Abstract
QPT-1 was discovered in a compound library by high-throughput screening and triage for substances with whole-cell antibacterial activity. This totally synthetic compound is an unusual barbituric acid derivative whose activity resides in the (−)-enantiomer. QPT-1 had activity against a broad spectrum of pathogenic, antibiotic-resistant bacteria, was nontoxic to eukaryotic cells, and showed oral efficacy in a murine infection model, all before any medicinal chemistry optimization. Biochemical and genetic characterization showed that the QPT-1 targets the β subunit of bacterial type II topoisomerases via a mechanism of inhibition distinct from the mechanisms of fluoroquinolones and novobiocin. Given these attributes, this compound represents a promising new class of antibacterial agents. The success of this reverse genomics effort demonstrates the utility of exploring strategies that are alternatives to target-based screens in antibacterial drug discovery.
Bacterial resistance to currently available therapeutic agents continues to be a growing problem (14, 21). Therefore, the identification of new antibacterial agents that use novel mechanisms of action remains an endeavor of great importance (25). Despite initial optimism among antibacterial discovery scientists in response to the publication of numerous bacterial genomes, it has become clear over the last decade that the use of target-based screening strategies to identify inhibitors of essential enzymes has not been successful in generating novel antibiotics (20). Although inhibitors of a number of attractive targets (5) have been identified (2, 6, 7, 9, 15, 26), their development into drugs has often been hindered by their lack of “whole-cell activity” (WCA), i.e., the inability to penetrate bacterial cells and/or maintain intracellular concentrations sufficient to inhibit growth. An alternate approach to target-based screening, called “reverse genomics” (also sometimes referred to as compound-driven target identification, chemical biology, or chemical genetics), is to screen for compounds with antibacterial WCA and to use this phenotype to determine their mechanisms of action (MOAs) by various biochemical and genetic approaches. Here we report on the discovery and subsequent biological and chemical characterization of PNU-286607, subsequently renamed QPT-1, which is the first member of a structurally novel class of bacterial topoisomerase II (TopoII) inhibitors (3).
MATERIALS AND METHODS
Strains.
All strains used in these studies were from the Pfizer (formerly Pharmacia) collection.
Generation of resistant strains.
Either ethyl methanesulfonate-mutagenized Staphylococcus aureus cultures prepared as described previously (16) or untreated S. aureus cultures (for spontaneous resistance) were grown at 37°C to an optical density at 600 nm of 1.0 × 1010, and 100 μl was plated on Mueller-Hinton agar plates which contained 4 μg/ml PNU-286607 or ciprofloxacin. Individual resistant colonies were confirmed by restreaking the colonies and subsequent MIC analysis (see Tables 2 and 4). The gyrAB and parCE topoisomerase genes from these and other resistant strains were amplified by PCR and sequenced at the Pfizer (formerly Pharmacia) sequencing core facility.
TABLE 2.
Susceptibilities of wild-type (RN4220) and resistant S. aureus strains with and without complementing plasmids overexpressing wild-type or mutant gyrB to a series of compounds with known MOAsa
| Antibacterial group and agent | MIC (μg/ml) of strains
|
|||
|---|---|---|---|---|
| Wild type | 286607-R1 | D437N/pgyrB-wt | wt/pgyrB-D437N | |
| PNU-286607 (QPT-1) | 1 | 16 | 4 | 1 |
| Cell wall synthesis | ||||
| Ampicillin | 2 | 4 | 2 | 2 |
| Neomycin | 2 | 4 | 2 | 2 |
| RNA synthesis | ||||
| Rifampin | <00.625 | <00.625 | <00.625 | <00.625 |
| Protein synthesis | ||||
| Chloramphenicol | 8 | 8 | 8 | 8 |
| Erythromycin | 0.5 | 0.5 | 0.5 | 0.5 |
| Kanamycin | 2 | 4 | 2 | 2 |
| Linezolid | 1 | 1 | 1 | 1 |
| Spectinomycin | 64 | 64 | 64 | 64 |
| Streptomycin | 4 | 4 | 4 | 4 |
| Tetracycline | 0.125 | 0.125 | 0.125 | 0.125 |
| DNA synthesis | ||||
| Ciprofloxacin | 0.125 | 0.25 | 0.25 | 0.25 |
| Nalidixic acid | 32 | 64 | 32 | 32 |
| Norfloxacin | 0.5 | 2 | 0.5 | 0.5 |
| Novobiocin | 0.125 | 0.00125 | 0.625 | 0.125 |
Strain 286607-R1 is a representative RN4220 strain in which resistance to PNU-286607 (QPT-1), which mapped to a D437N mutation in the gyrB gene, was generated. D437N/pgyrB-wt, strain 286607-R1 transformed with a plasmid that expressed wild-type gyrB; wt/pgyrB-D437N, wild-type strain RN4220 transformed with a plasmid that expressed the D437N gyrB. The antibacterial agents tested are listed by target pathway of inhibition.
