Abstract
Synovial sarcomas are highly aggressive mesenchymal cancers that show modest response to conventional cytotoxic chemotherapy, suggesting a definite need for improved biotargeted agents. Progress has been hampered by the lack of insight into pathogenesis of this deadly disease. The presence of a specific diagnostic t(X;18) translocation leading to expression of the unique SYT-SSX fusion protein in effectively all cases of synovial sarcoma suggests a role in the etiology. Other nonspecific anomalies such as overexpression of Bcl-2, HER-2/neu, and EGFR have been reported, but their role in the pathogenesis remains unclear. Using gene targeting, we recently generated mice conditionally expressing the human SYT-SSX2 fusion gene from mouse endogenous ROSA26 promoter in chosen tissue types in the presence of Cre recombinase. These mice develop synovial sarcoma when SYT-SSX2 is expressed within myoblasts, thereby identifying a source of this enigmatic tumor and establishing a mouse model of this disease that recapitulates the clinical, histologic, immunohistochemical, and transcriptional profile of human synovial sarcomas. We review the genetics of synovial sarcoma and discuss the usefulness of genetics-based mouse models as a valuable research tool in the hunt for key molecular determinants of this lethal disease as well as a preclinical platform for designing and evaluating novel treatment strategies.
Introduction
Synovial sarcoma is a highly aggressive and distinct soft tissue sarcoma predominantly affecting young people, causing death in more than half of the affected children, adolescents, and young adults. It is a rare tumor, comprising 5% to 10% of all soft tissue sarcomas [83]. The 5- and 10-year survival rates have been reported as low as 36% and 20%, respectively [48]. Although the diagnostic and histologic features for synovial sarcoma are now well defined [83] and its molecular profile is beginning to be elucidated, the clinical pathogenesis of malignancy remains poorly understood. Surgical resection with or without adjuvant radiotherapy and/or doxorubicin-based chemotherapy are the mainstays of treatment.
Data regarding the 5-year survival rates for synovial sarcoma vary greatly, ranging from 36% to 76% [22]. Favorable prognostic indicators include low tumor grade, age younger than 15 years at the time of diagnosis, tumor size smaller than 5 cm, and location in the distal extremity [57, 74, 75]. In a prospective series of 112 patients with synovial sarcoma, tumor size 5 cm or greater and the presence of bone and neurovascular invasion were independent adverse predictors of distant recurrence and mortality [40]. No clear prognostic correlation has been demonstrated between the histologic subtype of synovial sarcoma and survival. Heavily calcified tumors reportedly have more favorable outcomes with 5-year survival reported as high as 82%, and tumors having at least 20% poorly differentiated histologic pattern have a worse prognosis [83]. Certain cytogenetic features of synovial sarcoma have also been reported to influence progression (discussed later).
Surgery remains the mainstay of treatment for synovial sarcoma. Obtaining optimal surgical margins is crucial for local tumor control, especially in intermediate- and high-grade tumors. Repeat resection may be required to achieve optimal margins. Wide surgical excision with margins equal to or greater than 1 cm or margins involving fascia is recommended. Obtaining adequate margins must be balanced with the goal of preserving maximal function and minimizing morbidity. In extremity tumors, limb-sparing procedures are preferred to amputation if a functional outcome is possible [40]. Considerations for using adjuvant radiation therapy for local treatment are not specific to synovial sarcomas per se but follow guidelines used for most adult soft tissue sarcomas. Two randomized, controlled trials suggest the usefulness and a trend toward survival benefit of doxorubicin-based chemotherapy for aggregated groups of soft tissue sarcomas [1, 63], whereas other studies suggest no such trend [54, 61]. Nevertheless, data specific to synovial sarcoma are sparse.
We begin with a brief background on the clinical and pathologic aspects of synovial sarcoma and then review the genetics and molecular aberrations associated with this disease with an emphasis on the unique and specific t(X;18) translocation and its implications on the disease process. We discuss various classic and emerging molecular biology techniques that have been instrumental in our quest to understand the molecular pathogenesis of this disease and conclude with the unique advantages and usefulness of genetically designed animal models using the example of a recently reported mouse model of synovial sarcoma.
Search Strategies and Criteria
We used the search terms “synovial sarcoma[ti]” or “SYT-SSX[ti]” to search the PubMed database and limit the terms to the title. We identified 1008 articles using the “synovial sarcoma” field tag, whereas 54 articles were identified using the “SYT-SSX” field tag (until August 2007). We excluded articles written in languages other than English and case studies without novel molecular or diagnostic observations. These criteria reduced the number articles to 85.
Clinical and Pathologic Features
Although the name synovial sarcoma connotes a histogenesis from synovial lining, a result of its predilection to arise near large joints, the ultrastructural and immunologic profile of the tumor cells is inconsistent with this notion [46, 71, 83]. The tissue of origin remains unknown despite efforts to identify its source using such strategies as microarray-based transcriptional signature comparison [24]. An alternate nomenclature of “carcinosarcoma” has been suggested based on frequent coexpression of epithelial and mesenchymal markers. The tumor is currently regarded as a neoplasm of “uncertain differentiation.”
Synovial sarcoma tumors are uniquely composed of two morphologically distinct cell types: spindle cells and epithelioid cells. The presence of the two cell types in varying proportions lends to the classification of tumors into three histologic subtypes that exist along a continuous spectrum: biphasic, monophasic (monophasic fibrous or rare monophasic epithelial), and poorly differentiated [83]. Light microscopy reveals the spindle cell component consists of small, uniform, and ovoid cells. The nuclei are pale, often with inconspicuous nucleoli. The cytoplasm is sparse, and vague cell borders lend to the appearance of overlapping nuclei. Monophasic fibrous-type tumors demonstrate predominantly spindle cells with no or minimal evidence of epithelioid differentiation. These spindle cells are packed into dense sheets with the occasional appearance of palisading nuclei. The tumor stroma involves scant collagen and occasional dense fibrosis as well as variable mast cell abundance, microcyst formation, and myxoid changes. Biphasic tumors contain a mélange of both spindle and epithelial cell components. The epithelial component reveals cells with similarly ovoid nuclei but demonstrates abundant cytoplasm. These cells often form glandular structures with lumina-containing epithelial mucin or papillary structures that resemble adenocarcinoma. The rare monophasic epithelial type is histologically indistinguishable from adenocarcinoma, requiring cytogenetic analysis for diagnosis. The poorly differentiated type of synovial sarcoma poses a diagnostic challenge and demonstrates common features of high-grade small round cell tumors: dense cellularity, numerous mitotic figures, and areas of necrosis.
Synovial sarcomas consistently stain positive for cytokeratin and epithelial membrane antigen [83]. Ninety percent of tumors are cytokeratin-positive with a stronger presence in the epithelial component than among spindle cells. Occasionally, cytokeratin antigens are expressed in monophasic fibrous-type synovial sarcoma. Unlike other spindle cell sarcomas, synovial sarcomas express cytokeratins 7 and 19, which helps to distinguish synovial sarcoma from primitive neuroectodermal tumors and malignant peripheral nerve sheathe tumors [23, 59]. Epithelial membrane antigens are more commonly present in poorly differentiated type tumors than cytokeratin [23]. Because cytokeratin and epithelial membrane antigen do not have perfect overlap, both are used to detect epithelial differentiation synovial sarcoma. Along with markers for epithelial differentiation, these tumors also express vimentin, suggesting mesenchymal differentiation and the antiapoptotic protein Bcl-2 [29, 59].
