Abstract
Insulators are DNA elements that divide chromosomes into independent transcriptional domains. The Drosophila genome contains hundreds of binding sites for the Suppressor of Hairy-wing [Su(Hw)] insulator protein, corresponding to locations of the retroviral gypsy insulator and non-gypsy binding regions (BRs). The first non-gypsy BR identified, 1A-2, resides in cytological region 1A. Using a quantitative transgene system, we show that 1A-2 is a composite insulator containing enhancer blocking and facilitator elements. We discovered that 1A-2 separates the yellow (y) gene from a previously unannotated, non-coding RNA gene, named yar for y-achaete (ac) intergenic RNA. The role of 1A-2 was elucidated using homologous recombination to excise these sequences from the natural location, representing the first deletion of any Su(Hw) BR in the genome. Loss of 1A-2 reduced yar RNA accumulation, without affecting mRNA levels from the neighboring y and ac genes. These data indicate that within the 1A region, 1A-2 acts an activator of yar transcription. Taken together, these studies reveal that the properties of 1A-2 are context-dependent, as this element has both insulator and enhancer activities. These findings imply that the function of non-gypsy Su(Hw) BRs depends on the genomic environment, predicting that Su(Hw) BRs represent a diverse collection of genomic regulatory elements.
Author Summary
Insulators are conserved genomic elements that define domains of independent transcription. One class of insulators in the Drosophila genome are defined by the binding of the Su(Hw) protein, with the gypsy insulator representing the classic Su(Hw)-dependent insulator. Su(Hw) associates with hundreds of non-gypsy regions distributed throughout the genome that differ in sequence and organization from the gypsy insulator. To gain insights into the role of Su(Hw) in genome organization, we defined the properties of the first non-gypsy Su(Hw) binding region identified, 1A-2. Our studies reveal differences in 1A-2 activity, depending on the context tested. We show that 1A-2 is an insulator in enhancer blocking studies but functions as a transcriptional activator within the natural genomic location. Our findings are reminiscent of properties of binding regions that associate with the vertebrate CTCF protein, which have defined insulator, activator, and repressor functions. Finally, our studies indicate that a noncoding RNA gene may contribute to independent transcriptional regulation in the genome.
Introduction
In eukaryotic genomes, neighboring genes often display distinct spatial and temporal patterns of transcription, even though intergenic distances are within the range of enhancer and silencer action. These observations suggest that constraints exist that limit promiscuous interactions between long distance regulatory elements and non-target promoters. Chromatin insulators represent one class of genomic elements that restrict enhancer and silencer action [1]–[5].
Insulators have been identified based on two functional properties. First, insulators prevent enhancer and silencer modulation of a promoter in a position-dependent manner, such that an enhancer or silencer is blocked only when the insulator is located between these elements and a promoter. Second, insulators protect gene expression from positive and negative chromosomal position effects associated with ectopic placement of genes within genomes, an activity referred to as barrier function. Sequences with one or both of these properties have been identified in most eukaryotic genomes and have been implicated in the regulation of diverse cellular processes, ranging from centromere function in yeast to imprinting in mammals [6],[7]. These observations imply that insulators are fundamental components of eukaryotic genomes.
One of the best-characterized insulators resides in the 5′ untranslated region of the Drosophila gypsy retrovirus. This versatile gypsy insulator blocks over twenty enhancers active in different tissues and developmental stages, prevents repressive effects caused by Polycomb group complexes and heterochromatin and protects an origin of DNA replication from chromosomal position effects [2],[5]. The gypsy insulator consists of a cluster of twelve repeats that are bound by the zinc finger Suppressor of Hairy-wing [Su(Hw)] protein [8]. At least three additional proteins are associated with the gypsy insulator, including Modifier of (mdg4) 67.2 (Mod67.2), Centrosomal Protein of 190 kD (CP190) and Enhancer of y2 [E(y)2]. In general, Mod67.2 and CP190 are required for enhancer and silencer blocking by the gypsy insulator, while E(y)2 has been shown to be required only for barrier function [9]–[13].
The Su(Hw) protein associates with hundreds of non-gypsy regions in the Drosophila genome that have a largely unknown function. The extensive co-localization of the four gypsy insulator proteins at non-gypsy regions has led to the proposal that these represent chromatin insulators. Yet, non-gypsy Su(Hw) binding regions are different in sequence and organization from the gypsy insulator, with the majority of BRs containing single Su(Hw) binding sites (BSs) [14]–[18]. This observation is striking, as at least four tightly spaced Su(Hw) sites from the gypsy insulator were required for robust enhancer blocking [19]–[21]. Direct tests of the non-gypsy BRs in transgene assays show that most, but not all, interfere with enhancer-activated transcription [15]–[18]. These findings imply that non-gypsy regions contain additional elements that assist the insulator function of Su(Hw).
The first non-gypsy Su(Hw) BR identified, named 1A-2, is a cluster of two Su(Hw) BSs located in cytological region 1A (Figure 1). Here we investigated the properties of 1A-2, using two strategies. First, we employed a quantitative transgene system to define the 1A-2 sequences required for enhancer blocking. Second, we performed homologous recombination to establish lines carrying a deletion of 1A-2 at the natural genomic location, representing the first deletion of a non-gypsy Su(Hw) BR in the Drosophila genome. Effects of the loss of these sequences on gene expression in the 1A region were determined, leading to the discovery that 1A-2 contributes to transcriptional activation of a novel, non-coding RNA gene. Taken together, our studies demonstrate that 1A-2 has both activator and insulators properties, depending on the context tested. These findings imply that properties of non-gypsy Su(Hw) BRs are influenced by the genomic environment, predicting that Su(Hw) BRs represent a diverse collection of elements with distinct regulatory functions.
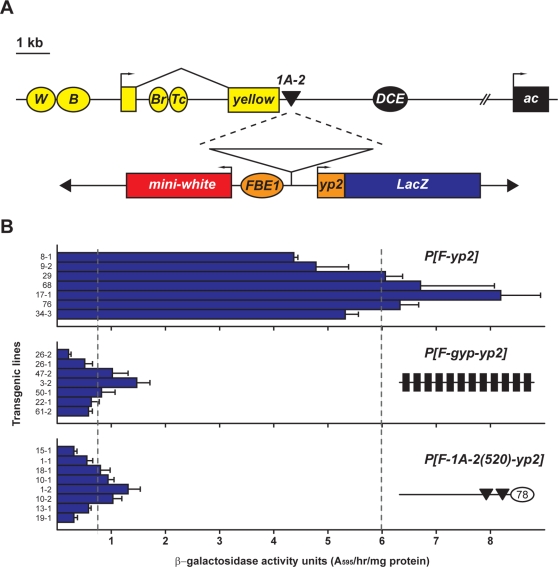
Figure 1. The 1A-2 insulator.
