Abstract
We have previously identified a cellular protein kinase activity termed TAK that specifically associates with the HIV types 1 and 2 Tat proteins. TAK hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II in vitro in a manner believed to activate transcription [Herrmann, C. H. & Rice, A. P. (1995) J. Virol. 69, 1612–1620]. We show here that the catalytic subunit of TAK is a known human kinase previously named PITALRE, which is a member of the cyclin-dependent family of proteins. We also show that TAK activity is elevated upon activation of peripheral blood mononuclear cells and peripheral blood lymphocytes and upon differentiation of U1 and U937 promonocytic cell lines to macrophages. Therefore, in HIV-infected individuals TAK may be induced in T cells following activation and in macrophages following differentiation, thus contributing to high levels of viral transcription and the escape from latency of transcriptionally silent proviruses.
The HIV types 1 and 2 (HIV-1 and HIV-2) encode closely related transcriptional activator proteins termed Tat-1 and Tat-2, respectively, that are essential for efficient viral replication (reviewed in ref. 1). To stimulate transcription, Tat proteins bind directly to the TAR RNA element that forms at the 5′ end of viral transcripts. The Tat activation domain is then positioned to activate the cellular RNA polymerase II (RNAP II) complex, predominantly acting at the level of transcriptional elongation. A number of cellular factors have been proposed to interact with the Tat activation domain and mediate transcription function, including TATA box-binding protein (2), Tat-SF1 (3), TFIIH (4, 5), and a cellular protein kinase activity termed TAK (Tat-associated kinase) (6, 7).
We have obtained substantial evidence that TAK may be an important Tat cofactor. In extensive analyses of wild-type and mutant Tat proteins, there is a precise correlation with in vivo function of the activation domains of Tat-1, Tat-2, and related equine infectious anemia virus Tat protein and the ability to bind TAK in vitro in glutathione S-transferase (GST) “pull-down” assays or in vivo as detected in coimmunoprecipitations (7, 8). TAK is a serine/threonine kinase that can cooperatively hyperphosphorylate the carboxyl-terminal domain (CTD) of the large subunit of RNAP II in vitro (7). Additionally, TAK phosphorylates the carboxyl terminus of Tat-2 in vivo and in vitro (6, 8); TAK does not phosphorylate Tat-1, probably because the carboxyl-terminal sequence of Tat-1 is unrelated to Tat-2. In kinase reactions, TAK also phosphorylates a 42-kDa polypeptide that appears to be a TAK subunit (8). Partial purification of TAK from HeLa cell nuclear extracts indicates that its native molecular mass is approximately 110 kDa. Characterization of TAK indicates that it is unrelated to previously identified cellular CTD kinases for which antisera are available (8, 9). As discussed previously (7, 8), it is possible that TAK is related to Drosophila pTEFb, a CTD kinase involved in transcriptional elongation (10).
The CTD is a heptad repeat consisting of the consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser, which is present at the carboxyl terminus of the large subunit of RNAP II. The underphosphorylated CTD (CTDa form) is found preferentially in RNAP II preinitiation complexes, whereas the hyperphosphorylated CTD (CTDo form) is found in actively elongating RNAP II complexes (11). Therefore, it is believed that phosphorylation of the CTD is a mechanism to convert RNAP II to a productively elongating complex. In addition to binding to TAK, Tat can associate with the basal transcription factor TFIIH, which is a CTD kinase (4, 5). Thus, Tat may activate transcription by the recruitment or activation of at least two cellular kinases, perhaps resulting in a cascade of CTD phosphorylation. In support of this notion, the CTD is required for Tat function in vivo (8, 12).
We have investigated here whether TAK is regulated in cells relevant to HIV infection. The results indicate that TAK activity is induced in activated peripheral blood mononuclear cells (PBMCs) and in peripheral blood lymphocytes (PBLs), as well as in promonocytic cell lines stimulated to differentiate to macrophages. Additionally, we show that the 42-kDa TAK subunit is a previously identified protein kinase named PITALRE. PITALRE is related to the cyclin-dependent family of protein kinases (CDKs) and was not previously recognized as a CTD kinase (13).
MATERIALS AND METHODS
Cells and Plasmids.