The Genetics of Synovial Sarcoma
Cytogenetics
Synovial sarcomas demonstrate a specific t(X;18)(p11.2;q11.2) translocation [56, 72, 81]. The breakpoint-associated genes are SYT, on chromosome 18, and SSX (SSX1, SSX2, or rarely SSX4) on the X chromosome [9, 10, 16, 69, 70]. This discovery led to the isolation of SYT-SSX fusion transcripts. The translocation is specific to synovial sarcoma and has been detected in both the spindle and epithelial component of the tumor [5, 34]. Although the SYT-SSX transcript is almost always detected in the tumor cells of synovial sarcoma, the reciprocal SSX-SYT is frequently undetectable [56]. As a result of the high frequency and specificity, detection of SYT-SSX using reverse transcriptase-polymerase chain reaction (RT-PCR) or fluorescence in situ hybridization is used as a diagnostic technique that is particularly useful in identifying monophasic fibrous or poorly differentiated synovial sarcomas [30]. The presence of this unique translocation product also suggests its central role in the etiology.
Translocation Partners
The SYT gene, also known as SS18 or SSXT, is located on chromosome 18q11 and encodes a 387-amino acid protein. SYT is ubiquitously expressed in both humans and mice and has 11 exons encompassing approximately 70 kb. The presence of canonical CpG islands and the absence of TATA elements in the promoter region of SYT are similar to the promoter region of housekeeping genes [12, 14]. The SYT protein (Fig. 1A) contains a novel 54-amino acid N-terminal domain designated as the SNH domain (SYT N-terminal homology domain) that is conserved in a wide variety of species [9, 12, 18]. The SNH domain interacts with the SWI/SNF ATPase BRM and BRGI, suggesting a role in chromatin remodeling [31, 60]. This domain has also been implicated in interaction of SYT with the acute leukemia-associated AF10, a putative transcriptional factor [11]. A recent study also demonstrated the N-terminal of SYT interacts with mSin3A, involved in transcriptional repression [32]. Other known regions of functional importance within the SYT protein are three SH2-binding motifs, one SH3-binding motif, and a C-terminal QPGY domain rich in glutamine, proline, glycine, and tyrosine that is similar to the transcriptional activation region within the EWS/FUS/TLS family of proteins [17, 79]. The SH2- and SH3-binding motifs are implicated in mediating protein-protein interactions in signal transduction [38, 58].
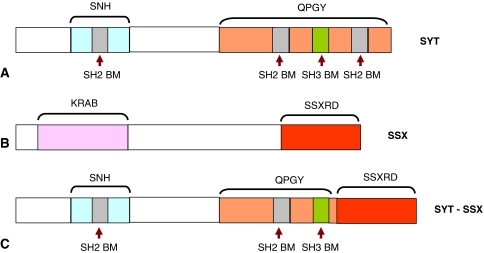
Fig. 1A−C.
A diagram illustrates the proteins involved in synovial sarcoma-specific t(X;18) translocation: (A) SYT protein; (B) SSX protein; and (C) SYT-SSX fusion protein, which retains almost the entire SYT except the last eight amino acids, which are replaced with the last 78 amino acids of SSX. SNH = SYT N-terminal homology domain; QPGY = glutamine-, proline-, glycine-, and tyrosine-rich domain; SH2 BM = Src homology 2 binding motif; SH3 BM = Src homology 3 binding motif; KRAB = Kruppel-associated box; SSXRD = SSX repression domain. Although SYT, SSX, and the SYT-SSX fusion proteins are localized to the nucleus, there is no nuclear localization signal or DNA-binding domain associated with these proteins.
Although the SYT lacks a typical DNA-binding domain and nuclear localization signal, the protein is localized in the nucleus and is believed to act as a transcriptional activator [7, 18]. SYT also plays a role in regulating cell adhesion through interaction with p300 histone acetyl transferase that appears independent of transcriptional activation [20]. Recently, a knockout mouse was generated for SYT that was lethal at the early implantation stage precluding further analysis [13]. A conditional knockout of this gene in specific cell/tissue types using the Cre-LoxP system may provide more valuable insight into its function.
Human SSX genes constitute a gene family with nine identified members [27]. Members of the SSX gene family have been categorized as “cancer testes” (CT) antigen, immunogenic antigens with expression in normal testes and various cancers [26, 27, 80]. All members of the SSX gene family share a high degree of similarity in nucleic acid and protein sequences [26]. Of the nine known members of the SSX gene family, only three, SSX1, SSX2, and very rarely SSX4, participate in the characteristic SYT-SSX translocation in synovial sarcoma, all encoding a 188-amino acid protein (Fig. 1B). Analysis of gene structure has revealed the presence of 10 exons, eight of which are used by all SSX genes [26, 27, 80].
The N-terminal of SSX protein contains the Kruppel-associated box (KRAB domain) (Fig. 1B) that is frequently seen in zinc finger proteins and is believed to mediate transcriptional repression [10, 41, 45]. The C-terminal of SSX protein contains another transcriptional repression domain (SSXRD domain) [41]. The SSX protein, like SYT, is localized to the nucleus but lacks any DNA-binding domain or nuclear localization signal. The SSXRD domain is responsible for the nuclear localization of this protein, interaction with polycomb group proteins implicated in gene repression, and association with histones [18, 19, 35, 73]. The C-terminal of SSX proteins also interacts with the Lim homeobox protein LHX4, a putative transcriptional factor that is deregulated in leukemias [15]. These suggest SSX, in association with other proteins, predominantly repress target genes.
The Chimeric Product
The presence of SYT-SSX transcripts in virtually all cases of synovial sarcoma suggests a crucial role of the fusion protein in the etiology of this tumor. Supporting this notion, studies by Nagai et al. [49] have demonstrated constitutive expression of human SYT-SSX1 in rat 3Y1 fibroblast-promoted growth rate in culture, anchorage-independent growth in soft agar, and formation of tumors with appearance similar to human synovial sarcoma on implantation of the transformed 3Y1 fibroblasts in nude mice. The N-terminal 181 amino acids of the fusion protein were required for this transforming activity. Nagai et al. [49] also demonstrated an interaction between SYT-SSX1 protein and the chromatin remodeling factor hBRM/hSNFα mediated through the N-terminal 181 amino acids of SYT-SSX1 and the region between amino acids 156 and 205 of hBRM/hSNFα. Overexpression of this interacting domain of hBRM/hSNFα inhibited the anchorage-independent growth in SYT-SSX1-expressing rat 3Y1 fibroblast, suggesting transformation by SYT-SSX requires interaction with hBRM/hSNFα.