TABLE 4.
In vitro antibacterial activities of (±)-PNU-286607 (QPT-1), purified enantiomers, and ciprofloxacin against multiple bacterial isolates
| Organism (no. of strains) | Agent | MIC (μg/ml)
|
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| S. aureus, methicillin susceptible (20) | PNU-286607 | 0.5-1 | 1 | 1 |
| (−)-Enantiomer | 0.25-0.5 | 0.5 | 0.5 | |
| (+)-Enantiomer | >64 | >64 | >64 | |
| Ciprofloxacin | 0.06-0.5 | 0.125 | 0.25 | |
| S. aureus, methicillin resistant (20) | PNU-286607 | 1-4 | 1 | 2 |
| (−)-Enantiomer | 0.25-1 | 0.5 | 0.5 | |
| (+)-Enantiomer | 64->64 | >64 | >64 | |
| Ciprofloxacin | 0.06-32 | 0.25 | 32 | |
| S. aureus, ciprofloxacin resistant (20) | PNU-286607 | 0.5-2 | 1 | 1 |
| (−)-Enantiomer | 0.25-1 | 0.5 | 0.5 | |
| (+)-Enantiomer | 64->64 | 64 | >64 | |
| Ciprofloxacin | 4->64 | 16 | >64 | |
| S. pneumoniae, penicillin susceptible, intermediate, or resistant (20) | PNU-286607 | 2-8 | 4 | 8 |
| (−)-Enantiomer | 1-4 | 2 | 4 | |
| (+)-Enantiomer | >64 | >64 | >64 | |
| Ciprofloxacin | 0.5-4 | 1 | 2 | |
| S. pneumoniae, ciprofloxacin resistant (17) | PNU-286607 | 1-8 | 4 | 8 |
| (−)-Enantiomer | 1-4 | 2 | 4 | |
| (+)-Enantiomer | >64 | >64 | >64 | |
| Ciprofloxacin | 2->64 | 32 | 64 | |
Molecular biology.
Genomic DNA was isolated from overnight cultures of S. aureus grown in brain heart infusion medium with a FastDNA kit (Qbiogene, Carlsbad, CA). Isolation was performed essentially as instructed by the manufacturer; however, the initial cell pellet was resuspended in 200 μl water containing 100 μg/ml lysostaphin (Sigma), and the mixture was incubated for 30 min at 37°C before we proceeded with the recommended protocol. DNA minipreps were performed with QIAprep spin miniprep kits from Qiagen (Valencia, CA). When plasmid DNA was isolated from S. aureus, 2 μl of 10 mg/ml lysostaphin (Sigma) was added to the cultures, resuspended in the P1 buffer supplied with the kit, and then incubated at 37°C for 30 min prior to addition of the P2 buffer supplied with the kit. Subsequent steps were identical to the manufacturer's instructions. To construct plasmids overexpressing either the wild-type or the D437N gyrB gene, the genes were amplified by PCR (under standard conditions, except for an increase in the final MgCl2 concentration to 3 mM) from corresponding genomic DNA with the oligonucleotides 5′-GCCTGCAGATGGTGACTGCATTGTCAGA-3′ (sense; the PstI site is underlined) and 5′-gcGCATGCGTCGACCAAGAGTTCCTCCTTCAAAA-3′ (antisense; the SalI and SphI sites are underlined) and cloned into the PstI and SphI sites of pSK265-TX3, a staphylococcus-specific pSK265 vector (13) containing a tetracycline-regulated promoter. Transformation of the plasmids, whose sequences were confirmed, was performed as described previously (23). The overexpression of GyrB from transformed strains was achieved by the addition of 100 ng/ml doxycycline.
MIC determination.
The MIC of the drug necessary to inhibit bacterial growth was determined by broth microdilution according to the standards of the National Committee for Clinical Laboratory Standards (18).
Macromolecular synthesis assay.