Monolayer cultures of HeLa cells and suspension cultures of U1 and U937 were maintained in Dulbecco’s modified Eagle’s and RPMI 1640 medium, respectively, supplemented with 10% fetal calf serum (FCS), penicillin, and streptomycin. To obtain PBMCs, heparinized blood was drawn from healthy HBV- and HIV-seronegative donors and diluted 1:1 with PBS, layered onto Ficoll/Paque (Pharmacia), and centrifuged at 1,200 rpm for 30 min at 20°C in a Sorvall 6000 (14). The mononuclear layer was removed and washed twice with cold PBS, and cells were resuspended in RPMI medium containing 10% FBS. PBLs were obtained from PBMCs after monocyte depletion using plastic adherence. Lymphocytes were 96% pure as determined by flow cytometry. For activation, cells were adjusted to 1 × 106/ml, phytohemagglutinin (PHA) was used at a final concentration of 1 μg/ml (Burroughs Wellcome), and phorbol 12-myristate 13-acetate (PMA) was used at 1 ng/ml (Sigma). HIV-1 p24 in culture supernatants was measured using a Coulter ELISA kit (15). Nuclear extracts and whole cell extracts from HeLa cells were prepared as described (7).
Standard PCR techniques using oligonucleotide primers based on the published sequence of PITALRE (13) were used to insert the full-length PITALRE cDNA into the FLAG-CMV-2 vector (Eastman Kodak). The entire protein-coding region of the FLAG-PITALRE cDNA was sequenced. For transfections of plasmids in HeLa cells, a liposome method was performed as recommended by the manufacturer (LipofectAMINE reagent; Life Technologies, Gaithersburg, MD). Recombinant adenoviruses were constructed that express the 72-residue HIV-1 Tat protein or 72-residue pro18IS mutant protein (16). Infections in HeLa cells (10-cm culture dishes) were carried out at a multiplicity of infection of 20, using an absorption period of 45 min in 2 ml of medium.
Immunoprecipitations, Immunoblots, Phosphopeptide Mapping, and Depletions of Nuclear Extracts.
An antiserum against the HIV-1 Tat protein was obtained from the AIDS Research and Reference Reagent Program (donated by B. Cullen, Duke University Medical Center, Durham, NC). Commercial antisera were obtained against the FLAG epitope (M2 monoclonal antibody, Eastman Kodak), CDK7 (Santa Cruz Biotechnology), and CDK2 (Santa Cruz Biotechnology). PITALRE antisera were obtained that were raised against carboxyl-terminal residues 353–372, which are not found in other CDKs (Santa Cruz Biotechnology) or raised against a GST-PITALRE fusion protein (Biodesign International, Kennebunkport, ME). Immunoprecipitations and immunoblots using enhanced chemiluminescence were performed by standard techniques as described (8). For GST “pull-down” assays, GST fusion proteins were incubated with a HeLa nuclear extract and washed extensively as described (7). Phosphopeptide maps using V8 protease were performed as described (8). Depletions of nuclear extracts were carried out with CDK7 and PITALRE antibodies and GST-Tat fusion proteins as described (9).
Kinase Assays.
For kinase reactions of immunoprecipitates, whole cell extracts were prepared and immunoprecipitations were performed as described (8). For reactions with GST-Tat fusion proteins attached to glutathione-Sepharose beads, whole cell extracts were prepared and binding and washing conditions were performed as described (7). The immune or bead complexes were incubated for 60 min at room temperature in 50 mM Tris⋅HCl (pH 7.4), 10 mM MgCl2, 5 mM DTT, 2.5 mM MnCl2, 200 ng GST-CTD (17), 5 μM or 10 μM ATP, and 5 μCi [γ-32P]ATP (3,000 Ci/mmol; 1 Ci = 37 GBq; New England Nuclear). To ensure that reactions were quantitative, pilot kinase assays were performed with different amounts of extracts. Where applicable (Figs. 1, 2, and 4), reactions were carried out with amounts of cell extracts that were in the linear range of assays for the kinase activities. Kinase assays were separated on SDS/polyacrylamide gels and were quantified with a Molecular Dynamics PhosphorImager.
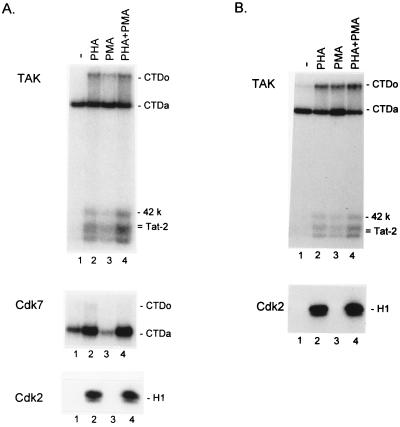
Figure 1.
PHA and PMA induction of TAK activity in PBMCs and PBLs. PBMCs (A) and PBLs (B) were isolated and incubated for 48 hr in medium alone or medium containing PHA and/or PMA as indicated. Kinase assays for TAK, CDK7, and CDK2 were performed as described in Material and Methods, and products were analyzed on a 9% SDS/polyacrylamide gels. The CTDo and CTDa, 42-kDa, and Tat-2 substrates of TAK are indicated; the CTDo and CTDa substrates of CDK7 and the H1 substrate of CDK2 are also indicated.