The SYT-SSX chimeric protein usually has the last eight amino acids of SYT replaced by the 78 amino acids from the C-terminal of SSX (Fig. 1C). Although almost the entire SYT protein is retained in the fusion protein, overexpression of wild-type SYT did not induce transformation in culture (unlike SYT-SSX), suggesting the requirement of the SSX-derived regions for transformation [49]. Although the loss of the N-terminal region of the fusion protein leads to loss of anchorage-independent growth, the loss of 34 amino acids at the C-terminal (derived from SSX) did not compromise the colony-forming activity of the transfected rat 3Y1 fibroblast [49]. This suggests the remainder of the SSX-derived region (amino acids 111 to 154) confers transforming potential to the fusion protein. Furthermore, the loss of one SH2-binding motif of SYT in the fusion protein may have implications in the oncogenic activity because mutant SYT protein lacking the last eight amino acids act as dominant-negative to wild-type SYT function in cell adhesion [20]. Therefore, the gain of the C-terminal SSX region and the loss of C-terminal SYT may both contribute uniquely to the oncogenic potential of the SYT-SSX fusion protein.
The SYT-SSX fusion protein contains the putative transcriptional activation regions of the SYT along with the repression domain of SSX, raising interesting possibilities as to how the fusion protein may induce transformation. Because both parts do not have a DNA-binding domain or nuclear localization signal, binding partners may play a crucial role. The transforming action may be predominantly a result of misexpression or mistargeting of the SSXRD domain or, alternatively, the SNH domain. Which domain has the dominant effect in the context of oncogenic potential of the fusion protein remains to be elucidated, although one study suggests SSXRD acts dominant over SYT in nuclear localization of the fusion protein [18].
Clinical Correlates of Translocation Type
The type of translocation product, SYT-SSX1 or SYT-SSX2, has been proposed to influence the histology and prognosis of the tumor. SYT-SSX1 subtype is associated with a biphasic histology and a higher Ki-67 index, whereas SYT-SSX2 is associated with the monophasic spindle cell subtype [3, 37, 39, 42, 43, 53]. A multiinstitutional retrospective study suggested better survival associated with SYT-SSX2 subtype compared with SYT-SSX1 [39]. Tumors with simple karyotypes and SYT-SSX2 fusion had fewer metastases than tumors showing complex karyotypes together with SYT-SSX1 [56]. However, despite the correlation with rates of metastases, Guillou et al. [25] did not find SYT-SSX subtype strongly predicted survival.
Molecular Abnormalities
The presence of SYT-SSX transcripts in synovial sarcoma is unique and there is evidence for their etiologic role. However, very little is known about the pathogenesis of this disease. The suggested transcriptional regulatory role of SYT, SSX, and the SYT-SSX fusion protein indicates its oncogenic effect is primarily mediated through transcriptional deregulations. RT-PCR and microarray-based transcriptional profiling studies on synovial sarcomas have revealed (1) differentially expressed genes correlating with synovial sarcoma such as IGF2, OLFM1, TLE2, CNTAP1, DRPLA, EGFR, ZIC2, ATSV, NSP, and so on [2, 52]; (2) differentially expressed genes correlating with the histologic subtype of synovial sarcoma such as ERBB2, several keratin encoding genes, ELF3, TGFβ1, ANX4, and so on associated with the biphasic subtype [2, 50]; and (3) differentially expressed genes correlating with the SYT-SSX subtype (SYT-SSX1 versus SYT-SSX2) such as FOXC1 and NCAM1 [21].
Complementary to microarray analysis, immunologic detection has also identified overexpressed proteins in synovial sarcomas such as several members of the cytokeratins, vimentin, BCL2, HGF, C-MET, IGF-1R, SALL2, KIT, PDGFR-β, and TLE1 [47, 51, 77, 78, 84]. Although such immunologic detection lacks the power of microarray analysis in terms of number of candidates investigated, it has the advantage of looking at the final product (protein) as opposed to transcripts in microarray and RT-PCR analysis. This is especially important for proteins or pathways that have considerable posttranslational regulation in which changes in protein levels occur in the absence of considerable transcriptional changes such as the p53 tumor suppressor. Such expression-based analysis provides not only insight into the biology of the disease, but also potential new diagnostic and prognostic markers. Along with expression analysis, surveys for mutations in certain well-known tumor-associated genes have revealed mutations in p53, MDM2, H-ras, APC, PTEN, and so on [55, 65, 66]. However, such analysis has not been able to establish any strong correlation of specific mutations with overall survival.
Although considerable information exists on differential expression of genes and mutations and although study of these individual genetic/molecular aberrations in isolation provides new insight, putting them in a more global context of aberrant pathways is required. This is important because certain crucial therapeutically vulnerable elements of a pathway may lie upstream or downstream to the primary molecular aberration, precluding its identification in expression/mutation-based analyses. Several studies have suggested involvement of the architecture-regulating ephrin pathway and signaling pathways such as Wnt, IGF, ERBB2, HGF/MET, and β-catenin in synovial sarcoma [2, 4, 47, 62, 78, 82, 84]. New studies continue to provide further insight into how such pathways are involved in certain facets of the tumor phenotype. For example, IGF2 protects cells from anoikis allowing transformation [76]. Similarly, SYT-SSX2 induces nuclear recruitment of β-catenin that seems independent of the canonical Wnt pathway [62]. Some of these pathways may be more general to the oncogenic phenotype, whereas others are more unique to the tumor. The potential for targeted therapies capitalizing on newly identified aberrant pathways in synovial sarcomas is discussed later.
Model Systems to Study Synovial Sarcoma
Tumor Models
Basic investigations into pathogenesis of synovial sarcoma, like most other tumors, have relied on human tumors, cell lines, and xenograft systems as study models. Each of these systems has its own unique advantages and disadvantages. Although studying human tumors offers the most “authentic” model, it has limited use as a result of our inability to experimentally manipulate the system. Cell lines derived from tumors have high manipulability and are ideal for in vitro investigations. Cell lines are mostly pure tumor cells as opposed to tumors, which are composed of tumor cells, stroma, blood vessels, and so on and offer an “enriched” source of tumor cells for study. For this reason, however, a cell line in a Petri dish behaves very differently from a real tumor. Xenograft models, obtained by injecting immunodeficient mice with tumor cells, have certain advantages over human tumors and cell lines as a model system. The tumors in xenograft models develop in vivo and can also be manipulated at several levels, which makes them well suited for studying tumor biology and drug evaluation. However, xenograft models have several drawbacks such as the lack of a natural immune response in the immunodeficient mice that may alter the biology of the tumor considerably. Furthermore, xenograft models are generated by injecting tumors cells, the final phenotype in the tumor pathway, thereby precluding study of any early tumor-inducing pathways.
Mouse Model
An ideal model would be one in which the pathogenic pathway can be recapitulated from tumor induction all the way to advanced disseminated disease in an immunocompetent animal genetically and physiologically similar to humans. The mouse is an ideal organism to model human diseases as a result of its physiological and genetic similarity with humans and our ability to manipulate its genome in a precise and specific way. Gene targeting allows us to introduce specific alterations to the mouse genome. Such genetic alterations could be activating or inactivating specific genes [8]. A major advancement in transgenics has been the introduction of Cre-LoxP-based conditional systems. Cre recombinase is a Type I topoisomerase from bacteriophage P1 that catalyzes site-specific recombination of DNA between LoxP sites [68]. The LoxP recognition element is a 34-bp sequence consisting of two 13-bp inverted repeats flanking an 8-bp spacer region that confers directionality. Recombination products depend on the location and relative orientation of the LoxP sites. DNA between directly repeated LoxP sites will be excised while DNA between opposing LoxP sites will be inverted.