A total of 13.5 ml of brain heart infusion medium was inoculated with 1.5-ml overnight cultures of S. aureus laboratory strain UC9218 and grown for 45 min at 37°C and 250 rpm and then diluted 1:10, and 140 μl was distributed to a 96-well microtiter plate. Five microliters of serially diluted (30×) PNU-286607 was added to achieve the final desired concentration, and the cells were incubated at 37°C for 20 min and 800 rpm on a Thermomixer. 14C-radiolabeled precursors (Amersham) were distributed to the wells such that each compound tested was mixed with precursors for each macromolecular pathway, as follows: DNA, 5 μl of a 1:2 dilution of thymidine (50 μCi/ml; catalog no. CFA-219); RNA, 5 μl of a 1:25 dilution of uridine (50 μCi/ml; catalog no. CFB-51); protein, 7.5 μl of undiluted l-leucine (50 μCi/ml; catalog no. CFB-183); and cell wall, 5 μl of a 1:2 dilution of [14C]d-alanine (200 μCi/ml; catalog no. ARC-1618). The final volume of the premixture was adjusted to 10 μl with dimethyl sulfoxide (DMSO), with the final DMSO concentration being no greater than 0.5%. Five microliters of each radioactive precursor was added to the appropriate wells, and the plates were incubated at 37°C and 800 rpm for 80 min. A total of 100 μl of each of the reaction mixtures was added to filter plates containing 100 μl ice-cold 50% trichloroacetic acid (TCA), mixed at 800 rpm for 1 min, and incubated on ice for 60 min. The incorporated counts were harvested with a Packard FilterMate-196 harvester by using UniFilter GF/B filter plates. The filter plates were prewashed with 5% TCA, and the samples were filtered through and washed four times with ice-cold 5% TCA and then 10% ethanol. After the plates were dried, the bottoms of the plates were sealed and 40 μl MicroScint scintillation fluid was added to each well. The tops of the plates were then sealed with MultiScreen sealing tape and counted on a TriLux apparatus.
In vitro topoisomerase assays.
Escherichia coli DNA gyrase and human TopoII were purchased as kits from Topogen (Columbus, OH). E. coli TopoIV was a gift from K. Marians at the Sloan-Kettering Cancer Center (Cornell University, New York, NY). DNase I and proteinase K were purchased from Invitrogen (Rockville, MD). Relaxed pBR322 and kinetoplast DNA were purchased from Topogen. The E. coli DNA gyrase assay was performed according to the instructions of the manufacturer, as follows: 1 U of gyrase was incubated with 0.5 μg of relaxed pBR322 in a reaction volume of 30 μl at 37°C for 30 min in incubation buffer (35 mM Tris HCl, pH 7.5, 24 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 1.8 mM spermidine, 1 mM ATP, 6.5% glycerol, 0.1 mg/ml bovine serum albumin [BSA]). Samples were analyzed by gel electrophoresis in a 0.8% agarose gel in 1× TBE (Tris-borate-EDTA; prepared without ethidium bromide [EtBr]). Bands were visualized by staining with EtBr (0.5 μg/ml in 1× TBE) for 15 min, followed by UV analysis on a Bio-Rad GelDoc apparatus. The 50% inhibitory concentrations (IC50s) were determined by analysis with GraphPad Prism software. The E. coli TopoIV enzyme was stored in 1× storage buffer (50 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl, 10 mM 2-mercaptoethanol, 40% glycerol). Before use, the E. coli TopoIV enzyme was diluted 1:10 in dilution buffer (50 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl, 10 mM 2-mercaptoethanol, 0.5 mg/ml BSA, 20% glycerol). The E. coli TopoIV decatanation assay was performed with 3.6 μg of kinetoplast DNA (Topogen) as the substrate in each reaction. The assay was run in 1× TopoIV assay buffer (10 mM HEPES-KOH, 2 mM magnesium acetate, 2 mM dithiothreitol, 4 mM KCl, 200 μM spermidine, 20 μg/ml BSA, 400 μM ATP). The final reaction volume was 20 μl, with a final DMSO or drug concentration of 10%. The reaction mixtures were assembled and incubated for 30 min at 37°C. After the incubation, 2 μl each of 10% sodium dodecyl sulfate and 1 mg/ml proteinase K (in H2O) was added, and the reaction mixtures were incubated for an additional 15 min at 37°C. After the second incubation, 2 μl of 10× loading dye (0.25% bromophenol blue, 50% glycerol) and 20 μl of phenol-chloroform-isoamyl alcohol was added to each reaction mixture, and the mixture was vortexed briefly and spun in a microcentrifuge at high speed for 1 min. Seventeen microliters of the upper blue aqueous phase was removed and loaded onto a 1% agarose gel containing 0.5 μg/ml EtBr. The human TopoII cleavage/decatanation assay was performed as described above for the TopoIV assay but was run in 1× cleavage buffer (30 mM Tris-HCl, pH 7.6, 3 mM ATP, 15 mM 2-mercaptoethanol, 8 mM MgCl2, and 60 mM NaCl) with 4 U of human TopoII (Topogen).
Eukaryotic proliferation assay.