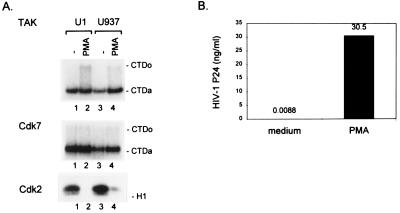
Figure 2.
PMA induction of TAK activity in U1 and U937 promonocytic cell lines. U1 and U937 cells were incubated in medium alone or medium containing PMA for 48 hr. (A) Kinase assays for TAK, CDK7, and CDK2 activities were performed as described in Fig. 1. (B) Amount of p24 in culture supernatants was measured by ELISA.
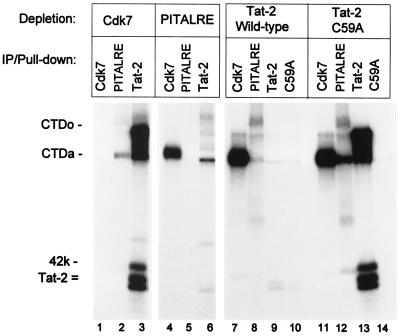
Figure 4.
Depletion of TAK activity from nuclear extracts with PITALRE antibodies. A HeLa cell nuclear extract was incubated as described in Materials and Methods with either antiserum against CDK7 or PITALRE or with GST-Tat-2 (wild type) or GST-C59A (Tat-2 mutant). Kinase assays for CDK7, PITALRE, or TAK (GST-Tat-2 and GST- C59A) remaining in extracts were then performed and analyzed on a 9% SDS/polyacrylamide gel.
RESULTS
Induction of TAK Activity in PBMCs and PBLs by PMA and PHA.
To investigate regulation of TAK in cells relevant to HIV infection, PBMCs from HBV- and HIV-seronegative donors were isolated and incubated for 48 hr in the presence of medium alone or medium containing PHA, PMA, or both PHA and PMA. Extracts were then prepared, and TAK levels were quantitatively measured by an in vitro kinase assay. In this assay, a GST fusion to the wild-type 99 residue Tat-2 protein was used to bind TAK from cell extracts. TAK bound to the Tat-2 protein phosphorylates the 42-kDa TAK subunit, the Tat-2 protein, and recombinant CTD added to reactions (7). In this assay, two forms of phosphorylated GST-Tat-2 are produced that migrate differently on SDS/polyacrylamide gels (6). Importantly, TAK hyperphosphorylates the recombinant CTD substrate to generate the CTDo form, as detected by significantly slower migration in SDS/polyacrylamide gels relative to the hypophosphorylated CTDa form. Transcription complexes containing the CTDo form of RNAPII are believed to be competent for elongation (11). To evaluate the specificity of TAK regulation, an antiserum against the CDK7 subunit of the TFIIH complex and an antiserum against the CDK2 protein were used to measure the levels of these kinase activities.
TAK levels were clearly elevated in extracts from PBMCs treated with PHA, PMA, or PHA + PMA, as indicated by phosphorylation of the CTD, 42-kDa, and Tat-2 substrates (Fig. 1A). As quantified by generation of the CTDo form of the CTD substrate, TAK levels were elevated 4.3-fold by PHA treatment, 3.1-fold by PMA treatment, and 4.9-fold by PHA + PMA treatment. In contrast, CDK7 activity was not significantly altered by either PHA or PHA + PMA treatment (1.4-fold and 0.8-fold inductions, respectively) and was reduced approximately 3-fold by PMA treatment. CDK2 activity was induced by PHA and PHA + PMA but not by PMA alone, as expected, because PHA induces T cell division whereas PMA does not. We obtained similar results as those shown in Fig. 1A from PBMCs isolated from three other donors. Because PHA and PMA treatment are known to activate HIV-1 LTR transcription in PBMCs (18), induction of TAK may be involved in stimulation of viral transcription by these agents.
Because T cells are a primary target of HIV infection, we examined TAK regulation in PBLs that were prepared from PBMCs by monocyte depletion by plastic adherence (see Materials and Methods). PBLs were incubated for 48 hr in medium alone or medium containing PHA, PMA, or both PHA and PMA. Extracts were prepared and assayed for relative levels of TAK and CDK2 (Fig. 1B). TAK levels were elevated by these agents in lymphocytes depleted of monocytes, indicating that monocytes were not largely responsible for induction of TAK observed in PBMCs. As quantified by generation of the CTDo product, TAK levels were induced 2.9-fold by PHA, 2.9-fold by PMA, and 3.5-fold by PHA + PMA. As expected, CDK2 levels were induced by PHA or PHA + PMA but not by PMA alone.