A. Top: The cuticle pigmentation yellow (y) gene contains two exons (yellow rectangles) and four tissue specific enhancers (ovals marked W for wing, B for body, Br for bristle and Tc for tarsal claw). The proneural achaete (ac) gene contains one exon (black rectangle). The tissue-specific enhancer in the upstream regulatory region is shown (oval marked DCE for dorsocentral enhancer). 1A-2 Su(Hw) BR is located downstream of the y gene, separating this gene from the ac regulatory region. Bottom: Structure of the FBE1-yp2-LacZ transgene used to define the sequences of 1A-2 required for enhancer blocking. Subregions of 1A-2 were inserted between the FBE1 enhancer and yp2 promoter. Effects of transcriptional activation were determined through enzymatic assay that tested β-galactosidase activity. The mini-white (w) gene was used for identification of transgenic flies in germ line transformation. B. β-galactosidase activity associated with transgenic lines carrying transposons derived from FBE1-yp2-LacZ. Each bar represents the average activity units (aau) for independent insertion lines corresponding to the indicated transposon (right). When pertinent, a cartoon is shown that represents the structure of sequences included in the FBE1-yp2-LacZ transgene. The gypsy Su(Hw) BS are shown as black rectangles, the 1A-2 Su(Hw) BS are shown as black triangles, a novel 1A-2 element is shown as an oval carrying 78 bp. Assays were completed on extracts isolated from females representing at least three independent crosses. Error bars indicate standard deviation. The vertical dashed line on the left represents the aau value for flies carrying P[F-gyp-yp2], while the vertical dashed line on the right represents the aau value for flies carrying P[F-yp2].
Results
1A-2 Is a Composite Insulator
The Su(Hw) BR 1A-2 is a 520 bp element that contains two Su(Hw) BSs [18] and a CP190 BS [9]. Previous studies using qualitative transgene assays demonstrated that 1A-2 blocked enhancer-activated transcription in a position-dependent manner, a key feature of insulator activity [17],[18]. We employed the quantitative Fat Body Enhancer (FBE)1-yolk protein (yp)2 -LacZ transgene to define DNA sequences required for 1A-2 enhancer blocking (Figure 1), a system previously used to characterize properties of the gypsy insulator [20],[22]. A reporter transgene was constructed wherein full length 1A-2(520) was inserted between FBE1 and the yp2 promoter. Multiple P[F-1A-2(520)-yp2] transgenic lines with single insertions were established. Quantitative β-galactosidase activity assays were completed to define the level of yp2 promoter activity. Protein extracts were isolated from adult females representing several independent lines, and multiple assays were undertaken to establish an average activity unit (aau) for each transgene (Figure 1). We found that transgenic P[F-1A-2(520)-yp2] females had low levels of yp2 expression (aau 0.86), similar to levels in P[F-gyp-yp2] females (aau 0.75) and significantly lower than levels found in the control P[FBE1-yp2] females (aau 5.97). We conclude that 1A-2 blocks FBE1, extending the enhancer blocking effects of 1A-2 to a new enhancer-promoter pair.
The minimal sequences required for 1A-2 insulator function were determined by generation of transgenic lines carrying transposons with insertion of subregions of 1A-2 between FBE1 and yp2-LacZ (Figure 2). P[F-1A-2(157)-yp2] females showed a strong enhancer block (aau 0.62). As this subregion lacks the CP190 BS [9], these findings indicate that direct CP190 binding is not required for insulator function. 1A-2(157) was further divided into two parts, one containing the two Su(Hw) BSs, 1A-2(79), and one containing the remaining sequences, 1A-2(78). Transgenic P[F-1A-2(79)-yp2] females showed a two-fold weaker enhancer block than 1A-2(157) (aau 1.29, P = 0.02), whereas P[F-1A-2(78)-yp2] females showed high yp2 activity levels, close to those obtained for the control P[F-yp2] females (aau 5.9 versus 5.97). These data suggest that 1A-2(78) contributes to the blocking effectiveness of the 1A-2 Su(Hw) BSs, but cannot itself block enhancer-promoter interactions.
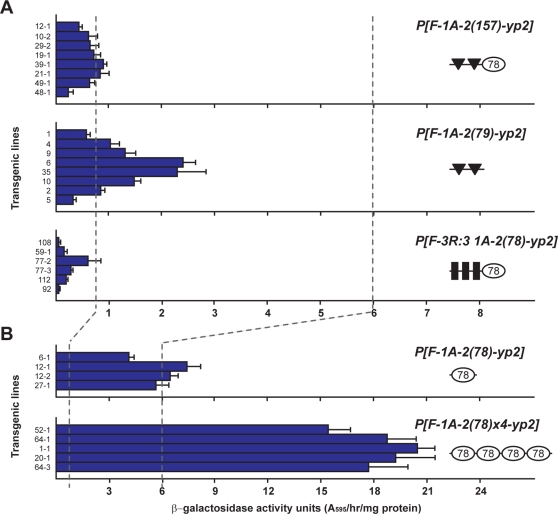
Figure 2. 1A-2 contains enhancer blocking and facilitator elements.
(A,B) β-galactosidase activity of independent transgenic lines carrying a derivative of the FBE1-yp2-LacZ transgene. The name and structure of the insertion is shown on the right, including the 3′ 157 bp 1A-2 fragment (P[F-1A-2(157)-yp2]), the 79 bp fragment with only two Su(Hw) BSs (P[F-1A-2(79)-yp2]), the 3′ 78 bp fragment combined with a Su(Hw) BR containing three copies of site 3 from the gypsy insulator (P[F-3R:3 1A-2(78)-yp2]), the 1A-2 78 bp fragment (P[F-1A-2(78)-yp2]) and four copies of the 3′ 1A-2 78 bp fragment (P[F-1A-2(78)×4-yp2]). Error bars indicate the standard deviation (n = 3). The vertical dashed line on the left represents the aau value for flies carrying P[F-gyp-yp2], while the vertical dashed line on the right represents the aau value for flies carrying P[F-yp2]. In B, the X-axis scale is increased three fold. Symbols are as described in Figure 1.
We considered two possibilities to account for the contributions made by 1A-2(78). First, these sequences might contain a binding site(s) for a second insulator protein that cooperates with the Su(Hw) BSs for insulator function. Second, 1A-2(78) might improve the activity of the Su(Hw) BSs, perhaps by increasing in vivo association. We reasoned that if 1A-2(78) contained a binding site for a novel insulator protein, then insulator effects might require a reiterated element, as observed previously when individual binding sites for other insulator proteins were tested [23],[24]. To this end, we generated P[F-1A-2 (78×4)-yp2] that carried four copies of 1A-2(78) inserted between FBE1 and the yp2 promoter. Surprisingly, these transgenic females had higher yp2 activity than the control P[F-yp2] females (aau 18.78 versus 5.97 aau, P = 6.3×10−8). Transgenic P[F-1A-2(78×4)-yp2] males showed no yp2 activity (data not shown). Based on the retained transcriptional specificity of the P[F-1A-2 (78×4)-yp2] transgene, we conclude that 1A-2(78) is not a general transcriptional enhancer but improves the activity of FBE1. These data imply that 1A-2(78) may possess a general activity that facilitates factor association. To test this postulate, we determined whether 1A-2(78) restored enhancer blocking to a synthetic Su(Hw) BR containing three reiterated gypsy BSs (3R:3) that was previously shown to be inactive in this transgene system [20]. Supporting a facilitator function of 1A-2(78) we found that transgenic P[F- 3R:3-1A-2(78)-yp2] females had low yp2 activity (aau 0.22). These studies show that in the presence of 1A-2(78), 3R:3 became a strong insulator. As previous findings suggest that the effectiveness of enhancer blocking by the Su(Hw) protein is limited by the in vivo accessibility of Su(Hw), we conclude 1A-2(78) is a facilitator that may improve transcription factor binding to chromosomes.
