this study are consistent with the reported literature and stress the importance of the proteasome as an active regulator of proinflammatory cytokines and their receptors.
ERK1/2, p38 and JNK participate in the activation of the AP-1 transcription factor.31 This would explain the results represented in Fig. 5, which show that proteasomal inhibition increased the activation of AP-1.
We are currently investigating the role of the proteasome in the regulation of receptors involved in the inflammatory process, like TREM-1, Toll-like receptors and NOD proteins, in immune cells and their regulation in in vitro and in vivo inflammatory models.
Our observations are important because both inflammatory and autoimmune diseases, such as rheumatoid arthritis, intestinal inflammatory disease, multiple sclerosis and asthma, are often associated with the active participation of monocytic cells and with the deregulated expression and biosynthesis of proinflammatory cytokines and their receptors, which influence a plethora of cellular functions.32,33 Therefore, proteasomal inhibition might be an anti-inflammatory strategy for these diseases.34 In conclusion, proteasomal inhibition may offer a means of regulating proinflammatory cytokines and their receptors, as well as the NF-κB and AP-1 transcription factors, which participate in the pathophysiology of inflammatory diseases.
Acknowledgments
We thank Drs Rosa Elena Navarro Hernández and Ana Soledad Sandoval Rodríguez for innumerable helpful suggestions and assistance. This work was supported by FOFOI/IMSS Grant 2005/1/I156.
References
- 1.Bjarne Ø, Eirik B. Role of monocytes in atherogenesis. Physiol Rev. 2003;3:1069–112. doi: 10.1152/physrev.00005.2003. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway C. Innate immunity. N Engl J Med. 2000;343:338–44. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 3.Levine SJ. Mechanisms of soluble cytokine receptor generation. J Immunol. 2004;173:5343–8. doi: 10.4049/jimmunol.173.9.5343. [DOI] [PubMed] [Google Scholar]
- 4.Jablonska E, Jabionski J, Holownia A, et al. TNFRs and IL-6R transmembrane receptors expression and release of their soluble forms by neutrophils and mononuclear cells from cancer patients. Rocz Akad Med Bialymst. 2001;46:113–25. [PubMed] [Google Scholar]
- 5.Qureshi N, Vogel SN, Charles VW, Papasian CJ, Qureshi AA, Morrison DC. The proteasome, a central regulatory of inflammation and macrophage function. Immunol Res. 2005;31:243–60. doi: 10.1385/IR:31:3:243. [DOI] [PubMed] [Google Scholar]
- 6.Kloetzel PM, Soza A, Stohwasser R. The role of the proteasome system and the proteasome activator PA28 complex in the cellular immune response. Biol Chem. 1999;380:293–7. doi: 10.1515/BC.1999.040. [DOI] [PubMed] [Google Scholar]
- 7.Chao-Huang Y, Yu-Chung Y, Ruscetti SK, Kirken RA, Dai RM, Li CCH. Involvement of the ubiquitin–proteasome pathway in the degradation of nontyrosine kinase-type cytokine receptors of IL-9, IL-2, and erythropoietin. J Immunol. 2000;165:6372–80. doi: 10.4049/jimmunol.165.11.6372. [DOI] [PubMed] [Google Scholar]
- 8.Aixin Y, Malek TR. The proteasome regulates receptor-mediated endocytosis of interleukin-2. J Biol Chem. 2001;276:381–5. doi: 10.1074/jbc.M007991200. [DOI] [PubMed] [Google Scholar]
- 9.Adams J. Proteasome inhibition: a novel approach to cancer therapy. Trends Mol Med. 2002;8:S49–S54. doi: 10.1016/s1471-4914(02)02315-8. [DOI] [PubMed] [Google Scholar]
- 10.Karin M, Greten RF. NF-kB: Linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 11.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–73. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 12.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–6. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 13.Sundstrom C, Nilsson K. Stablishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–77. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 14.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruger NJ. The Bradford method for protein quantitation. Methods Mol Biol. 1994;32:9–15. doi: 10.1385/0-89603-268-X:9. [DOI] [PubMed] [Google Scholar]