Induction of TAK Activity in U1 and U937 Cells by PMA.
We were interested in regulation of TAK in promonocytic cell lines in which PMA is known to induce differentiation to macrophages and also induce cellular factors involved in transcription of the HIV-1 LTR (19). We examined the U1 cell line that contains an integrated HIV-1 provirus that can be activated transcriptionally by PMA, as well as the parental U937 cell line. Cultures of U1 and U937 cells were incubated in the presence or absence of PMA for 48 hr, extracts were prepared, and levels of TAK, CDK7, and CDK2 were measured (Fig. 2A). Measurement of p24 production in the supernatant of U1 cells indicated that PMA treatment induced viral replication more than 3,000-fold (Fig. 2B). Quantitation of the CTDo product demonstrated that PMA elevated TAK levels 3.9-fold in U1 cells and 2.8-fold in U937 cells, suggesting that induction of TAK by PMA may be involved in the transcriptional activation of the HIV-1 LTR in these cell lines (19). We note that our assay may underestimate TAK activity in U1 cell extracts, because these extracts contain endogenous Tat-1 protein, which may bind TAK and prevent its binding to the GST-Tat-2 fusion protein. CDK7 levels were essentially unchanged by PMA treatment of U1 and U937 cells (1.1-fold and 1.3-fold induction, respectively). CDK2 levels were reduced by PMA in both cell lines as expected, because PMA causes the cells to stop proliferation, enlarge, and differentiate to macrophages (20). We failed to observe induction of TAK by PMA in the transformed T cell lines Jurkat and CEM (data not shown). However, TAK levels in these cell lines are high relative to PBMCs, PBLs, and U1/U937 cells. Therefore, it is possible that in transformed T cell lines TAK activity is deregulated and expressed at a constitutively high level.
The CDK Family Member PITALRE Is a Component of TAK.
Although our characterization of TAK suggested that it is unrelated to previously described CTD kinases, it was possible that TAK is related to a known human kinase that had not been evaluated as a CTD kinase. A feature of many members of the cyclin-dependent family of protein kinases is their ability to hyperphosphorylate the CTD; CDKs that have been shown to be such CTD kinases include CDC2 (21), CDK2 (7), CDK7 (22), and CDK8 (23). Examination of CDK family members described in the literature revealed a 42-kDa kinase that was a candidate for a TAK subunit. This protein, named PITALRE for its PSTAIRE-like motif, was isolated using PCR primers designed to amplify CDK-related proteins (13). The cyclin partner of PITALRE has not yet been identified, and no cell cycle regulation or specific function has been ascribed to this kinase. PITALRE appears to be ubiquitously expressed in human tissues, although its expression in lymphoid tissue has not yet been examined (13). The 42-kDa PITALRE kinase autophosphorylates in kinase reactions (13, 24), which is similar to the phosphorylation of the 42-kDa TAK subunit. Like TAK, PITALRE also does not phosphorylate histones in vitro (7, 13). We therefore investigated whether PITALRE might be related to TAK.
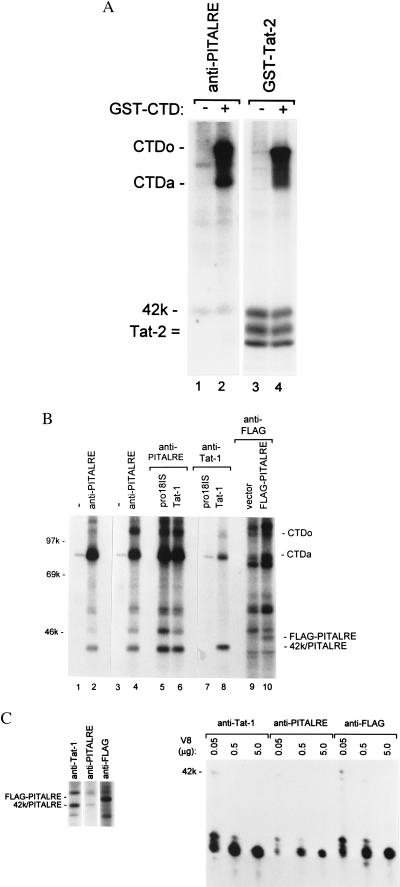
Using antibodies against PITALRE, we performed kinase assays of immunoprecipitations from a HeLa nuclear extract (Fig. 3A). In agreement with previous studies, the 42-kDa PITARLE kinase was autophosphorylated in our kinase reaction. The 42-kDa PITALRE autophosphorylation product comigrated on the SDS/polyacrylamide gel with the 42-kDa TAK subunit that was phosphorylated in a TAK kinase reactions (compare lanes 1 and 2 with 3 and 4). When recombinant CTD was added to the PITALRE kinase reaction, the hyperphosphorylated CTDo form was generated (lane 2), suggesting that PITALRE is a CTD kinase.