MC9 (ATCC CRL-8306) cells were maintained as a suspension culture with between 2 × 105 cells/ml and 1.6 × 106 cells/ml by dilution into fresh Dulbecco modified Eagle medium (catalog no. 23800-014; Gibco) containing 10% heat-inactivated fetal bovine serum and 10% conditioned medium from concanavalin A-stimulated rat splenocytes (catalog no. 40115; Becton Dickinson). The cells were maintained in T75 vented cap culture flasks at 37°C in 5% CO2 in air. The doubling time for MC9 cells is approximately 27 h. Twofold serial dilutions of the test compounds and the controls were prepared in duplicate in U-bottom 96-well microtiter plates by the addition of a test or a control compound from 10 mM DMSO stock solutions to 50 μl of MC9 medium. At the time of the assay, the viability of the MC9 cells was assessed in 100 μl of phosphate-buffered saline by trypan blue exclusion. A total of 10,000 viable cells was added to each well in 50 μl of medium to give a final volume of 100 μl. The microtiter plates were incubated at 37°C in 5% CO2 in air for 72 h. To determine the percentage of viable cells in proliferation, 20 μl of a soluble tetrazolium compound, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) (catalog no. G4000; Promega), containing an electron-coupling reagent (phenyl methylsulfonate [PMS]) was added to each well, and the plates were incubated for 3 h at 37°C in 5% CO2. Following incubation, the absorbance of the formazan product resulting from the bioreduction of the PMS-MTS solution was measured at 490 nm. The value of the absorbance is directly proportional to the number of respiring cells present in the culture. Absorbance values were corrected for the background absorbance, and the data were expressed as a percentage of the total loss of viability resulting from treatment with 10 μM cycloheximide.
In vivo efficacy and pharmacokinetic studies.
All animal procedures were performed in compliance with the Animal Welfare Act regulations (9 CFR, Parts 1, 2, and 3) and with the Guide for the Care and Use of Laboratory Animals (11) and were completed under the guidelines established by the Pharmacia and Upjohn Institutional Animal Care and Use Committee. The 50% effective doses (ED50s) were determined as follows. Briefly, all CF-1 female mice (weight, 20 g; Harlan Sprague-Dawley, Indianapolis, IN) were given sufficient bacteria intraperitoneally to kill 90 to 100% of the untreated mice. Thawed bacterial cultures were suspended in brain heart infusion broth which contained 4% (wt/vol) dried brewer's yeast (Champlain Industries Inc., Clifton, NJ). At least five dosage levels of antibiotics were used for each ED50 determination. One treatment group of mice (six infected animals) was used for each antibiotic dosage level. Antibiotic was administered either orally or subcutaneously at 1 and 5 h postinfection. The mice were monitored for 1 week, and ED50s were calculated by probit analysis. For pharmacokinetic analyses, all mice were dosed with a volume of 0.2 ml for both the intravenous and the oral routes. Samples were taken from mice treated intravenously at 0, 2, 15, 30, 60, 120, 240, and 480 min; and samples were taken from mice treated orally at 0, 20, 30, 60, 120, 240, 360, and 480 min. The vehicle for the oral formulation was pH 4.5 acetate-buffered, 20% DMSO-methylcellulose suspension. The vehicle for the intravenous formulation was 20% hydroxypropyl beta-cyclodextrin (pH 7 phosphate buffer). The extent of binding of PNU-286607 to mouse plasma was determined by ultracentrifugation. PNU-286607 demonstrated moderate binding (70%), with the (−)- and (+)-enantiomers showing similar levels of protein binding in mouse plasma. Pharmacokinetic analysis was performed by noncompartmental analysis using Watson Laboratory Information Management System (Thermo, Inc., Philadelphia, PA). The mean concentration-time data (three mice per time point) were used for calculation of the values of the pharmacokinetic parameters.
RESULTS
Screening, triage, and discovery of PNU-286607.
Antibacterial compounds were selected via a series of high-throughput MIC screens of the Pharmacia Research Compound Collection (consisting of ∼250,000 compounds), resulting in the identification of several thousand bacterial compounds with WCA. These compounds were then triaged on the basis of the following criteria (listed in order of priority): activity against both E. coli and S. aureus at 32 μg/ml or better, amenability to medicinal chemistry, a low to moderate level of serum binding, a lack of antifungal activity, a high degree of purity, and an adequate inventory for follow-up. The first prioritization effort led to the identification of 14 compounds deemed worthy of MOA determination, and one of these was PNU-286607.
Mechanism of action.
Table 1 highlights the overall antibacterial activity profiles of racemic PNU-286607 and its individual enantiomers (which were isolated and characterized as described below after MOA analysis was performed with the racemic mixture). In general, PNU-286607 exhibited a broad spectrum of antibacterial activity and had activity against both gram-positive and gram-negative organisms, including methicillin-resistant and quinolone-resistant strains (Table 1). Like many antibiotics, this compound is actively effluxed from gram-negative bacteria (compare the MICs for E. coli and Haemophilus influenzae wild types and those for the acrAB- and tolC pump-knockout strains), but unlike the fluoroquinolones, it is not a substrate for NorA efflux in S. aureus. Also of note in Table 1 is the observation of a modest effect of the addition of serum on the antibacterial activity of PNU-286607 against S. aureus UC9218. At 4× the MIC, the frequency of spontaneous resistance to PNU-286607 was found to be 1 in 109 for both S. aureus and Streptococcus pneumoniae and 1 in 107 for H. influenzae (data not shown).