The y-ac Intergenic Region Contains a Novel, Non-Coding RNA Gene
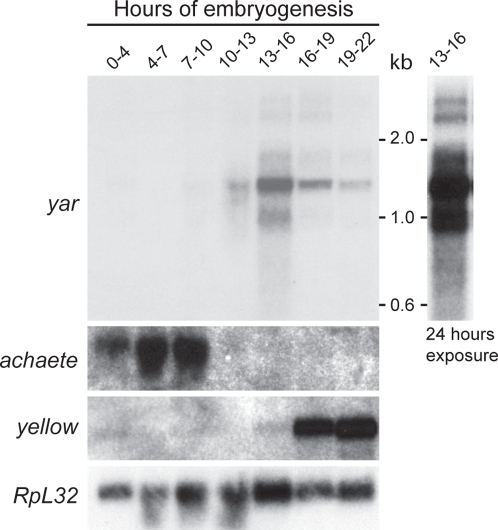
As a first step in defining the role of 1A-2 within the y-ac region, we evaluated whether the existing annotation reflected the transcriptional potential of this region. These analyses were motivated by the recent studies showing widespread transcription in intergenic regions of the Drosophila genome [25]. A search of the NCBI databases uncovered a small, novel, processed EST of ∼400 nt that was transcribed from the y-ac intergenic sequences. Sequences corresponding to this EST are located ∼1.4 kb downstream of the y termination signal and transcribed in the same direction as the y and ac genes. Northern analyses of embryonic polyA+ RNA using a radiolabeled probe representing the intergenic EST identified a family of RNAs, with the most abundant species sized at ∼1.6 kb (Figure 3). Accumulation of these RNAs began ∼7 hours after the start of embryogenesis, in agreement with the expression profile obtained using tiling array studies of embryonic RNAs [25]. These data suggest that the y-ac intergenic region contains a previously uncharacterized gene, which we call yar, for y-ac intergenic RNA. Activation of genes in the 1A locus is temporally in an order following chromosomal position, such that ac, then yar and then y is transcribed.
Figure 3. Transcription of yar is regulated during embryogenesis.
Northern analysis of five µg of polyA+ RNA isolated from staged embryo collections. The blot was probed with a yar cDNA, and two exposures are shown: 3 hours (left) and 24 hours (right). The blot was stripped and reprobed with DNA sequences corresponding to y, ac and the constitutively active RpL32 gene that served as a loading control.
The structure of the yar RNAs was defined using rapid amplification of cDNA ends (RACE, Figure 4). Sequence analysis of the 5′ RACE products revealed three discrete transcription start sites within an ∼200 bp region, with the most distal RNA starting ∼1.2 kb downstream of the y gene. Each putative start site showed weak homology to Drosophila transcriptional control elements [26], with two having a partial match to the TATA consensus sequence located 17 to 35 bp upstream of the start site. Sequence analysis of the 3′ RACE products identified multiple splice variants, each ending in a common exon that contained an unconventional polyadenylation signal sequence AAATACA, previously estimated to be present in ∼3% of Drosophila genes [27], that was located 12 bp upstream of the string of As in the RACE products. Predicted translation of the yar RNAs indicated that no transcript would encode a protein of more than 75 amino acids, implying that yar is a non-coding RNA gene.
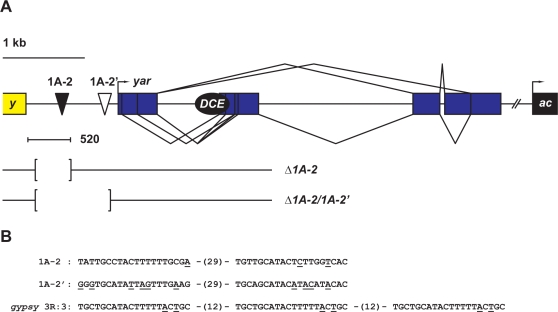
Figure 4. Detailed structure of control elements in the region separating the y and ac genes.
A. The region separating the y and ac genes contains a cluster of two strong Su(Hw) BS (black triangle, 1A-2) and two weak Su(Hw) BS (white triangle, 1A-2′). 1A-2′ is located 85 bp upstream of the most upstream yar transcription start site (bent arrow). The alternative splicing pattern is shown: thin lines represent introns and blue rectangles indicate exons. The ac DCE (black oval) resides within yar. The limits of the original 520 bp 1A-2 insulator are shown as a bracketed line marked 520. The extent of the regions deleted in the yΔ1A-2 (Δ1A-2) and yΔ1A-2/Δ1A-2′ (Δ1A-2/1A-2′) flies are shown, where the bracketed regions were removed. B. The sequence of the Su(Hw) BSs in 1A-2 (top), 1A-2′ (middle), and the synthetic gypsy insulator (3R:3) (bottom), with the numbers indicating the distance of separation between BSs. The nucleotides different from the genomic Su(Hw) consensus BS [14] are underlined.
Loss of 1A-2 Does Not Alter Adult Phenotypes Generated by y and ac Expression
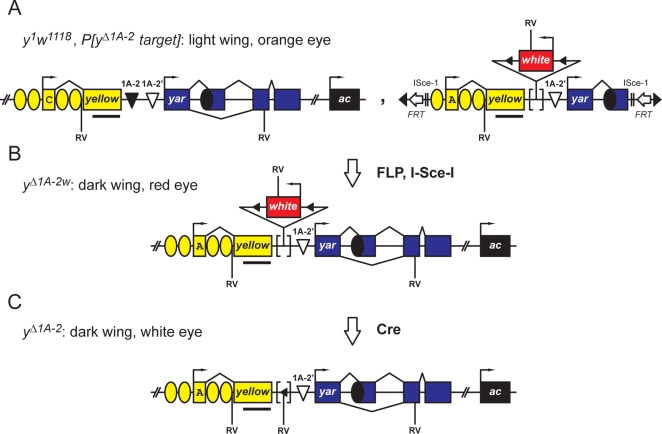
Ends out gene targeting was used to delete 1A-2 from the y-ac region (Figures 4, 5). Gene targeting is a two step processes that requires establishment of transgenic flies that carry a transposon with the replacement gene, followed by the introduction of endonucleases to stimulate homologous recombination between the replacement gene and its endogenous homologue. To delete 1A-2, we constructed P[yΔ1A-2 target]. This transposon carried a modified y gene, wherein 1A-2 was replaced by the hypomorphic whs gene that was flanked by loxP sites (Figure 5). Transgenic lines were established in a y1 w1118 background, where the endogenous y gene carried a mutation of the translation start codon, and the endogenous w gene carried a deletion of the promoter. P[yΔ1A-2 target] flies had orange eyes and dark pigmentation of all cuticle structures except the wing, as the y gene lacked the wing enhancer. To stimulate recombination, transgenic y1 w1118; P[yΔ1A-2 target] males were crossed to females carrying the heat shock (hs)-FLP recombinase and the hs-I-SceI endonuclease transgenes and progeny of this cross were heat shocked to produce the endonucleases. Over 100 resulting females were crossed to y1 w1118 males and homologous recombinants were identified among the offspring of this cross in two ways. First, flies were screened for dark wings, as recombination at the endogenous y1 gene would reconstitute a wild type y transcription unit with all enhancers, whereas progeny with ectopic insertions of the replacement y gene would produce flies with lightly colored wings due to the absent wing enhancer. Second, we conducted genetic analyses to determine whether the w+ phenotype was linked to the X chromosome. Five putative homologous recombination lines were established based on dark wing pigmentation. Further genetic analyses showed that in one line, XGL339-23-38, the w marker mapped to the X chromosome, suggesting a correct targeting event. Southern analyses confirmed the structure of the y gene in these flies (Figure S1). This targeted allele was named, yΔ1A-2w.