Figure 3.
PITALRE is the 42-kDa TAK subunit. (A) PITALRE antibodies or a GST-Tat-2 protein was used to bind PITALRE or TAK from a HeLa nuclear extract. Kinase reactions were performed with or without the addition of recombinant CTD, and the products were analyzed on a 9% SDS/polyacrylamide gel. The CTDo and CTDa, 42-kDa, and GST-Tat-2 reaction products are indicated. (B) Antibodies against PITALRE, Tat-1, FLAG epitope, or a nonimmune rabbit serum (−) were used for immunoprecipitations as indicated. Kinase reactions were performed with immunoprecipitates from: a HeLa nuclear extract (lanes 1 and 2), a HeLa whole cell extract (lanes 3 and 4), a whole cell extract of HeLa cells infected with a recombinant adenovirus that expresses the wild-type Tat-1 protein (lanes 6 and 8) or the pro18IS mutant protein (lanes 5 and 7), a whole cell extract of HeLa cells transfected with FLAG-CMV-2 vector (lane 9), and a whole cell extract of HeLa cells transfected with CMV-FLAG-PITALRE expression plasmid (lane 10). Reaction products were analyzed on a 9% SDS/polyacrylamide gel. The 42-kDa TAK/PITALRE products and the FLAG-PITALRE 45-kDa product are indicated. (C) Preparative immunoprecipitations using antibodies against Tat-1, PITALRE, and FLAG epitope were performed as described in Fig. 3B for lanes 8, 4, and 10, respectively, and kinase reactions were carried out. The preparative reaction products were separated on a 9% SDS/polyacrylamide gel (Left). The 42-kD/PITALRE products and FLAG-PITALRE product were excised from the gel and digested with the indicated amount of V8 protease. The digestion products were analyzed on a 15% SDS/polyacrylamide gel.
To further investigate the relationship between PITALRE and TAK, additional kinase reactions were performed with immunoprecipitates using antibodies against PITALRE, Tat-1, or a FLAG epitope-tagged PITALRE (Fig. 3B). For expression of Tat-1 proteins, HeLa cells were infected with a recombinant adenovirus that expresses either the wild-type Tat-1 protein or the transactivation-defective pro18IS Tat-1 mutant protein (25). The epitope-tagged PITALRE was expressed in HeLa cells by transfection of an expression plasmid.
Kinase reactions with PITALRE antibodies generated the CTDo and 42-kDa PITLARE products in immunoprecipitates from a HeLa nuclear extract (lane 2), a HeLa whole cell extract (lane 4), or whole cell extracts from HeLa cells expressing either the wild-type or pro18IS Tat-1 proteins (lanes 5 and 6). Kinase reactions with Tat antibodies generated the CTDo and 42-kDa TAK products in the immunoprecipitate from the wild-type but not mutant Tat-1 protein (lanes 7 and 8), because only the wild-type protein can associate with TAK in vivo (8). Kinase reactions with the FLAG antibodies generated a 32P-labeled 45-kDa product in cells transfected with the FLAG-tagged PITALRE expression plasmid but not cells transfected with the CMV-FLAG parent plasmid (lanes 9 and 10). This 45-kDa product differs in apparent molecular mass from the 42-kDa PITALRE and TAK kinase products by the size of the FLAG epitope and, therefore, represents autophosphorylation of the FLAG-tagged PITALRE kinase.
The kinase reaction with the FLAG-PITALRE immunoprecipitates did not generate a readily detectable CTDo product (lane 10). We were unable to observe generation of the CTDo in several other experiments with the overexpressed FLAG-tagged PITALRE protein. PITALRE has been observed to exist in multiple protein complexes in cell extracts (24). Although the monomer 42-kDa PITALRE protein is capable of autophosphorylation, complexes containing proteins in addition to the 42-kDa PITALRE protein have significantly greater activity for phosphorylation of a retinoblastoma protein substrate (24). It seems likely, therefore, that when overexpressed from the transfected plasmid, FLAG-PITALRE may be present in immunoprecipitates predominantly as a monomer or other complexes that are capable of autophosphorylation but largely inactive for CTDo phosphorylation.