TABLE 1.
In vitro antibacterial activities of PNU-286607 (QPT-1) and purified enantiomers against selected organisms as revealed by the MIC required to inhibit bacterial growth
| Organism | MIC (μg/ml)
|
||
|---|---|---|---|
| PNU-286607 (QPT-1) | (+)-Enantiomer | (−)-Enantiomer | |
| Staphylococcus aureus UC9218a | 1 | 128 | 0.25 |
| Staphylococcus aureus UC9218 + 25% serum | 2 | 128 | 0.5 |
| Staphylococcus aureus UC9218 + 50% serum | 4-8 | >128 | 1 |
| Staphylococcus aureus UC12673b | 1 | >64 | 0.5 |
| Staphylococcus aureus UC9213c | 1 | >64 | 0.5 |
| Staphylococcus aureus RN4220 | 1 | NTd | NT |
| Staphylococcus aureus RN4220 (norA negative) | 1 | NT | NT |
| Staphylococcus epidermidis UC12084c | 1 | >64 | 0.5 |
| Streptococcus pneumoniae UC9912 | 4-8 | >128 | 2 |
| Streptococcus pneumoniae UC15142 | 4 | >128 | 2 |
| Streptococcus pyogenes UC152 | 2 | >64 | 1 |
| Enterococcus faecalis UC9217 | 4 | >128 | 2 |
| Enterococcus faecium UC12712 | 8 | >64 | 4 |
| Haemophilus influenzae UC30063 | 0.5 | >128 | 0.125 |
| Haemophilus influenzae 31809 (acrAB negative) | <0.06 | NT | NT |
| Moraxella catarrhalis UC3067 | 2 | >64 | 1 |
| Escherichia coli UC6674 | 4-8 | >128 | 2 |
| Escherichia coli UC31701 (acrAB negative) | 0.125 | NT | NT |
| Escherichia coli UC31708 (tolC negative) | <0.06 | NT | NT |
| Klebsiella pneumoniae UC12081 | NT | >64 | >64 |
| Pseudomonas aeruginosa UC6676 | NT | >64 | >64 |
Methicillin-susceptible, in vivo test strain.
Methicillin- and ciprofloxacin-resistant strain.
Methicillin resistant.
NT, not tested.
Characterization of the PNU-286607 MOA began with an assessment of its effects on global biosynthetic pathways. Analysis of the effect of this compound on macromolecular synthesis pathways, as revealed by measurement of the incorporation of radiolabeled precursors, showed that PNU-286607 specifically inhibits DNA synthesis in S. aureus (Fig. 1). This was confirmed by comparative microarray analysis of S. aureus and E. coli treated with a compendium of known classes of antibiotics with well-defined MOAs. The transcript profiles for both S. aureus and E. coli treated with PNU-286607 were closely related to those of other known DNA synthesis inhibitors (ciprofloxacin, norfloxacin, and novobiocin) but did not show any similarity to those of drugs which inhibit other pathways (data not shown). To genetically identify the target of inhibition of PNU-286607, laboratory-generated resistant strains of S. aureus were analyzed for their susceptibilities to a panel of compounds with well-characterized MOAs. Resistant strains were found to have an increase in sensitivity to novobiocin as well as a slight (twofold) decrease in susceptibility to norfloxacin, with no significant change in susceptibility to any other class of antibacterial tested (Table 2). These results suggest that PNU-286607 may inhibit the same enzymes or enzymes related to the enzymes that norfloxacin and novobiocin inhibit, namely, the topoisomerases, which are required for DNA replication and repair. Bacteria utilize two of these types of enzymes, DNA gyrase (encoded by gyrAB) and TopoIV (encoded by parCE): each enzyme consists of a dimer containing an A and a B subunit (10). Accordingly, gyrAB and parCE were amplified by PCR from several resistant isolates and sequenced. Most of these isolates were found to contain single point mutations in the gyrB gene that resulted in changes of the aspartic acid at residue 437 to either asparagine or valine; one isolate had a gyrB point mutation which resulted in the replacement of the alanine at residue 439 with a serine. This is in contrast to the classical mutations conferring resistance to the fluoroquinolones (which occur at the S79 or the D83 residue of ParC in gram-positive bacteria and the S83 or the D87 residue