Figure 5. Ends-out targeting strategy to generate deletions of 1A-2 at the endogenous 1A locus.
A. Transgenic flies were generated that carried the mutant y1 allele (structure shown where C indicates the mutation of the translation ATG start) at the endogenous X chromosome location (left) and the P[yΔ1A-2 target] transposon on a different chromosome (right) that carries a y gene lacking the wing enhancer, but encodes a wild type RNA (A indicates the presence of the correct translation ATG start). In this transposon, the y gene, flanked by FRT sites (white arrows) and I-SceI sites, is within a P transposon (inverted black triangles). Other symbols representing the y, yar and ac genes are as described in Figure 1. Transgenic flies y1 w1118, P[yΔ1A-2 target] had a light wing color and orange eyes. B. FLP and I-SceI enzymes catalyzed replacement of the y1 allele at the endogenous locus, with yΔ1A-2w, in which the 1A-2 insulator is substituted by the whs gene (raised triangle) inserted between loxP sites (black arrowheads on raised triangle). The recombinant yΔ1A-2w flies had dark wings and red eyes. C. Cre recombinase deleted the whs gene, leaving behind a single loxP site to form yΔ1A-2. In the case of the yΔ1A-2, the remaining loxP site was mutated, forming a new EcoRV site (RV), whereas in the similarly derived yΔ1A-2/Δ1-A2′ flies a wild type loxP site remained. The bar under the y gene indicates the probe used in the Southern analyses (see Figure S1).
We reasoned that if 1A-2 was an insulator in the y-ac locus, then deletion of 1A-2 would release constraints on the y and ac enhancers, causing changes in gene expression that would alter cuticle pigmentation and bristle number in yΔ1A-2w relative to wild type flies [28],[29]. However, we found that adult phenotypes of yΔ1A-2w flies were indistinguishable from wild type flies. In yΔ1A-2w, the whs gene replaced 1A-2. To rule out the possibility that this gene served as a surrogate insulator by carrying a promoter that captured the y and ac enhancers, yΔ1A-2w flies were crossed to flies carrying a source of Cre recombinase to remove the whs gene. Southern and PCR analyses confirmed the structure of y gene in yΔ1A-2 flies (Figure S1). Again, the cuticle and bristle phenotypes of the yΔ1A-2 flies were indistinguishable from wild type. Taken together, these data imply that 1A-2 is not an insulator at the endogenous genomic location.
Within the y-ac intergenic region, we identified a second cluster of Su(Hw) binding sites, which we called 1A-2′. These sites differ from the Su(Hw) consensus sequence at multiple highly conserved positions (Figure 4). Electrophoretic mobility shift assays demonstrated that 1A-2′ had ∼3-fold lower affinity for Su(Hw) than 1A-2 (data not shown). Even so, we considered it possible that weaker 1A-2′ Su(Hw) BR might provide a redundant function with 1A-2 to define regulatory interactions in the y-ac region. For this reason, we generated a second targeting vector, P[yΔ1A-2/Δ1A-2′ target], wherein the whs gene replaced an ∼1.0 kb deletion that encompassed both 1A-2 and 1A-2′. Following the procedure described above, six putative homologous recombinant lines were identified based on dark wing pigmentation. Further genetic analyses showed that one of these lines, XGL426-41-4, had marker linkage to the X chromosome. This allele was named yΔ1A-2/Δ1A-2′w. Flies from this line were used to obtain a derivative line lacking the whs gene, producing yΔ1A-2/Δ1A-2′. Southern and PCR analyses confirmed the structure of the y gene resulting from these targeting events (Figure S1). Comparison of adult phenotypes in yΔ1A-2/Δ1A-2′ and wild type flies showed that the cuticle color and bristle number were indistinguishable, suggesting that 1A-2′ did not compensate for 1A-2.
yar Expression Is Lowered in the Absence of 1A-2
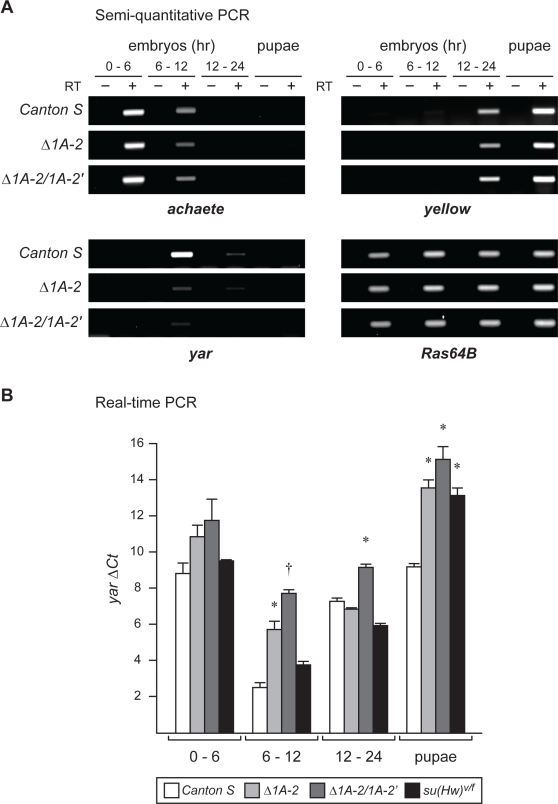
We postulated that changes in y and/or ac gene expression might occur, but that these differences may not be readily observed in analyses of adult phenotypes. For this reason, levels of RNA accumulation were quantified using reverse transcriptase PCR (Figure 6). Total RNA was isolated from staged collections of Canton S (wild type), yΔ1A-2 and yΔ1A-2/Δ1A-2′ embryos and pupae, representing the developmental periods where the y and ac genes are maximally expressed [30]. Following conversion to cDNA, templates were amplified using primers against y, ac, yar, and Ras64B, a constitutively expressed RNA [16],[31]. In a first set of experiments, PCR products obtained from cDNA amplification in the linear range were run on an agarose gel and visualized by ethidium bromide staining (Figure 6A semi-quantitative, Figure S2). These studies revealed that the loss of 1A-2 and 1A-2′ did not change the timing or level of y and ac RNA accumulation, consistent with the lack of phenotypic changes. In contrast, amplification of yΔ1A-2 and yΔ1A-2/Δ1A-2′ cDNA showed reduced yar levels relative to Canton S, without a temporal change. These findings indicate that 1A-2 and 1A-2′ are required for yar expression.