To determine the relationship between the 32P-labeled 45-kDa FLAG-PITALRE product and the 32P-labeled 42-kDa products from PITALRE and Tat immunoprecipitations, preparative kinase reactions were performed and these 32P-labeled products were isolated from an SDS/polyacrylamide gel. V8 protease digestion were carried out to compare phosphopeptide maps of these 32P-labeled polypeptides (Fig. 3C). The phosphopeptide maps were identical between the PITALRE, TAK, and FLAG-PITALRE products, demonstrating that they are closely related polypeptides. Because the FLAG-PITALRE was expressed from a cloned gene and its phosphopeptide map is identical to that of the 42-kDa TAK subunit in a Tat-1 immunoprecipitation, we conclude that the PITALRE protein is the 42-kDa TAK subunit.
Antibodies Against PITALRE Can Deplete TAK Activity from Nuclear Extracts.
If PITALRE is a TAK subunit, antibodies against PITALRE should be able to deplete TAK from nuclear extracts and, likewise, a GST-Tat fusion protein should be able to deplete PITALRE activity. HeLa cell nuclear extracts were incubated with antibodies against CDK7 or PITALRE or with a GST fusion to wild-type Tat-2 or a transactivation-defective Tat-2 mutant (C59A). The PITALRE antibodies were raised against carboxyl-terminal residues 353–372, a region with no apparent homology to other CDKs or known human proteins. The levels of CDK7, PITALRE, or TAK activity remaining in extracts were then measured by kinase assays (Fig. 4). The CDK7 antibodies and the GST-Tat-2 mutant protein C59A were unable to deplete either PITALRE (lanes 2 and 12) or TAK (lanes 3 and 13). Autophosphorylation of the 42-kDa PITALRE protein was apparent in lanes 2 and 12 in a longer exposure of the gel. However, anti-PITALRE antibodies depleted both PITALRE and TAK activities (lanes 5 and 6). Likewise, the wild-type GST-Tat-2 fusion depleted both PITALRE and TAK activities (lanes 8 and 9). These results agree well with our conclusion that PITALRE is a TAK subunit.
We note that under the conditions of this experiment, the wild-type GST-Tat-2 protein appeared not to associate with CDK7, the kinase component of the basal transcription factor TFIIH complex. Additionally, although CDK7 antibodies were effective in depleting CDK7 activity from the nuclear extract (lane 1), the CDK7 antibodies did not measurably reduce the level of CTD kinase activity that associated with the wild-type GST-Tat-2 protein (lane 3). Although others have shown that TFIIH is capable of specific association with Tat proteins under some conditions (4, 5), the results shown in Fig. 4 agree with our previous study in which an immunoblot analysis indicated that TFIIH does not associate with GST-Tat fusions under these conditions (9).
Specific Binding of Wild-Type Tat Proteins to PITALRE.
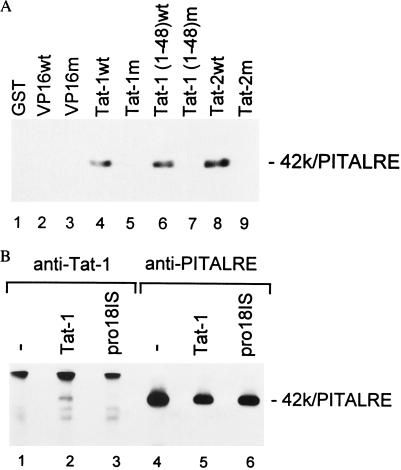
We next examined whether PITALRE associates with Tat proteins with the same specificity as previously demonstrated for TAK (6–8). In vitro binding of PITALRE in a HeLa nuclear extract to GST fusion proteins was analyzed by immunoblotting (Fig. 5A). PITALRE clearly associated with the wild-type Tat-1 and Tat-2 proteins, as well as the wild-type activation domain of Tat-1 (lanes 4, 6, and 8). However, PITALRE did not associate with transactivation-defective Tat-1 (pro18IS) and Tat-2 (C59A) proteins or the activation domain of Tat-1 containing the pro18IS mutation (lanes 5, 7, and 9). PITALRE did not associate with the GST protein, the wild-type VP16 activation domain, or a mutant VP16 activation domain (lanes 1–3). Thus, PITALRE and TAK bind in vitro to GST-Tat fusion proteins with a similar specificity that correlates with Tat transactivation function in vivo.
Figure 5.