of GyrA in gram-negative bacteria [10]) or novobiocin (to which resistance maps to the N-terminal ATP-binding cleft of gyrB [8]). No mutations conferring resistance to PNU-286607 were found in the gyrA or parC gene. Complementation studies were conducted to confirm the source of the PNU-286607 resistance. The gyrB genes encoding either the wild-type subunit or the subunit with the D437N mutation were cloned into overexpression vectors and transformed into both parent and mutant strains. No effect on the MIC was seen for either strain bearing an overexpressed copy of the endogenous gyrB (data not shown); but overexpression of the wild-type GyrB in the mutant background resulted in the reduction of the MIC to levels near those for the wild-type, while overexpression of the mutant GyrB in a wild-type background did not affect the susceptibility to PNU-286607 (Table 2). Likewise, the novobiocin-hypersusceptible and slightly norfloxacin-resistant phenotypes were rescued by complementation by the wild-type gyrB in the mutant strain, but no change in the MICs was observed when mutant gyrB was overexpressed in the wild-type background. No significant difference in response to any other compounds was observed between the complemented strains, indicating that the D437N mutation conferred the resistance to PNU-286607 and that the wild-type phenotype is dominant. To further define the distinction in the MOAs between PNU-286607 and the fluoroquinolones, the MICs of PNU-286607 and a representative of the class, ciprofloxacin, were determined against several S. aureus mutant strains which had been generated by the spontaneous selection for resistance to either compound plus several classical clinical isolates whose genomes have been sequenced. As shown in Table 3, mutations conferring resistance to ciprofloxacin did not affect the susceptibilities of the strains to PNU-286607 and ciprofloxacin retained activity against the PNU-286607-resistant strain, indicating that the MOAs of PNU-286607 and ciprofloxacin are distinct. Further demonstration of the lack of cross-resistance between PNU-286607 and the fluoroquinolones was shown when the activity of PNU-286607 was tested against multiple strains of ciprofloxacin-resistant S. aureus and S. pneumoniae, a number of which contained the classical resistance mutations described above. As shown in Table 4, PNU-286607 and its active enantiomer exhibited potent activity against ciprofloxacin-resistant isolates.
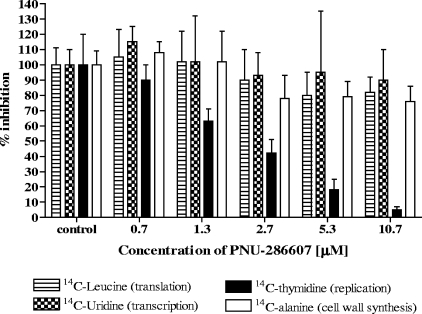
FIG. 1.
PNU-286607 (QPT-1) specifically inhibits DNA synthesis in a S. aureus macromolecular synthesis assay. The dose-dependent reduction of [14C]thymidine but not other radiolabeled precursors of macromolecular synthesis occurs upon treatment of growing S. aureus cells (strain UC9218) with PNU-286607. The MIC for this strain is 1 μg/ml (2.6 μM) (Table 1).
TABLE 3.
Susceptibilities of S. aureus strains with known topoisomerase mutations to PNU-286607 and ciprofloxacin
| Strain | Topoisomerase gene sequence
|
MIC (μg/ml)
|
||||
|---|---|---|---|---|---|---|
| gyrA | gyrB | parC | parE | PNU-286607 | Ciprofloxacin | |
| RN4220a | wtb | wt | wt | wt | 1 | 0.25 |
| SAUR 1015-01c | S84L | wt | S80F | S189R | 0.5 | 64 |
| COLd | wt | V28A | wt | wt | 1 | 0.125 |
| MW2d | wt | wt | R400C | wt | 1 | 0.25 |
| USA300d | S84L | wt | S80Y | wt | 1 | 8 |
| ATCC 29213 | wt | wt | wt | wt | 1 | 0.125 |
| ATCC 29213, ciprofloxacin resistant | D83N | wt | S80F | wt | 1 | 8 |
| ATCC 29213, quinoline pyrimidine trione resistant | wt | D437N | wt | wt | 8 | 0.5 |
The sequence was determined at Pfizer.
wt, wild type.
Methicillin-resistant S. aureus clinical isolate; the sequence was determined at Pfizer.
The sequence was obtained from www.tigr.org.
In vitro activity and selectivity.