Figure 6. Loss of 1A-2 and 1A2′ reduces yar RNA accumulation during embryogeneis.
A. Ethidium bromide stained PCR products obtained from semi-quantitative PCR to evaluate ac, y, yar and Ras 64B RNA levels in wild type (Canton S), yΔ1A-2 (Δ1A-2) and yΔ1A-2/Δ1A-2′ (Δ1A-2/Δ1A-2′). Ras64B is constitutively expressed and serves as a control. The minus (−) RT lanes control for genomic DNA contamination. Different stages of embryonic and mixed pupal RNA were analyzed. B. Quantitative real time PCR (Q-PCR) was used to determine levels of yar mRNA accumulation from RNAs isolated during development from wild type and mutant lines. Individual transcript levels defined by Q-PCR were normalized to Ras64B for amount of input cDNA (ΔCT). A larger ΔCT indicates a reduction in RNA. Error bars indicate standard deviation of values obtained from analyses of three independently isolated RNAs. Significant changes in RNA accumulation relative to wild type are as indicated (*, P = <0.01; †, P<0.001, Student's two-tailed t-test).
Quantitative real time PCR analyses (Q-PCR) were undertaken to test the semi-quantitative results (Figure 6B, Figure S3). In these studies, we included analysis of scute (sc) RNA accumulation, the gene downstream of ac. A cycle threshold (CT) for each primer set was determined and a corresponding ΔCT was calculated, using the Ras64B CT for standardization. These analyses identified a significant increase in ΔCT for yar within yΔ1A-2 and yΔ1A-2/Δ1A-2′ samples, relative to Canton S. These data correspond to a 7- and 25-fold decrease in yΔ1A-2 embryonic and pupal yar RNA respectively and a 32- and 41-fold decrease in yΔ1A-2/Δ1A-2′ embryonic and pupal yar RNA (Figure 6B, Figure S3). These data suggest that within the context of the y-ac genomic region, 1A-2 and 1A-2′ serve as an enhancer of the non-coding yar gene.
To determine whether the Su(Hw) contributes to expression of genes in the 1A region, we quantified of y, yar, ac and sc RNAs in a su(Hw)v/su(Hw)f mutant background, using Q-PCR. The su(Hw)v allele carries a promoter deletion and the su(Hw)f allele carries a point mutation that produces a full-length protein with an inactivate finger 10 [32]. We found that only the level of pupal yar RNA was significantly changed in the su(Hw)v/f mutant background, associated with an ∼21-fold decrease (Figure 6B, Figure S3). These data indicate that Su(Hw) makes positive contribution to the normal low level of pupal yar transcription. The absence of expression changes in embryonic RNA may be confounded by an ability of Su(Hw)f to bind to 1A-2 in early embryos. Previous studies have shown that disruption of Su(Hw) zinc finger 10 limits chromosome accessibility, without altering DNA recognition [32]. It is possible that in early embryos, 1A-2 is in a more accessible chromatin structure, thereby allowing Su(Hw)f to bind 1A-2 and activate yar, but that this property is lost during development. We are unable to test yar expression in a su(Hw) null background, as complete loss of Su(Hw) blocks oogenesis. Regardless, our data imply that Su(Hw), along with contributions made by other proteins associated with 1A-2, function as an activator of yar transcription during development.
Discussion
Prevailing models of gypsy insulator function predict that the gypsy insulator establishes independent transcriptional domains through cooperation with genomic insulators defined by non-gypsy Su(Hw) BRs. Recent findings indicate that the sequence and organization of non-gypsy BSs differ from the Su(Hw) BR in the gypsy retrovirus [14]–[16]. These observations imply that properties of non-gypsy BRs may be distinct from those of the gypsy insulator. We defined the properties of 1A-2, to gain insights into mechanisms of Su(Hw) insulator action.
Enhancer Blocking by 1A-2 Requires Su(Hw) BSs and a Facilitator
We used the quantitative FBE1-yp2-LacZ reporter system to define the sequence requirements for enhancer blocking by 1A-2(520). Prior application of this system demonstrated that at least four gypsy Su(Hw) sites were needed for robust blocking [20]. Here, we show that 1A-2(157) provided as strong an enhancer block as the gypsy insulator (Figures 1, 2). A fragment containing only the Su(Hw) BRs [1A-2(79)] reconstituted a weaker enhancer block than 1A-2(157), but had a greater blocking capacity than the synthetic insulators made from reiterated copies of BS3 of the gypsy insulator [20]. While we do not know the reason for the more robust blocking, we note that these regions differ in sequence and distance of separation from Su(Hw) sites (Figure 4). Blocking effectiveness does not appear to be due to differences in DNA recognition, as the in vitro binding constants for Su(Hw) for the 1A-2 and gypsy BSs are similar [16]. Our experiments revealed that 1A-2 contains a second regulatory element located in 1A-2(78). When these sequences were positioned next to the inactive, synthetic Su(Hw) BR (3R:3), a functional insulator was reconstituted (Figure 2B). These data are consistent with previous findings that Su(Hw) chromosome association is limited [32]. Taken together, we propose that 1A-2 is a composite insulator that contains an enhancer blocking and a facilitator function that may improve Su(Hw) chromosome association. Further, we predict that in vivo effectiveness of enhancer blocking by the Su(Hw) protein is related to the accessibility of Su(Hw) BSs. If single or small clusters of Su(Hw) BSs are located in genomic regions of open chromatin, then these regions will demonstrate enhancer blocking, as defined in transgene assays. This proposal implies that genomic context greatly influences the properties of non-gypsy Su(Hw) BRs.
A Novel Non-Coding RNA Gene Separates the y and ac Genes in the 1A Locus
1A-2 is located between the independently regulated y and ac genes. Chromatin immunoprecipitation studies demonstrated that 1A-2 is associated with Su(Hw), Mod67.2 and E(y)2 in vivo [12],[16],[18], suggesting that this element binds a complex competent for establishing a genomic insulator. Based on these properties, we postulated that 1A-2 was responsible for the regulatory independence of the y and ac genes in the 1A locus [16]. As a first step in testing this proposal, we investigated transcription in the y-ac region to evaluate the current accuracy of the genomic annotation of this region. These studies identified a previously unannotated gene, yar, located ∼1.2 kb downstream of the y gene and ∼3.0 kb upstream of ac. Multiple, differentially spliced, polyA+ RNAs are encoded by yar, with the largest translation product predicted to be 75 amino acids, indicating that this is a non-coding RNA gene. Emerging data suggest that non-coding RNAs are abundant in eukaryotes and have a wide repertoire of biological functions, ranging from structural components in protein complexes to regulatory molecules involved in transcription and translation [33]–[35]. It is unknown whether yar has a function. As flies carrying a large genomic deletion that removes sequences upstream of y and extends downstream of ac (y− ac−) are viable and fertile, yar is a non-essential gene.