Specific association of PITALRE with Tat proteins in vitro and in vivo. (A) GST-fusion protein-binding assays. Binding assays were performed as described in Materials and Methods with a HeLa nuclear extract and GST protein (lane 1) or GST fusions to the VP16 wild-type (wt) activation domain (lane 2), a mutant VP16 activation domain (F442S+F475A) (9) (lane 3), Tat-1 wt 86R protein (lane 4), mutant Tat-1 pro18IS 86R protein (lane 5), wt Tat-1 48R activation domain (lane 6), mutant Tat-1 48R pro18IS activation domain (lane 7), wild-type Tat-2 99R protein, and mutant Tat-2 99R C59A protein. The products were separated on a 9% SDS/polyacrylamide gel, and the presence of PITALRE was evaluated by an immunoblot using anti-PITALRE antibodies. Coomassie blue staining of a gel run in parallel with that shown demonstrated that all GST-fusion proteins were present at equivalent levels in pull-downs. (B) Immunoprecipitation immunoblots. HeLa cells were mock-infected or infected with a recombinant adenovirus expressing the wild-type Tat-1 72R protein or the Tat-1 transactivation-defective pro18IS 72R mutant. Extracts were prepared at 48 hr postinfection, and immunoprecipitations were performed with an antiserum against Tat-1 or PITALRE as indicated. Immunoprecipitates were separated on an 8% SDS/polyacrylamide gel, and the presence of PITALRE was evaluated by an immunoblot with antibodies against PITALRE.
We also investigated whether PITALRE associates with Tat in vivo. HeLa cells were infected with a recombinant adenovirus that expresses either a wild-type Tat-1 protein or the transactivation-defective pro18IS Tat-1 protein (Fig. 5B). Immunoprecipitations were performed with antibodies against Tat-1 (lanes 1–3) or antibodies against PITALRE (lanes 4–6). The presence of PITALRE in immunoprecipitates was evaluated by immunoblotting using a PITALRE antiserum. As expected, PITALRE was detected in all immunoprecipitations with the PITALRE antiserum (lanes 4–6). However, PITALRE was detected in Tat immunoprecipitations only in cells expressing the wild-type Tat-1 protein (lane 2) and not in cells expressing the mutant pro18IS (lane 3). We note that as measured in immunoblots, the pro8IS protein was expressed at slightly lower levels than wild type Tat-1 from the adenovirus vectors. However, in several repeats of the experiment shown in Fig. 5B we observed that wild-type Tat-1 but not pro18IS is associated with PITALRE. In a reciprocal experiment using a Tat-1 antiserum for the immunoblot, we observed that wild-type Tat-1 was present in immunoprecipitates using the PITALRE antiserum (data not shown). Using coimmunoprecipitations and GST pull-down assays, we have also performed experiments that indicate the FLAG epitope-tagged PITALRE protein expressed in COS or HeLa cells specifically associates with the Tat-1 and Tat-2 proteins (data not shown). We conclude from these experiments that similar to the in vitro binding with GST-Tat fusion proteins, PITALRE specifically associates in vivo with wild-type Tat proteins and not transactivation-defective Tat proteins.
DISCUSSION
Using a number of experimental approaches, we demonstrated that the CDK-related kinase known as PITALRE is a subunit of TAK, a Tat-associate kinase. When autophosphorylated in kinase reactions, the 42-kDa TAK subunit has an identical phosphopeptide map as that of natural PITALRE and a cloned epitope-tagged PITALRE. In nuclear extracts, antibodies against PITALRE can deplete TAK activity and, likewise, a GST-Tat fusion protein can deplete PITALRE kinase activity. PITALRE associates with Tat proteins in vitro and in vivo with the same specificity previously demonstrated for TAK that precisely correlates with Tat transactivation function. We also failed to observe cell cycle regulation of TAK activity in HeLa cells (unpublished result), which is similar to previous work with PITALRE in which no cell cycle regulation was observed (13).
Because partially purified TAK has a native molecular mass of approximately 110 kDa (8), it is likely that the 42-kDa PITALRE protein is the TAK catalytic subunit and active TAK contains one or more additional polypeptides. It is possible that some polypeptides normally associated with PITALRE in vivo were removed during our previous purification without inactivating a functional TAK in kinase assays, because PITALRE has been observed to associate with several unidentified polypeptides whose sizes are 48, 80, 95, 98, and 155 kDa (13, 24). It is probable that one of these polypeptides is the cyclin partner of PITALRE; one or more of these polypeptides might be other regulatory subunits such as an assembly factor or inhibitory factor. PITALRE may exist in vivo in more than one distinct protein complex, with different functional properties associated with each complex, and Tat may associate with only a specific PITALRE complex. The identification of PITALRE-associated proteins in the future will allow investigations of these possibilities. Additionally, the identification of PITALRE as a TAK subunit will allow detailed investigations of the role of TAK in Tat function and the mechanisms of TAK regulation.