We have recently developed a mouse model of synovial sarcoma by inducing expression of the human SYT-SSX2 fusion protein within myoblasts of skeletal muscle lineage in mice by using the technique of conditional gene expression [28]. In our model, we have introduced the human SYT-SSX2 coding sequence in the ROSA26 locus [28] on chromosome 6 under the control of the ubiquitously active ROSA promoter [28, 85]. We introduced a LoxP-flanked transcriptional stop signal preceding the SYT-SSX2 coding sequence to make its expression conditional (Fig. 2). This simple strategy allows us to express SYT-SSX2 at any desired tissue or cell type by delivering Cre at the desired spatiotemporal domain. Several systems have been reported in the literature for the controlled delivery of Cre. The most widely used method is generating a mouse line that expresses Cre from the promoter of a gene whose expression pattern matches the desired spatiotemporal domain of Cre delivery. This could be achieved by either replacing an endogenous gene whose expression pattern matches the desired pattern of Cre with the Cre cDNA or placing the Cre cDNA at the 3′-UTR of the gene such that expression of Cre is driven through an internal ribosomal entry site (IRES).
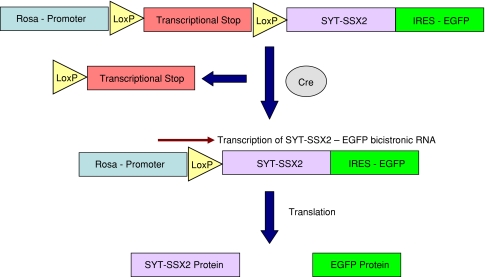
Fig. 2.
A diagram illustrates the conditional expression of SYT-SSX2 in the mouse model of synovial sarcoma. The transcriptional stop signal includes the Pgk-Neo selection cassette followed by a polyadenylation signal that prevents transcription of downstream SYT-SSX2. Cre recombines the two LoxP sites placed in the same orientation and removes the transcriptional stop and allows transcription through the ROSA promoter. Transcription leads to formation of a bicistronic SYT-SSX2-IRES-EGFP message that is then translated into two separate proteins, SYT-SSX2 and EGFP. This strategy allows expression of SYT-SSX2 within investigator-specified tissue based on delivery of the Cre recombinase.
The mouse model for synovial sarcoma is generated by breeding mice harboring the conditional SYT-SSX2 construct in ROSA locus to Myf5-Cre mice that expresses Cre in progenitors of skeletal muscles called myoblasts [28]. The tumors generated (Fig. 3) resemble human synovial sarcoma based on histologic appearance, immunohistochemistry, SYT-SSX2 expression in all tumor cells, and transcriptional profile [28]. By linking expression of SYT-SSX2 to expression of an enhanced green fluorescent protein (EGFP) through IRES (Fig. 2), we were able to mark all tumors based on green fluorescence (Fig. 3E). This facilitates identification of small primary tumors or metastasis on necropsy. Using this model system, we have been able to gain some new insights into the pathogenesis of this disease that highlights the usefulness of such model systems. An interesting observation is the generation of synovial sarcoma-like tumors by expressing the fusion protein in myoblast, thereby suggesting skeletal muscle lineage could be a source of this disease in humans. This is the first experimental evidence for a potential cell of origin for this mysterious tumor. Another intriguing observation was identification of both monophasic and biphasic subtypes in the mouse tumors by expressing SYT-SSX2, which has implications in the epidemiologic observation of correlation between tumor histology and translocation subtype. On preliminary examination of mouse tumors, there appears to be a correlation between tumor size and histology so that larger, more advanced tumors have biphasic histology, whereas more incipient tumors preferentially have monophasic histology. This suggests the tumors undergo a mesenchymal to epithelial transformation as they progress such that larger tumors have more epithelial component and the rate of this transformation is different between those harboring an SYT-SSX2 type and those with SYT-SSX1 type of translocation. Saito et al. [64] recently demonstrated the SYT-SSX fusion protein prevents the repression of E-cadherin, leading to aberrant overexpression of E-cadherin, an important determinant of epithelial differentiation; SYT-SSX1 demonstrated a stronger derepression of E-cadherin than SYT-SSX2, which further supports the notion that the correlation of tumor histology with fusion subtype is a reflection of the rate of tumor progression from a mesenchymal to an epithelial differentiation.
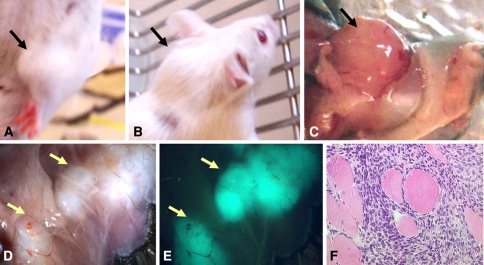
Fig. 3A–F.
Photographs show the tumors in the mouse model of synovial sarcoma: (A) a tumor in the right forelimb; (B) a tumor arising in the neck; (C) gross features of a large tumor in the left forelimb; and (D) small tumors in the thoracic cage. (E) Small tumors in the thoracic cage (shown in D) are easily identified by fluorescence. (F) Histology on a representative mouse synovial sarcoma shows the presence of trapped skeletal muscle lending further proof of a myogenic origin. Consistent expression of the EGFP fluorescent protein within the mouse synovial sarcoma-like tumors is apparent by the intense green fluorescence and demonstrates the expression of SYT-SSX2 within these tumors. Absence of fluorescence in the surrounding tissue suggests only the tumors express SYT-SSX2.
Along with basic biologic investigations, a mouse model is also a valuable tool for clinical investigations. One intriguing clinically relevant issue is whether there is any correlation between cell of origin and prognosis or drug response. It is possible synovial sarcoma (and other tumors) could have more than one source such that the clinical behavior of the tumor is influenced by its origin. The most attractive clinical use of such model systems, however, is to use them as preclinical platforms to evaluate new drugs or therapeutic strategies. Better trials with more statistical power can be easily designed, especially given the high penetrance of this model, and results from such preclinical trials can be extrapolated to the human case with a much greater degree of confidence than using either cell culture or xenograft models.
Discussion
Synovial sarcoma is a rare but aggressive tumor. The urgent need for a novel tumor-specific therapeutic strategy arises from synovial sarcoma’s moderate chemosensitivity and the fact that most patients who die of synovial sarcoma succumb to distant rather than local disease. The lack of complete understanding of the pathogenesis of this disease leave researchers steps shy of targeted therapy. As we discuss in this review, decades of investigation into the molecular pathogenesis of this disease have begun to provide considerable insight into the molecular and genetic aberrations associated with this disease.
Our study is limited by the search engine used and the search strategies. We did not include other databases (e.g., EMBASE, which covers European journals better than PubMed). We also excluded non-English language articles and limited our search to articles with the search terms in the title (and not the text). These strategies may have excluded important reports, but we presume the concepts would appear in articles searchable via PubMed and titles.