1A-2 Is Required for Expression of a Non-Coding RNA Gene
Having re-defined the transcriptional profile in the 1A locus, we tested the function of 1A-2 and a second, weaker Su(Hw) BR, 1A-2′, on gene regulation, using gene targeting to delete these elements. Our studies represent the first deletional analysis of any non-gypsy Su(Hw) BR in the Drosophila genome. Two targeted deletion lines, yΔ1A-2 and yΔ1A-2/Δ1A-2′ were established (Figure 4). Levels of y, ac, sc and yar RNA accumulation during development were studied using quantitative PCR. We find that loss of 1A-2 and 1A2′ has no effect on the timing and level of y, ac or sc RNAs relative to the wild type control (Figure S3), but strongly reduced yar RNA (Figure 6). These data suggest that the effects of loss of 1A-2 are limited to local changes of gene expression, implying that these sequences are not a chromatin insulator at the endogenous location. Instead, our data indicate that 1A-2 may be an activator of yar expression, consistent with other studies that have suggested a role for Su(Hw) in gene activation [36]–[38]. These data, coupled with genetic studies on the effects of the loss of Su(Hw) on expression of genes adjacent to Su(Hw) BRs [16], demonstrate that Su(Hw) BRs have diverse functions in the genome.
The complexity of the transcriptional effects associated with Su(Hw) BRs is reminiscent of regions in mammalian genomes that bind the versatile regulatory protein CTCF. High throughput genomic analyses have identified hundreds of CTCF binding sites within the mouse and human genomes [7], [39]–[41]. Although many of these sequences possess enhancer blocking activity [39],[42],[43], CTCF has been implicated in transcriptional activation [44]–[46], repression [47]–[50], and chromosome pairing [44],[51],[52]. These observations suggest that, similar to the non-gypsy Su(Hw) BRs, genomic context will have an important influence on the properties of CTCF BSs within a given region.
The mechanism(s) used to maintain transcriptional autonomy in the 1A locus are unclear. The discovery of yar provides an alternative explanation to the need for a chromatin insulator. Based on the developmental timing displayed by the 1A genes, we postulate that activation of yar transcription may cause inactivation of ac through transcriptional interference. Similarly, activation of y may repress yar transcription. Although yΔ1A-2 and yΔ1A-2/Δ1A-2′ flies show reduced yar expression, transcription is not abolished, suggesting that the remaining yar activity may be sufficient to turn off ac. Alternatively, other mechanisms can be considered that might influence enhancer preference, including selectivity of enhancers for certain classes of promoters [53],[54], the presence of promoter targeting sequences that direct enhancer action [55],[56], or promoter tethering elements that capture enhancers [57]. Further experiments to define the properties of DNA elements within the 1A locus will resolve how transcriptional independence is achieved.
Materials and Methods
Fly Stocks and Crosses
Flies were raised at 25°C, 70% humidity on standard corn meal/agar medium. Description of the alleles used can be found at http://flybase.bio.indiana.edu.
Construction of FBE1-yp2 -LacZ Reporter Genes
The FBE1-yp2 -LacZ fusion gene [20] carried a BglII site, positioned at −335 relative to the transcription start site (TSS) that was used for insertion of tested 1A-2 fragments. Resulting transgenes were inserted into a P element transformation vector, generating P[F-1A-2(520)-yp2] with the full length 1A-2, P[F-1A-2(157)-yp2] with a 157 bp region of 1A-2, P[F-1A-2(79)-yp2] with two 1A-2 Su(Hw) binding sites, P[F-1A-2(78)-yp2] with the 78 bp 3′ region, P[F-1A-2(78×4)-yp2] with four tandem repeats of the 1A-2 78 bp element and P[F-3R:3(78)-yp2] with a hybrid insertion between a cluster of three tandem repeats of the gypsy Su(Hw) binding sites [nucleotides 732–759 [58]], as described in [20] and the 78 bp element. P transposons were injected into the host y1w67c23 strain or w1118 (Genetic Services, Inc, Cambridge, MA). Transgenic lines were analyzed by Southern and PCR analyses to determine the number and integrity of the transposons. Lines with single transposon insertions were used in subsequent analyses.
β-Galactosidase Spectrophotometric Assay
The yp2 promoter activity was assessed using quantitative β-galactosidase assays, performed essentially as previously described [20]. Each transgenic line was assayed using extracts isolated from three different matings. Each extract was assayed in duplicate, and the error between these samples was less than 10%. Average promoter activity and standard deviation were determined using the statistical analysis feature of the Microsoft Excel program.
Ends out Gene Targeting
Two targeting transposons were constructed for gene targeting, using pW25 [59]–[61]. This vector has multi-cloning site, NotI-SphI-Acc65I-Stop-lox-whs-lox-Stop-AscI-BsiWI. The lox sites are in direct orientation, permitting removal of the whs transformation marker by Cre recombinase. P[yΔ1A-2 target] (XGL339) was used to target an ∼0.43 kb deletion encompassing 1A-2 alone, whereas P[yΔ1A-2/Δ1A-2′ target] (XGL426) was used to target an ∼1.03 kb deletion that included 1A-2 and 1A-2′. These targeting transposons were generated in a two-step procedure. First, a 6.6 kb yellow fragment (−1842 to +4796 relative to the yTSS) was PCR amplified, using primers carrying the BsiWI and AscI sites and cloned into pW25 to make XGL235. This fragment contains the yellow transcription unit and the body enhancer, but lacks the wing enhancer. Second, PCR primers containing NotI sites generated a 3 kb fragment (y+5234 to y+8184 relative to the yTSS) to make P[yΔ1A-2 target] or a 3.5 kb fragment (y+5826 to y+9318 relative to the yTSS) to make P[yΔ1A-2/Δ1A-2′ target]. In all cases, PCR fragments were sequenced to confirm appropriate amplification. For targeting, we generated transgenic lines in a y1 w1118 mutant background. Gene targeting followed the procedure outlined in [59]. A combination of Southern and PCR analyses identified correctly targeted events. To remove the whs gene, red-eyed males carrying a targeted deletion event were crossed to females carrying Cre recombinase, as described in [62]. The white-eyed flies were collected and used to establish homozygous stocks. Deletion events were confirmed by PCR amplification and sequence analyses.