- 16.Šafránek R, Ishibashi N, Oka Y, Shirouzu K, Holeček M. Modulation of inflammatory response in sepsis by proteasome inhibition. Int J Exp Pathol. 2006;87:369–72. doi: 10.1111/j.1365-2613.2006.00490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuschieri J, Gourlay D, Garcia I, Jelacic S, Maier RV. Implications of proteasome inhibition: an enhanced macrophage phenotype. Cell Immunol. 2004;227:140–7. doi: 10.1016/j.cellimm.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Van Kerkhof P, Vallon E, Strous GJ. A method to increase the number of growth hormone receptors at the surface of cells. Mol Cell Endocrinol. 2003;201:57–62. doi: 10.1016/s0303-7207(02)00434-3. [DOI] [PubMed] [Google Scholar]
- 19.Hui-yen JS, Duong TT, Yeung RSM. TNF-α is necessary for induction of coronary artery inflammation and aneurysm formation in an animal model of Kawasaki disease. J Immunol. 2006;176:6294–301. doi: 10.4049/jimmunol.176.10.6294. [DOI] [PubMed] [Google Scholar]
- 20.Sims J, Gayle M, Slack J, et al. Interleukin 1 signaling occurs exclusively via the type I receptor. Proc Natl Acad Sci USA. 1993;90:6155–9. doi: 10.1073/pnas.90.13.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggarwal BB, Eessalu TE. Effect of phorbol esters on down-regulation and redistribution of cell surface receptors for tumour necrosis factor-α. J Biol Chem. 1987;262:16450–5. [PubMed] [Google Scholar]
- 22.Ding AH, Sanchez E, Srimal S, Nathan CF. Macrophages rapidly internalize their tumour necrosis factor receptors in response to bacterial lipopolysaccharide. J Biol Chem. 1989;264:3924–9. [PubMed] [Google Scholar]
- 23.Peiretti F, Canault M, Bernot D, et al. Proteasome inhibition activates the transport and the ectodomain shedding of TNF-α receptors in human endothelial cells. J Cell Sci. 2005;118:1061–70. doi: 10.1242/jcs.01696. [DOI] [PubMed] [Google Scholar]
- 24.Levine JS, Adamik B, Hawari FI, et al. Proteasome inhibition induces TNFR1 shedding from human airway epithelial (NCI-H292) cells. Am J Physiol Lung Cell Mol Physiol. 2005;289:233–43. doi: 10.1152/ajplung.00469.2004. [DOI] [PubMed] [Google Scholar]
- 25.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 26.Kriehuber E, Bauer W, Charbonnier AS, et al. Balance between NF-κB and JNK/AP-1 activity controls dendritic cell life and death. Blood. 2005;106:75–183. doi: 10.1182/blood-2004-08-3072. [DOI] [PubMed] [Google Scholar]
- 27.Johnston CJ, Holm BA, Gelein R, Finkelstein JN. Postnatal lung development: immediate-early gene responses post ozone and LPS exposure. Inhal Toxicol. 2006;18:875–83. doi: 10.1080/08958370600822466. [DOI] [PubMed] [Google Scholar]
- 28.Qureshi N, Perera PY, Shen J, et al. The proteasome as a lipopolysaccharide-binding protein in macrophages: differential effects of proteasome inhibition on lipopolysaccharide-induced signaling events. J Immunol. 2003;171:1515–25. doi: 10.4049/jimmunol.171.3.1515. [DOI] [PubMed] [Google Scholar]
- 29.Wei Q, Xia Y. Proteasome inhibition down-regulates endothelial nitric-oxide synthase phosphorylation and function. J Biol Chem. 2006;281:21652–9. doi: 10.1074/jbc.M602105200. [DOI] [PubMed] [Google Scholar]
- 30.Mitsuhashi S, Shima H, Tanuma N, et al. Protein phosphatase type 2A, PP2A, is involved in degradation of gp130. Mol Cell Biochem. 2005;269:183–7. doi: 10.1007/s11010-005-3089-x. [DOI] [PubMed] [Google Scholar]
- 31.Karin M. Inflammation-activated protein kinases as targets for drug development. Proc Am Thorac Soc. 2005;2:386–90. doi: 10.1513/pats.200504-034SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang Liu S, Mark AB. NF-κB activation as a patholoical mechanism of septic shock and inflammation. Am J Physiol Lung Cell Mol Physiol. 2006;290:L622–45. doi: 10.1152/ajplung.00477.2005. [DOI] [PubMed] [Google Scholar]
- 33.Garton KJ, Gough PJ, Raines EW. Emerging roles for ectodomain shedding in the regulation of inflammatory responses. J Leukoc Biol. 2006;79:1105–16. doi: 10.1189/jlb.0106038. [DOI] [PubMed] [Google Scholar]
- 34.Elliott PJ, Zollner TM. Proteasome inhibition: a new anti-inflammatory strategy. J Mol Med. 2003;81:235–45. doi: 10.1007/s00109-003-0422-2. [DOI] [PubMed] [Google Scholar]