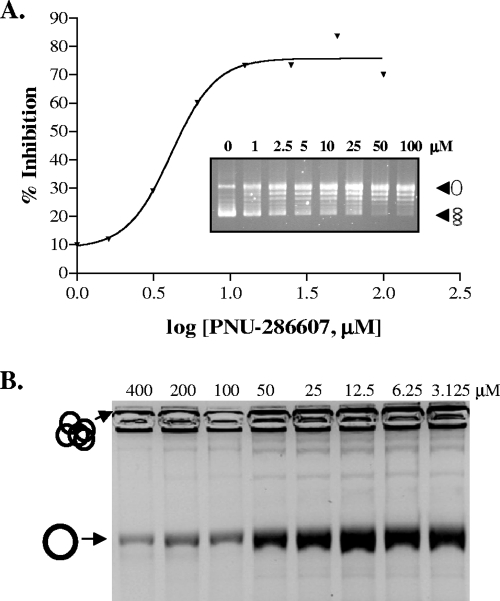
Having genetically identified gyrB as the target of inhibition of PNU-286607, we then sought to confirm its ability to inhibit topoisomerases in vitro. Using standard gel-based supercoiling and decatenation assays, we found that the IC50 against purified E. coli DNA gyrase was 9 μM and that that against E. coli TopoIV was 30 μM (Fig. 2). By way of comparison, the IC50s of both ciprofloxacin and novobiocin against these enzymes range from 0.1 to 0.5 μM and 1 to 10 μM, respectively (data not shown). The inhibition of topoisomerase activity by PNU-286607 is selective for bacteria, as no activity was seen against purified human TopoII (tested up to 200 μM; data not shown). In addition, the compound did not exhibit any activity in a eukaryotic cell proliferation assay at concentrations up to 500 μM, which is in contrast to the activities of some other marketed antibacterials, such as linezolid, whose IC50 was 30 μM, suggesting that it is nontoxic to eukaryotic cells (data not shown).
FIG. 2.
PNU-286607 (QPT-1) activity in vitro against DNA gyrase (A) and TopoIV (B). Purified E. coli DNA gyrase (A) or TopoIV (B) was added to relaxed plasmid DNA (indicated by open circles) (A) or concatameric DNA substrate (indicated by multiple circles at the top of the gel) (B) in the presence of increasing amounts of PNU-286607 (QPT-1), and the relative activity of each enzyme was determined by gel electrophoresis and visualization by staining with EtBr. The IC50s of PNU-286607 (QPT-1) against gyrase and TopoIV are 9 μM and 30 μM, respectively.
In vivo efficacy.
PNU-286607 was examined for potential efficacy against susceptible bacteria in a relevant animal model of infection. Remarkably, it was found that orally administered PNU-286607 showed appreciable efficacy (ED50, 19.5 ± 5 mg/kg of body weight) against methicillin-resistant S. aureus UC9213 in a lethal systemic mouse infection model. For comparison, orally administered ciprofloxacin provides an ED50 of 10 ± 2.5 mg/kg in this model. Subcutaneously administered PNU-286607 afforded an ED50 of 12.5 ± 5 mg/kg. In addition, no adverse effects were observed with doses up to 200 mg/kg. Taken together, this suggests a reasonable level of oral bioavailability and safety for PNU-286607 in the mouse. Subsequent pharmacokinetic analysis of the compound showed that it had remarkably favorable properties (Table 5).
TABLE 5.
Pharmacokinetic parametersa of PNU-286607 (QPT-1) in plasma from mice
| Treatment route | Dose (mg/kg) | t1/2 (h) | Tmax (hr) | Cmax (μg/ml) | AUC0-∞ (μg·hr/ml) | CL (liter/kg/h) | Vss (liter/kg) | F (%) |
|---|---|---|---|---|---|---|---|---|
| Intravenous | 21.6 | 1.22 | 15.7 | 31 | 0.697 | 0.623 | ||
| Oral | 20.3 | 0.927 | 1 | 15.7 | 27.6 | 94.7 |
AUC0-∞ area under the plasma drug concentration-time curve from 0 h to infinity; Cmax, maximum concentration observed in plasma; CL, plasma clearance; F, oral bioavailability; Tmax, time at which the maximum concentration in plasma was first observed; t1/2, elimination half-life; Vss, volume of distribution at steady state.
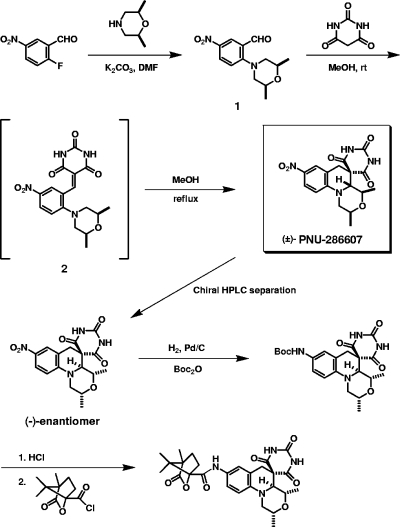
Structure determination, synthesis, and elucidation of the bioactive absolute configuration.