Rapid Amplification of cDNA Ends (RACE)
The structures of the yar RNAs were determined using RACE of total RNA isolated from 6–12 hour CS embryos. In the 3′-RACE experiments, 5 µg of RNA were reverse transcribed using the adaptor oligo-dT primer (3′-RACE kit, Invitrogen), and cDNA was amplified using a yar specific primer (1 µM) and the abridged universal primer (80 nM, Invitrogen). Several products were identified by agarose gel electrophoresis, gel purified and cloned into the TOPO vector (Invitrogen). Sequencing and BLAST search identified three yar splice variants that shared a common distal exon and poly-A signal. In the 5′-RACE experiments, 5 µg of RNA were reverse transcribed with a yar specific primer (100 nM), purified over a S.N.A.P column (Invitrogen) to remove unincorporated nucleotides and primers, and C-tailed at 4° for 2 hours, using terminal deoxynucleotidyl transferase. Tailed cDNAs were amplified with nested yar specific primers (400 nM) and an abridged anchor primer (400 nM, Invitrogen). PCR products were directly cloned into the TOPO vector. Forty-eight clones were analyzed by restriction digestion, revealing nine classes of insert. At least one representative of each class was sequenced. BLAST analyses of these data identified ten alternative splice variants and three alternative start sites. Both the 3′-RACE and 5′-RACE were performed on two independent RNA isolations. Gene-specific primer sequences are available upon request.
Northern and Real-Time PCR Analyses
RNA was isolated from staged embryos collected from cages of wild type (CS) flies, using the NaDodSO4/phenol technique [63]. Five µg of oligo-dT selected polyA+ RNA was used in northern analyses and hybridized with radiolabeled fragments corresponding to y (a ClaI-BglII fragment, representing +2466 to +4815 relative to the yTSS), yar (EST DN154052, 418 bp ) and ac (a PCR fragment representing +115 to +531 relative to the acTSS). Hybridization with sequences corresponding to the ribosomal gene, RpL32, served as a loading control. For real-time PCR experiments, RNA was isolated from embryos and pupae from three lines: CS, yΔ1A-2 line XGL339-23-38, yΔ1A-2/Δ1A-2′ line XGL426-41-4. RNA isolation and real-time PCR analyses were performed as described in [16]. PCR primers amplified 100–200 bp fragments. y primers flanked the intron. yar primers were in the invariant fourth exon, to ensure quantification of all transcripts. Primer sequences are available upon request. Duplicate or triplicate reactions were performed and averaged, with the difference among the replicates no greater than 0.5 cycle threshold (CT). At least three independent experiments were performed for each primer set from two independent RNA samples. The expression level of each gene was determined using Ras64B as an internal control (ΔCT). The fold change in expression of each gene relative to the wild type (CS) value was determined with the ΔΔCT method.
Supporting Information
Southern analysis of y-ac locus in homologous recombinant lines. Genomic DNA was isolated from ten flies, digested with EcoRV (NEB) and run on a 1% agarose gel. Flies analyzed were the parental y1w1118 line, the P[yΔ1A-2 target] or P[yΔ1A-2/1A-2′] transgenic (TG) lines, homologous recombinants carrying the whs gene (yΔ1A-2w and yΔ1A-2/1A-2′w), and homologous recombinants deleted for whs gene (yΔ1A-2 and yΔ1A-2/1A-2′). DNAs were transferred to Nytran and hybridized with a 32P-labeled probe made with ClaI to BglII fragment of y gene (black bar, Figure 5). The probe recognizes an endogenous band of 7.6 kb in y1w1118 flies, and transgene band of 4.5 kb. Correct recombination events removed the endogenous band. Excision of whs gene with Cre recombinase lead to appearance of a new EcoRV site at the LoxP element in yΔ1A-2 line (3 kb band). A similar event did not occur in the yΔ1A-2/1A-2′ line, therefore a smaller band is seen due to the ∼1.0 kb deletion of the Su(Hw) BSs (6.7 kb band).
(5.15 MB TIF)
Definition of parameters for semi-quantitative PCR analyses. Indicated volumes of cDNA were used as a template for amplification by the ac, yar, y and Ras64B primers for the number of cycles shown at the right. Ethidium-stained PCR products from each input were analyzed. These studies demonstrated that at the cycle number shown, each primer set produced an increasing amount of product with increasing input. In the semi-quantitative PCR reactions shown in Figure 6, 1 µl of template was used for the given number of cycles.
(2.17 MB TIF)
Analysis of RNA accumulation from 1A region genes in wild type and mutant backgrounds. Quantitative real time PCR (Q-PCR) was used to determine levels of y, ac and sc mRNA accumulation from RNAs isolated during development from wild type and mutant lines. Individual transcript levels defined by Q-PCR were normalized to Ras64B for amount of input cDNA (ΔCT). A larger ΔCT indicates a reduction in RNA. Error bars indicate standard deviation of values obtained from analyses of three independently isolated RNAs. No significant changes in RNA accumulation relative to wild type were detected.
(10.6 MB TIF)
Acknowledgments
We thank Lori Wallrath and the Geyer laboratory for critically reading this manuscript. We thank Jinsil Kim, Bill Chen and Jonathan Zuk for technical assistance. We thank Yikang Rong for providing pW25 and Kent Golic for providing the 70FLP and 70I-SceI flies.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by a National Institutes of Health grant (GM42539) to PKG.
References
- 1.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 3.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 4.Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr Opin Genet Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Dorman ER, Bushey AM, Corces VG. The role of insulator elements in large-scale chromatin structure in interphase. Semin Cell Dev Biol. 2007;18:682–690. doi: 10.1016/j.semcdb.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engel N, Bartolomei MS. Mechanisms of insulator function in gene regulation and genomic imprinting. Int Rev Cytol. 2003;232:89–127. doi: 10.1016/s0074-7696(03)32003-0. [DOI] [PubMed] [Google Scholar]
- 7.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, et al. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corces VG, Geyer PK. Interactions of retrotransposons with the host genome: the case of the gypsy element of Drosophila. Trends in Genetics. 1991;7:86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- 9.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. Embo J. 2001;20:2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 12.Kurshakova M, Maksimenko O, Golovnin A, Pulina M, Georgieva S, et al. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol Cell. 2007;27:332–338. doi: 10.1016/j.molcel.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 13.Georgiev PG, Gerasimova TI. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol Gen Genet. 1989;220:121–126. doi: 10.1007/BF00260865. [DOI] [PubMed] [Google Scholar]
- 14.Adryan B, Woerfel G, Birch-Machin I, Gao S, Quick M, et al. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 2007;8:R167. doi: 10.1186/gb-2007-8-8-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos E, Ghosh D, Baxter E, Corces V. Genomic organization of gypsy-like chromatin insulators in Drosophila melanogaster. Genetics. 2006 doi: 10.1534/genetics.105.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, et al. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol Cell Biol. 2006;26:5983–5993. doi: 10.1128/MCB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golovnin A, Birukova I, Romanova O, Silicheva M, Parshikov A, et al. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development. 2003;130:3249–3258. doi: 10.1242/dev.00543. [DOI] [PubMed] [Google Scholar]
- 18.Parnell TJ, Viering MM, Skjesol A, Helou C, Kuhn EJ, et al. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc Natl Acad Sci U S A. 2003;100:13436–13441. doi: 10.1073/pnas.2333111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spana C, Corces VG. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes & Development. 1990;4:1505–1515. doi: 10.1101/gad.4.9.1505. [DOI] [PubMed] [Google Scholar]