The most important concept to come out of the studies reviewed is the detection of a specific and unique t(X;18) translocation and expression of a derivative SYT-SSX fusion gene in this disease that not only provided us with a sensitive and reliable diagnostic marker, but also has the potential to be a prime candidate for targeted therapy in this disease. A recently generated mouse model demonstrated the SYT-SSX fusion protein indeed acts as an oncogene in this tumor. However, our current knowledge is very limited regarding tumorigenic events downstream to the expression of SYT-SSX. Although emerging pathway analyses are beginning to shed light into how SYT-SSX may induce transformation, the underlying mechanism is still largely a mystery. Future investigations will ask more focused mechanistic questions that are built on previous observations. We have now entered into an exciting phase of research into this disease in which the availability of considerable background information based on previous work and newly available tools such as more powerful microarray platforms, better analysis methods, advanced genetically designed mouse models, and so on should greatly expedite our understanding of this disease. However, some of these advanced research tools are still evolving and have certain limitations that must be identified and justified. In the case of the mouse model described here, synovial sarcoma cells often harbor a plethora of genetic and molecular aberrations as discussed in this review. It is still not clear whether the mouse model recapitulates all of these anomalies. In cancer, more than one genetic aberration often collaborates to induce transformation and the progress toward the final clinicopathologic phenotype of the tumor. Although theoretically not impossible, this is often very difficult to achieve in mouse model, mostly as a result of our incomplete understanding of the tumorigenic process. Therefore, rather than replacing classic methods, the mouse model is more of a complimentary research tool.
Several unanswered biologic questions are closely linked to the prospect of targeted therapy. One of them is whether the continuous presence of SYT-SSX is required for the maintenance of the tumor phenotype. As we have seen with the mouse model, expression of SYT-SSX2 appears sufficient to initiate the cascade of events leading to tumor formation. However, we still do not know whether SYT-SSX is required for tumor progression after its induction. If this is the case, then SYT-SSX offers the most tantalizing drug target because it is specific to tumor cells. The contribution of the variance in the SSX domain to tumor biology and prognosis is also unclear. Regardless, the SYT-SSX fusion protein does serve as a promising target for tumor-specific therapy such as immunotherapy. Cytotoxic CD8+ T lymphocytes specific to the SYT-SSX protein have been identified in patients with synovial sarcoma and induce a specific response by in vitro stimulations [67]. Recently, the first Phase 1 pilot trial of a SYT-SSX-derived junction peptide vaccine in six patients with synovial sarcoma was carried out. The procedure was well tolerated and one patient appeared to have stabilization of disease during the vaccination period [36]. Using inhibitors of SYT-SSX function or RNA interference-based targeted downregulation of SYT-SSX is another potentially effective strategy that should be explored.
Downstream effectors of SYT-SSX-induced tumorigenesis could also serve as potential drug targets. Several pathways and proteins that seem correlated with synovial sarcoma could prove worthy targets after further validation. Bcl-2 is an antiapoptotic protein that prevents or delays programmed cell death and is overexpressed in 79% to 94% of synovial sarcomas. Expression is limited to the spindle cells in biphasic synovial sarcomas, which may be related to a role of Bcl-2 in protecting cells from apoptosis before terminal differentiation [43]. The increased expression of Bcl-2 in synovial sarcoma is not the result of gene amplification or rearrangement but may be the result of transcriptional activation and/or enhanced protein stabilization [44]. Oblimersen (G3139, Genasense®; Genta Inc, Berkeley Heights, NJ) is an 18-base phosphorothiated antisense oligonucleotide complementary to the first six codons of the open reading frame of Bcl-2 mRNA. It has been designed to decrease Bcl-2 expression and therefore allow apoptosis of cancer cells, whether natural or chemotherapy-induced [33]. Experiments were performed to test the effect of Bcl-2 blockade by G3139 on the viability of the synovial cell line in the presence of doxorubicin [33]. The nontranslocation-containing SW982 had a dose-dependent decrease in viability after 24-hour incubation with doxorubicin. The viability was no less when the cells were first incubated with G3139 before the doxorubicin. However, the FU-SY-1 cell line, which showed a similar loss of viability after doxorubicin, had an augmented and dose-dependent loss of viability and increase in apoptosis when G3139 was added to the doxorubicin. Therefore, it appeared G3139 increased cell death only when the cell line had a high level of Bcl-2 expression and/or it contained the translocation. A study to test the benefit of the addition of G3139 to standard chemotherapy (doxorubicin and ifosfamide) on the neoadjuvant response rate of high-risk patients with synovial sarcoma is in development.
Epidermal growth factor receptors (EGFRs) are overexpressed in synovial sarcomas compared with other soft tissue sarcomas. Therefore, members of the EGF family made up of several tyrosine kinases are appealing targets in the treatment of synovial sarcoma. Although a Phase 2 trial of the EGFR inhibitor gefitinib for patients with locally advanced or metastatic synovial sarcomas that overexpress EGFR has been launched in Europe [6], preliminary data have not demonstrated a major response. Nonetheless, other therapeutic agents are in development targeting this pathway. Similarly, targets within other pathways involved in synovial sarcoma could be evaluated as potential targets. The mouse model we discussed here should be a useful tool for rapid preclinical evaluation of such new therapeutic modalities.
Along with new therapeutic strategies, there is a need for better prognostic determinants. The correlation of the fusion subtype, secondary genetic events, and histology with prognosis should be further addressed. In this regard as well, the mouse model will be valuable as a research tool in which much better controlled studies can be designed to answer such questions. Another intriguing issue is whether there are multiple cells of origin for this cancer such that the cell of origin could influence the prognosis. One way to investigate this would be to induce expression of SYT-SSX2 in several different tissue types in mice and look for tumor induction. If synovial sarcoma-like tumors are generated by expressing the fusion protein in more than one cell type, then one could compare these tumors to look for differences in the clinical and biologic behavior of the tumor.
Despite relatively slow progress, emerging data regarding key molecular pathways involved in synovial sarcoma should lead to development of novel targeted therapy, a necessity because improvements with existing cytotoxic chemotherapy seem unlikely. Translation research using model organisms such as the one we have recently developed has a prominent place in this quest for better therapy.
Footnotes
Each author certifies that he or she has no commercial associations (eg, consultancies, stock ownership, equity interest, patent/licensing arrangements, etc) that might pose a conflict of interest in connection with the submitted article.