We demonstrated in this study that TAK activity can be induced by activation of cells relevant to HIV-1 infection. TAK is induced in PBMCs and PBLs stimulated with either PMA or PHA, and TAK is induced when the promonocytic cell lines U1 and U937 are stimulated by PMA to differentiate to macrophages. These agents are known to activate transcription of the HIV-1 LTR is these cells (18), suggesting that TAK, along with other transcription factors such as NF-κB, may be an important regulator of HIV-1 transcription in cells infected by HIV-1. Because HIV-infected individuals demonstrate elevated levels of T cell activation, increased production of proinflammatory cytokines, and altered monocyte differentiation (26), it is tempting to speculate that HIV-1 and HIV-2 may have targeted TAK as a Tat cofactor because of the importance of this cellular kinase in T cells and monocytes/macrophages. Induction of TAK in activated T cells may contribute to elevated levels of viral replication, whereas induction of TAK during differentiation of monocytes to macrophages may contribute to an escape from transcriptional latency of integrate proviruses.
Acknowledgments
This work was supported by National Institutes of Health Grant AI35381 and by the Center for AIDS Research, Baylor College of Medicine (AI36211). We gratefully acknowledge David Price and colleagues for proposing that pTEFb is a CDK-related protein and implying that it might be TAK (abstract published at Cold Spring Harbor Retrovirus Meeting, Cold Spring Harbor, NY, May 20–25, 1997).
Note Added in Proof
De Luca et al. have recently demonstrated that PITALRE is expressed at high levels in primary human lymphoid tissue (27). Zhu et al. have recently demonstrated that human pTEFb is related to TAK (28).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RNAP II, RNA polymerase II; TAK, Tat-associated kinase; GST, glutathione S-transferase; CTD, carboxyl-terminal domain; PBMC, peripheral blood mononuclear cells; PBL, peripheral blood lymphocytes; CDK, cyclin-dependent family of protein kinases; PHA, phytohemagglutinin; PMA, phorbol 12-myristate 13-acetate; CTDa and CTDo, underphosphorylated and hyperphosphorylated CTD, respectively.
References
- 1.Jones K A, Peterlin B M. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 2.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Nature (London) 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Sharp P A. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- 4.Parada C A, Roeder R G. Nature (London) 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Martinez L F, Mavankal G, Neveu J M, Lanes W S, Ivanov D, Gaynor R B. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrmann C H, Rice A P. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann C H, Rice A P. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Herrmann C H, Rice A P. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann C H, Gold M O, Rice A P. Nucleic Acids Res. 1996;24:501–508. doi: 10.1093/nar/24.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall N F, Peng J, Xie Z, Price D H. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 11.Dahmus M E. J Biol Chem. 1996;217:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto H, Sheline C T, Corden J L, Jones K A, Peterlin B M. Proc Natl Acad Sci USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grana X, De Luca A, Sang N, Fu Y, Claudio P P, Rosenblatt J, Morgan D O, Giordano A. Proc Natl Acad Sci USA. 1994;91:3834–3838. doi: 10.1073/pnas.91.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis D E, Tang D S, Adu-Oppong A, Schober W, Rodgers J R. J Immunol. 1994;153:412–420. [PubMed] [Google Scholar]
- 15.Lewis D E, Adu-Oppong A, Hollinger F B, Rosenblatt H M, Hanson I C, Reuben J M, Kline M W, Kozinetz C A, Shearer W T. Clin Diagnostic Lab Immunol. 1995;2:87–90. doi: 10.1128/cdli.2.1.87-90.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGrory W J, Bautista D S, Graham F L. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 17.Peterson S R, Dvir A, Anderson C W, Dynan W S. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- 18.Poli G, Fauci A S. In: Role of Cytokines in the Pathogenesis of HIV Infection. Aggarwal B B, Puri P K, editors. Cambridge, MA: Blackwell Science; 1994. p. 421. [Google Scholar]
- 19.Folks T M, Justement J, Kinter A, Schnittman S, Orenstein J, Poli G, Fauci A S. J Immunol. 1988;140:1117–1122. [PubMed] [Google Scholar]
- 20.Asiedu C, Biggs J, Lilly M, Kraft A S. Cancer Res. 1995;55:3716–3720. [PubMed] [Google Scholar]
- 21.Cisek L J, Corden J L. Nature (London) 1989;339:679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- 22.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H, Egly J M. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 23.Gold M O, Tassan J-P, Nigg E A, Rice A P, Herrmann C H. Nucleic Acids Res. 1996;24:3771–3777. doi: 10.1093/nar/24.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garriga J, Mayol X, Grana X. Biochem J. 1996;319:293–298. doi: 10.1042/bj3190293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice A P, Carlotti F. J Virol. 1990;64:1864–1868. doi: 10.1128/jvi.64.4.1864-1868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantaleo G, Fauci A S. Annu Rev Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- 27.De Luca A, Esposito V, Baldi A, Claudio P A, Caputi M, Pisano M M, Baldi F, Giordano A. J Cell Physiol. 1997;172:265–273. doi: 10.1002/(SICI)1097-4652(199708)172:2<265::AID-JCP13>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M, Price D. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]