- 20.Scott KC, Taubman AD, Geyer PK. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics. 1999;153:787–798. doi: 10.1093/genetics/153.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes & Development. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 22.Scott KS, Geyer PK. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO Journal. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes & Development. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geyer PK, Clark I. Protecting against promiscuity: the regulatory role of insulators. Cell Mol Life Sci. 2002;59:2112–2127. doi: 10.1007/s000180200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manak JR, Dike S, Sementchenko V, Kapranov P, Biemar F, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- 26.Gershenzon NI, Trifonov EN, Ioshikhes IP. The features of Drosophila core promoters revealed by statistical analysis. BMC Genomics. 2006;7:161. doi: 10.1186/1471-2164-7-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics. 2006;7:176. doi: 10.1186/1471-2164-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campuzano S, Balcells L, Villares R, Carramolino L, Garcia-Alonso L, et al. Excess function hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell. 1986;44:303–312. doi: 10.1016/0092-8674(86)90764-6. [DOI] [PubMed] [Google Scholar]
- 29.Morris JR, Chen J, Filandrinos ST, Dunn RC, Fisk R, et al. An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics. 1999;151:633–651. doi: 10.1093/genetics/151.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campuzano S, Carramolino L, Cabrera CV, Ruiz-Gomez M, Villares R, et al. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985;40:327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- 31.Mozer B, Marlor R, Parkhurst S, Corces V. Characterization and developmental expression of a Drosophila ras oncogene. Mol Cell Biol. 1985;5:885–889. doi: 10.1128/mcb.5.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn-Parnell EJ, Helou C, Marion DJ, Gilmore BL, Parnell TJ, et al. Investigation of the properties of non-gypsy Su(Hw) binding sites. Genetics in press. 2008 doi: 10.1534/genetics.108.087254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 34.Barrandon C, Spiluttini B, Bensaude O. Non-coding RNAs regulating the transcriptional machinery. Biol Cell. 2008;100:83–95. doi: 10.1042/BC20070090. [DOI] [PubMed] [Google Scholar]
- 35.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 36.Parkhurst SM, Corces VG. Interactions among the gypsy transposable element and the yellow and the suppressor of hairy-wing loci in Drosophila melanogaster. Mol Cell Biol. 1986;6:47–53. doi: 10.1128/mcb.6.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golovnin A, Melnick E, Mazur A, Georgiev P. Drosophila Su(Hw) insulator can stimulate transcription of a weakened yellow promoter over a distance. Genetics. 2005;170:1133–1142. doi: 10.1534/genetics.104.034587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akopov SB, Ruda VM, Batrak VV, Vetchinova AS, Chernov IP, et al. Identification, genome mapping, and CTCF binding of potential insulators within the FXYD5-COX7A1 locus of human chromosome 19q13.12. Mamm Genome. 2006;17:1042–1049. doi: 10.1007/s00335-006-0037-3. [DOI] [PubMed] [Google Scholar]
- 41.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 43.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 44.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 45.Vostrov AA, Taheny MJ, Quitschke WW. A region to the N-terminal side of the CTCF zinc finger domain is essential for activating transcription from the amyloid precursor protein promoter. J Biol Chem. 2002;277:1619–1627. doi: 10.1074/jbc.M109748200. [DOI] [PubMed] [Google Scholar]
- 46.Engel N, Thorvaldsen JL, Bartolomei MS. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum Mol Genet. 2006;15:2945–2954. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 47.Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene. 2002;21:3414–3421. doi: 10.1038/sj.onc.1205400. [DOI] [PubMed] [Google Scholar]
- 48.Lutz M, Burke Les J, Barreto G, Geoman F, Greb H, et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Research. 2000;28:1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Awad TA, Bigler J, Ulmer JE, Hu YJ, Moore JM, et al. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem. 1999;274:27092–27098. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- 50.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, et al. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 52.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 53.Merli C, Bergstrom DE, Cygan JA, Blackman RK. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- 54.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J, Levine M. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell. 1999;99:567–575. doi: 10.1016/s0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]
- 56.Lin Q, Wu D, Zhou J. The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Development. 2003;130:519–526. doi: 10.1242/dev.00227. [DOI] [PubMed] [Google Scholar]
- 57.Akbari OS, Bae E, Johnsen H, Villaluz A, Wong D, et al. A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development. 2008;135:123–131. doi: 10.1242/dev.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marlor RL, Parkhurst SM, Corces VG. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol. 1986;6:1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong WJ, Golic KG. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci U S A. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 62.Chen JL, Huisinga KL, Viering MM, Ou SA, Wu CT, et al. Enhancer action in trans is permitted throughout the Drosophila genome. Proc Natl Acad Sci U S A. 2002;99:3723–3728. doi: 10.1073/pnas.062447999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spradling AC, Mahowald AP. Identification and genetic localization of mRNAs from ovarian follicle cells of Drosophila melanogaster. Cell. 1979;16:589–598. doi: 10.1016/0092-8674(79)90032-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Southern analysis of y-ac locus in homologous recombinant lines. Genomic DNA was isolated from ten flies, digested with EcoRV (NEB) and run on a 1% agarose gel. Flies analyzed were the parental y1w1118 line, the P[yΔ1A-2 target] or P[yΔ1A-2/1A-2′] transgenic (TG) lines, homologous recombinants carrying the whs gene (yΔ1A-2w and yΔ1A-2/1A-2′w), and homologous recombinants deleted for whs gene (yΔ1A-2 and yΔ1A-2/1A-2′). DNAs were transferred to Nytran and hybridized with a 32P-labeled probe made with ClaI to BglII fragment of y gene (black bar, Figure 5). The probe recognizes an endogenous band of 7.6 kb in y1w1118 flies, and transgene band of 4.5 kb. Correct recombination events removed the endogenous band. Excision of whs gene with Cre recombinase lead to appearance of a new EcoRV site at the LoxP element in yΔ1A-2 line (3 kb band). A similar event did not occur in the yΔ1A-2/1A-2′ line, therefore a smaller band is seen due to the ∼1.0 kb deletion of the Su(Hw) BSs (6.7 kb band).
(5.15 MB TIF)
Definition of parameters for semi-quantitative PCR analyses. Indicated volumes of cDNA were used as a template for amplification by the ac, yar, y and Ras64B primers for the number of cycles shown at the right. Ethidium-stained PCR products from each input were analyzed. These studies demonstrated that at the cycle number shown, each primer set produced an increasing amount of product with increasing input. In the semi-quantitative PCR reactions shown in Figure 6, 1 µl of template was used for the given number of cycles.
(2.17 MB TIF)
Analysis of RNA accumulation from 1A region genes in wild type and mutant backgrounds. Quantitative real time PCR (Q-PCR) was used to determine levels of y, ac and sc mRNA accumulation from RNAs isolated during development from wild type and mutant lines. Individual transcript levels defined by Q-PCR were normalized to Ras64B for amount of input cDNA (ΔCT). A larger ΔCT indicates a reduction in RNA. Error bars indicate standard deviation of values obtained from analyses of three independently isolated RNAs. No significant changes in RNA accumulation relative to wild type were detected.
(10.6 MB TIF)