References
- 1.Adjuvant chemotherapy for localised resectable soft-tissue sarcoma of adults: meta-analysis of individual data. Sarcoma Meta-analysis Collaboration. Lancet. 1997;350:1647–1654. [PubMed]
- 2.Allander SV, Illei PB, Chen Y, Antonescu CR, Bittner M, Ladanyi M, Meltzer PS. Expression profiling of synovial sarcoma by cDNA microarrays: association of ERBB2, IGFBP2, and ELF3 with epithelial differentiation. Am J Pathol. 2002;161:1587–1595. [DOI] [PMC free article] [PubMed]
- 3.Antonescu CR, Kawai A, Leung DH, Lonardo F, Woodruff JM, Healey JH, Ladanyi M. Strong association of SYT-SSX fusion type and morphologic epithelial differentiation in synovial sarcoma. Diagn Mol Pathol. 2000;9:1–8. [DOI] [PubMed]
- 4.Barco R, Hunt LB, Frump AL, Garcia CB, Benesh A, Caldwell RL, Eid JE. The synovial sarcoma SYT-SSX2 oncogene remodels the cytoskeleton through activation of the ephrin pathway. Mol Biol Cell. 2007;18:4003–4012. [DOI] [PMC free article] [PubMed]
- 5.Birdsall S, Osin P, Lu YJ, Fisher C, Shipley J. Synovial sarcoma specific translocation associated with both epithelial and spindle cell components. Int J Cancer. 1999;82:605–608. [DOI] [PubMed]
- 6.Blay JY, Ray-Coquard I, Alberti L, Ranchere D. Targeting other abnormal signaling pathways in sarcoma: EGFR in synovial sarcomas, PPAR-gamma in liposarcomas. Cancer Treat Res. 2004;120:151–167. [DOI] [PubMed]
- 7.Brett D, Whitehouse S, Antonson P, Shipley J, Cooper C, Goodwin G. The SYT protein involved in the t(X;18) synovial sarcoma translocation is a transcriptional activator localised in nuclear bodies. Hum Mol Genet. 1997;6:1559–1564. [DOI] [PubMed]
- 8.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. [DOI] [PubMed]
- 9.Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM, Gusterson BA, Cooper CS. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7:502–508. [DOI] [PubMed]
- 10.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. Embo J. 1995;14:2333–2340. [DOI] [PMC free article] [PubMed]
- 11.de Bruijn DR, dos Santos NR, Thijssen J, Balemans M, Debernardi S, Linder B, Young BD, Geurts van Kessel A. The synovial sarcoma associated protein SYT interacts with the acute leukemia associated protein AF10. Oncogene. 2001;20:3281–3289. [DOI] [PubMed]
- 12.de Bruijn DR, Kater-Baats E, Eleveld M, Merkx G, Geurts Van Kessel A. Mapping and characterization of the mouse and human SS18 genes, two human SS18-like genes and a mouse Ss18 pseudogene. Cytogenet Cell Genet. 2001;92:310–319. [DOI] [PubMed]
- 13.de Bruijn DR, Peters WJ, Chuva de Sousa Lopes SM, van Dijk AH, Willemse MP, Pfundt R, de Boer P, Geurts van Kessel A. Targeted disruption of the synovial sarcoma-associated SS18 gene causes early embryonic lethality and affects PPARBP expression. Hum Mol Genet. 2006;15:2936–2944. [DOI] [PubMed]
- 14.de Bruijn DR, Peters WJ, de Sousa Lopes SM, van Dijk AH, Willemse MP, Pfundt R, de Boer P, van Kessel AG. Targeted disruption of the synovial sarcoma-associated SS18 gene causes early embryonic lethality and affects PPARBP expression. Hum Mol Genet. 2006;15:2936–2944. [DOI] [PubMed]
- 15.de Bruijn DR, van Dijk AH, Willemse MP, van Kessel AG. The C terminus of the synovial sarcoma-associated SSX proteins interacts with the LIM homeobox protein LHX4. Oncogene. 2008;27:653–662. [DOI] [PubMed]
- 16.de Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet. 1995;4:1097–1099. [DOI] [PubMed]
- 17.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. [DOI] [PubMed]
- 18.dos Santos NR, de Bruijn DR, Kater-Baats E, Otte AP, van Kessel AG. Delineation of the protein domains responsible for SYT, SSX, and SYT-SSX nuclear localization. Exp Cell Res. 2000;256:192–202. [DOI] [PubMed]
- 19.dos Santos NR, de Bruijn DR, van Kessel AG. Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosomes Cancer. 2001;30:1–14. [DOI] [PubMed]
- 20.Eid JE, Kung AL, Scully R, Livingston DM. p300 interacts with the nuclear proto-oncoprotein SYT as part of the active control of cell adhesion. Cell. 2000;102:839–848. [DOI] [PubMed]
- 21.Fernebro J, Francis P, Eden P, Borg A, Panagopoulos I, Mertens F, Vallon-Christersson J, Akerman M, Rydholm A, Bauer HC, Mandahl N, Nilbert M. Gene expression profiles relate to SS18/SSX fusion type in synovial sarcoma. Int J Cancer. 2006;118:1165–1172. [DOI] [PubMed]
- 22.Ferrari A, Gronchi A, Casanova M, Meazza C, Gandola L, Collini P, Lozza L, Bertulli R, Olmi P, Casali PG. Synovial sarcoma: a retrospective analysis of 271 patients of all ages treated at a single institution. Cancer. 2004;101:627–634. [DOI] [PubMed]
- 23.Folpe AL, Schmidt RA, Chapman D, Gown AM. Poorly differentiated synovial sarcoma: immunohistochemical distinction from primitive neuroectodermal tumors and high-grade malignant peripheral nerve sheath tumors. Am J Surg Pathol. 1998;22:673–682. [DOI] [PubMed]
- 24.Fukukawa C, Nakamura Y, Katagiri T. Molecular target therapy for synovial sarcoma. Future Oncol. 2005;1:805–812. [DOI] [PubMed]
- 25.Guillou L, Benhattar J, Bonichon F, Gallagher G, Terrier P, Stauffer E, Somerhausen Nde S, Michels JJ, Jundt G, Vince DR, Taylor S, Genevay M, Collin F, Trassard M, Coindre JM. Histologic grade, but not SYT-SSX fusion type, is an important prognostic factor in patients with synovial sarcoma: a multicenter, retrospective analysis. J Clin Oncol. 2004;22:4040–4050. [DOI] [PubMed]
- 26.Gure AO, Tureci O, Sahin U, Tsang S, Scanlan MJ, Jager E, Knuth A, Pfreundschuh M, Old LJ, Chen YT. SSX: a multigene family with several members transcribed in normal testis and human cancer. Int J Cancer. 1997;72:965–971. [DOI] [PubMed]
- 27.Gure AO, Wei IJ, Old LJ, Chen YT. The SSX gene family: characterization of 9 complete genes. Int J Cancer. 2002;101:448–453. [DOI] [PubMed]
- 28.Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. [DOI] [PubMed]
- 29.Hibshoosh H, Lattes R. Immunohistochemical and molecular genetic approaches to soft tissue tumor diagnosis: a primer. Semin Oncol. 1997;24:515–525. [PubMed]
- 30.Hiraga H, Nojima T, Abe S, Sawa H, Yamashiro K, Yamawaki S, Kaneda K, Nagashima K. Diagnosis of synovial sarcoma with the reverse transcriptase-polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol. 1998;7:102–110. [DOI] [PubMed]
- 31.Ishida M, Tanaka S, Ohki M, Ohta T. Transcriptional co-activator activity of SYT is negatively regulated by BRM and Brg1. Genes Cells. 2004;9:419–428. [DOI] [PubMed]
- 32.Ito T, Ouchida M, Ito S, Jitsumori Y, Morimoto Y, Ozaki T, Kawai A, Inoue H, Shimizu K. SYT, a partner of SYT-SSX oncoprotein in synovial sarcomas, interacts with mSin3A, a component of histone deacetylase complex. Lab Invest. 2004;84:1484–1490. [DOI] [PubMed]
- 33.Joyner DE, Albritton KH, Bastar JD, Randall RL. G3139 antisense oligonucleotide directed against antiapoptotic Bcl-2 enhances doxorubicin cytotoxicity in the FU-SY-1 synovial sarcoma cell line. J Orthop Res. 2006;24:474–480. [DOI] [PubMed]
- 34.Kasai T, Shimajiri S, Hashimoto H. Detection of SYT-SSX fusion transcripts in both epithelial and spindle cell areas of biphasic synovial sarcoma using laser capture microdissection. Mol Pathol. 2000;53:107–110. [DOI] [PMC free article] [PubMed]
- 35.Kato H, Tjernberg A, Zhang W, Krutchinsky AN, An W, Takeuchi T, Ohtsuki Y, Sugano S, de Bruijn DR, Chait BT, Roeder RG. SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J Biol Chem. 2002;277:5498–5505. [DOI] [PubMed]
- 36.Kawaguchi S, Wada T, Ida K, Sato Y, Nagoya S, Tsukahara T, Kimura S, Sahara H, Ikeda H, Shimozawa K, Asanuma H, Torigoe T, Hiraga H, Ishii T, Tatezaki SI, Sato N, Yamashita T. Phase I vaccination trial of SYT-SSX junction peptide in patients with disseminated synovial sarcoma. J Transl Med. 2005;3:1. [DOI] [PMC free article] [PubMed]
- 37.Kawai A, Woodruff J, Healey JH, Brennan MF, Antonescu CR, Ladanyi M. SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N Engl J Med. 1998;338:153–160. [DOI] [PubMed]
- 38.Koch CA, Anderson D, Moran MF, Ellis C, Pawson T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. [DOI] [PubMed]
- 39.Ladanyi M, Antonescu CR, Leung DH, Woodruff JM, Kawai A, Healey JH, Brennan MF, Bridge JA, Neff JR, Barr FG, Goldsmith JD, Brooks JS, Goldblum JR, Ali SZ, Shipley J, Cooper CS, Fisher C, Skytting B, Larsson O. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res. 2002;62:135–140. [PubMed]
- 40.Lewis JJ, Antonescu CR, Leung DH, Blumberg D, Healey JH, Woodruff JM, Brennan MF. Synovial sarcoma: a multivariate analysis of prognostic factors in 112 patients with primary localized tumors of the extremity. J Clin Oncol. 2000;18:2087–2094. [DOI] [PubMed]
- 41.Lim FL, Soulez M, Koczan D, Thiesen HJ, Knight JC. A KRAB-related domain and a novel transcription repression domain in proteins encoded by SSX genes that are disrupted in human sarcomas. Oncogene. 1998;17:2013–2018. [DOI] [PubMed]
- 42.Llombart-Bosch A, Lopez-Guerrero JA, Peydro-Olaya A. Synovial sarcoma (SS): new perspectives supported by modern technology. Arkh Patol. 2002;64:39–47. [PubMed]
- 43.Lopes JM, Nesland JM, Reis-Filho JS, Holm R. Differential Ki67 and bcl-2 immunoexpression in solid-glandular and spindle cell components of biphasic synovial sarcoma: a double immunostaining assessment with cytokeratin and vimentin. Histopathology. 2002;40:464–471. [DOI] [PubMed]
- 44.Mancuso T, Mezzelani A, Riva C, Fabbri A, Dal Bo L, Sampietro G, Perego P, Casali P, Zunino F, Sozzi G, Pierotti MA, Pilotti S. Analysis of SYT-SSX fusion transcripts and bcl-2 expression and phosphorylation status in synovial sarcoma. Lab Invest. 2000;80:805–813. [DOI] [PubMed]
- 45.Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ 3rd. Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA. 1994;91:4509–4513. [DOI] [PMC free article] [PubMed]
- 46.Miettinen M, Virtanen I. Synovial sarcoma—a misnomer. Am J Pathol. 1984;117:18–25. [PMC free article] [PubMed]
- 47.Motoi T, Ishida T, Kuroda M, Horiuchi H, Oka T, Matsumoto K, Nakamura T, Machinami R. Coexpression of hepatocyte growth factor and c-Met proto-oncogene product in synovial sarcoma. Pathol Int. 1998;48:769–775. [DOI] [PubMed]
- 48.Mullen JR, Zagars GK. Synovial sarcoma outcome following conservation surgery and radiotherapy. Radiother Oncol. 1994;33:23–30. [DOI] [PubMed]
- 49.Nagai M, Tanaka S, Tsuda M, Endo S, Kato H, Sonobe H, Minami A, Hiraga H, Nishihara H, Sawa H, Nagashima K. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci USA. 2001;98:3843–3848. [DOI] [PMC free article] [PubMed]
- 50.Nagayama S, Katagiri T, Tsunoda T, Hosaka T, Nakashima Y, Araki N, Kusuzaki K, Nakayama T, Tsuboyama T, Nakamura T, Imamura M, Nakamura Y, Toguchida J. Genome-wide analysis of gene expression in synovial sarcomas using a cDNA microarray. Cancer Res. 2002;62:5859–5866. [PubMed]
- 51.Nielsen TO, Hsu FD, O’Connell JX, Gilks CB, Sorensen PH, Linn S, West RB, Liu CL, Botstein D, Brown PO, van de Rijn M. Tissue microarray validation of epidermal growth factor receptor and SALL2 in synovial sarcoma with comparison to tumors of similar histology. Am J Pathol. 2003;163:1449–1456. [DOI] [PMC free article] [PubMed]
- 52.Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O’Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. [DOI] [PubMed]
- 53.Nilsson G, Skytting B, Xie Y, Brodin B, Perfekt R, Mandahl N, Lundeberg J, Uhlen M, Larsson O. The SYT-SSX1 variant of synovial sarcoma is associated with a high rate of tumor cell proliferation and poor clinical outcome. Cancer Res. 1999;59:3180–3184. [PubMed]
- 54.O’Byrne K, Steward WP. The role of chemotherapy in the treatment of adult soft tissue sarcomas. Oncology. 1999;56:13–23. [DOI] [PubMed]
- 55.Oda Y, Sakamoto A, Satio T, Kawauchi S, Iwamoto Y, Tsuneyoshi M. Molecular abnormalities of p53, MDM2, and H-ras in synovial sarcoma. Mod Pathol. 2000;13:994–1004. [DOI] [PubMed]
- 56.Panagopoulos I, Mertens F, Isaksson M, Limon J, Gustafson P, Skytting B, Akerman M, Sciot R, Dal Cin P, Samson I, Iliszko M, Ryoe J, Debiec-Rychter M, Szadowska A, Brosjo O, Larsson O, Rydholm A, Mandahl N. Clinical impact of molecular and cytogenetic findings in synovial sarcoma. Genes Chromosomes Cancer. 2001;31:362–372. [DOI] [PubMed]
- 57.Paulino AC. Synovial sarcoma prognostic factors and patterns of failure. Am J Clin Oncol. 2004;27:122–127. [DOI] [PubMed]
- 58.Pawson T, Gish GD. SH2 and SH3 domains: from structure to function. Cell. 1992;71:359–362. [DOI] [PubMed]
- 59.Pelmus M, Guillou L, Hostein I, Sierankowski G, Lussan C, Coindre JM. Monophasic fibrous and poorly differentiated synovial sarcoma: immunohistochemical reassessment of 60 t(X;18)(SYT-SSX)-positive cases. Am J Surg Pathol. 2002;26:1434–1440. [DOI] [PubMed]
- 60.Perani M, Ingram CJ, Cooper CS, Garrett MD, Goodwin GH. Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene. 2003;22:8156–8167. [DOI] [PubMed]
- 61.Pisters PW, Patel SR, Prieto VG, Thall PF, Lewis VO, Feig BW, Hunt KK, Yasko AW, Lin PP, Jacobson MG, Burgess MA, Pollock RE, Zagars GK, Benjamin RS, Ballo MT. Phase I trial of preoperative doxorubicin-based concurrent chemoradiation and surgical resection for localized extremity and body wall soft tissue sarcomas. J Clin Oncol. 2004;22:3375–3380. [DOI] [PubMed]
- 62.Pretto D, Barco R, Rivera J, Neel N, Gustavson MD, Eid JE. The synovial sarcoma translocation protein SYT-SSX2 recruits beta-catenin to the nucleus and associates with it in an active complex. Oncogene. 2006;25:3661–3669. [DOI] [PubMed]
- 63.Rosenberg SA. Prospective randomized trials demonstrating the efficacy of adjuvant chemotherapy in adult patients with soft tissue sarcomas. Cancer Treat Rep. 1984;68:1067–1078. [PubMed]
- 64.Saito T, Nagai M, Ladanyi M. SYT-SSX1 and SYT-SSX2 interfere with repression of E-cadherin by snail and slug: a potential mechanism for aberrant mesenchymal to epithelial transition in human synovial sarcoma. Cancer Res. 2006;66:6919–6927. [DOI] [PubMed]
- 65.Saito T, Oda Y, Kawaguchi K, Takahira T, Yamamoto H, Tanaka K, Matsuda S, Sakamoto A, Iwamoto Y, Tsuneyoshi M. PTEN and other tumor suppressor gene mutations as secondary genetic alterations in synovial sarcoma. Oncol Rep. 2004;11:1011–1015. [PubMed]
- 66.Saito T, Oda Y, Sakamoto A, Kawaguchi K, Tanaka K, Matsuda S, Tamiya S, Iwamoto Y, Tsuneyoshi M. APC mutations in synovial sarcoma. J Pathol. 2002;196:445–449. [DOI] [PubMed]
- 67.Sato Y, Nabeta Y, Tsukahara T, Hirohashi Y, Syunsui R, Maeda A, Sahara H, Ikeda H, Torigoe T, Ichimiya S, Wada T, Yamashita T, Hiraga H, Kawai A, Ishii T, Araki N, Myoui A, Matsumoto S, Umeda T, Ishii S, Kawaguchi S, Sato N. Detection and induction of CTLs specific for SYT-SSX-derived peptides in HLA-A24(+) patients with synovial sarcoma. J Immunol. 2002;169:1611–1618. [DOI] [PubMed]
- 68.Sauer B, Henderson N. Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 1989;17:147–161. [DOI] [PMC free article] [PubMed]
- 69.Shipley JM, Clark J, Crew AJ, Birdsall S, Rocques PJ, Gill S, Chelly J, Monaco AP, Abe S, Gusterson BA, et al. The t(X;18)(p11.2;q11.2) translocation found in human synovial sarcomas involves two distinct loci on the X chromosome. Oncogene. 1994;9:1447–1453. [PubMed]
- 70.Skytting B, Nilsson G, Brodin B, Xie Y, Lundeberg J, Uhlen M, Larsson O. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst. 1999;91:974–975. [DOI] [PubMed]
- 71.Smith ME, Fisher C, Wilkinson LS, Edwards JC. Synovial sarcoma lack synovial differentiation. Histopathology. 1995;26:279–281. [DOI] [PubMed]
- 72.Smith S, Reeves BR, Wong L, Fisher C. A consistent chromosome translocation in synovial sarcoma. Cancer Genet Cytogenet. 1987;26:179–180. [DOI] [PubMed]
- 73.Soulez M, Saurin AJ, Freemont PS, Knight JC. SSX and the synovial-sarcoma-specific chimaeric protein SYT-SSX co-localize with the human Polycomb group complex. Oncogene. 1999;18:2739–2746. [DOI] [PubMed]
- 74.Spillane AJ, A’Hern R, Judson IR, Fisher C, Thomas JM. Synovial sarcoma: a clinicopathologic, staging, and prognostic assessment. J Clin Oncol. 2000;18:3794–3803. [DOI] [PubMed]
- 75.Spurrell EL, Fisher C, Thomas JM, Judson IR. Prognostic factors in advanced synovial sarcoma: an analysis of 104 patients treated at the Royal Marsden Hospital. Ann Oncol. 2005;16:437–444. [DOI] [PubMed]
- 76.Sun Y, Gao D, Liu Y, Huang J, Lessnick S, Tanaka S. IGF2 is critical for tumorigenesis by synovial sarcoma oncoprotein SYT-SSX1. Oncogene. 2006;25:1042–1052. [DOI] [PubMed]
- 77.Tamborini E, Bonadiman L, Greco A, Gronchi A, Riva C, Bertulli R, Casali PG, Pierotti MA, Pilotti S. Expression of ligand-activated KIT and platelet-derived growth factor receptor beta tyrosine kinase receptors in synovial sarcoma. Clin Cancer Res. 2004;10:938–943. [DOI] [PubMed]
- 78.Terry J, Saito T, Subramanian S, Ruttan C, Antonescu CR, Goldblum JR, Downs-Kelly E, Corless CL, Rubin BP, van de Rijn M, Ladanyi M, Nielsen TO. TLE1 as a diagnostic immunohistochemical marker for synovial sarcoma emerging from gene expression profiling studies. Am J Surg Pathol. 2007;31:240–246. [DOI] [PubMed]
- 79.Thaete C, Brett D, Monaghan P, Whitehouse S, Rennie G, Rayner E, Cooper CS, Goodwin G. Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum Mol Genet. 1999;8:585–591. [DOI] [PubMed]
- 80.Tureci O, Chen YT, Sahin U, Gure AO, Zwick C, Villena C, Tsang S, Seitz G, Old LJ, Pfreundschuh M. Expression of SSX genes in human tumors. Int J Cancer. 1998;77:19–23. [DOI] [PubMed]
- 81.van de Rijn M, Barr FG, Collins MH, Xiong QB, Fisher C. Absence of SYT-SSX fusion products in soft tissue tumors other than synovial sarcoma. Am J Clin Pathol. 1999;112:43–49. [DOI] [PubMed]
- 82.Watanabe T, Tsuda M, Makino Y, Ichihara S, Sawa H, Minami A, Mochizuki N, Nagashima K, Tanaka S. Adaptor molecule Crk is required for sustained phosphorylation of Grb2-associated binder 1 and hepatocyte growth factor-induced cell motility of human synovial sarcoma cell lines. Mol Cancer Res. 2006;4:499–510. [DOI] [PubMed]
- 83.Weiss SW, Goldblum JR. Enzinger and Weiss’s Soft Tissue Tumors. 4th Ed. St Louis: Mosby, Inc; 2001.
- 84.Xie Y, Skytting B, Nilsson G, Brodin B, Larsson O. Expression of insulin-like growth factor-1 receptor in synovial sarcoma: association with an aggressive phenotype. Cancer Res. 1999;59:3588–3591. [PubMed]
- 85.Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. [DOI] [PMC free article] [PubMed]