The originally assigned structure of chemical library compound PNU-286607 was found to be grossly incorrect, which became readily apparent upon attempts to resynthesize the purported structure. Extensive nuclear magnetic resonance studies (data not shown) suggested an unusual barbituric acid derivative with the relative stereochemistry shown in Fig. 3, which was subsequently confirmed by X-ray crystallography (see Fig. S1 in the supplemental material). The tetracyclic structure of PNU-286607, which comprises a relatively flat tricyclic core with a tetrahydroquinoline ring system fused to form a morpholine residue and an appended spirocyclic barbituric acid moiety, was judged to be quite unique and was therefore designated QPT-1 (for quinoline pyrimidine trione, the first member of the class).
FIG. 3.
Synthesis, enantiomer isolation, and determination of the absolute configuration of the active isomer of PNU-286607 (QPT-1). DMF, dimethyl formamide; MeOH, methanol; Boc2O, di-tert-butyl dicarbonate.
The interesting ring framework of QPT-1, taken together with its three attendant stereogenic centers, suggested that the preparation of this compound with control of its relative stereochemistry would be a challenge. A careful examination of the literature drew our attention to the chemistry described by Reinhoudt and coworkers (28). In order to explore the applicability of the chemistry of Reinhoudt and colleagues to QPT-1, the requisite alkylidene precursor (compound 1) was needed (Fig. 3). To this end, 2-fluoro-5-nitrobenzaldehyde was reacted with cis-2,6-dimethylmorpholine to give the adduct, compound 1, at a high yield (3). Compound 1 was then reacted with barbituric acid in methanol at ambient temperature to furnish the alkylidene (compound 2) at a 56% yield after chromatographic purification (3). In a gratifying result, the alkylidene intermediate (compound 2) was then refluxed in methanol to provide racemic QPT-1 at a nearly quantitative yield (3). The relative stereochemistry was rigorously controlled in the desired sense during this process. There was no evidence for any diastereomeric impurities. The conversion of compound 1 to QPT-1 could also be accomplished in one step without the isolation of the alkylidene (compound 2) by simply conducting the condensation/rearrangement reaction at reflux temperature. Racemic QPT-1 was separated into its individual enantiomers by preparative chiral stationary-phase high-pressure liquid chromatography with a Chiralcel, Chiralpak, or Chirose column and various alcohol eluant systems. In this way, multigram quantities of the (+)- and (−)-antipodes were obtained. Proof of the absolute configuration of the bioactive (−)-enantiomer was obtained by use of a short reaction sequence, starting with reduction of the nitro group to the corresponding aniline. This aniline intermediate was found to be quite sensitive, so, in general, the aniline was isolated as the Boc (tert-butoxycarbonyl) derivative shown in Fig. 3. Subsequent removal of the protecting group, conversion to the corresponding amide derived from (−)-camphanic chloride, and single crystal X-ray analysis (see Fig. S2 in the supplemental material) confirmed that the absolute configuration of the (−)-enantiomer is as shown in Fig. 3. As shown in Tables 1 and 4, essentially all of the antibacterial activity resides in the (−)-enantiomer. In addition, it was also observed that the (−)-enantiomer had an IC50 of 2.4 μM against DNA gyrase, while the (+)-enantiomer was essentially inactive (IC50, >100 μM) (data not shown), which correlates well with the observed MICs for these compounds (Table 1). In addition, spontaneous resistance to the active enantiomer at 4× the MIC was reduced to less than 1 in 1010 organisms for S. aureus and S. pneumoniae and to 1 in 108 for H. influenzae (data not shown).
DISCUSSION
The number of antibiotics which retain clinical efficacy is predicted to decrease dramatically, yet new entities that show promise in clinical trials are alarmingly low in number (27). This dilemma is exacerbated by the recent departure of many major pharmaceutical companies from the area of therapeutic antibacterial development (19, 22, 24). The lack of success in finding novel antibiotics over the last decade has made the dwindling forces of the remaining antibacterial discovery scientists obliged to consider alternatives to target-based high-throughput screens. One such approach is reverse genomics, i.e., use of the antibacterial activities of WCA compounds to determine their MOAs by resistance mapping and other techniques. Recent advances in MOA technologies, such as error-prone PCR (4) and rapid sequencing of entire bacterial genomes (1, 17), make reverse genomics screening approaches even more feasible than they were in years past. Our implementation of such a strategy led to the discovery of a compound with a number of attractive features, most notably, activity against a broad spectrum of pathogenic bacteria, including multidrug-resistant strains, and oral in vivo efficacy. Remarkably, these properties were resident in the original hit from the file before any optimization by medicinal chemistry was initiated. Although QPT-1 targets the same enzymes as marketed antibiotics, such as fluoroquinolones and novobiocin, the potential for cross-resistance to these agents, promisingly, appears to be low due to the apparently distinct MOA revealed by genetic analysis (Table 2), which is supported by its activity against strains resistant to these compounds (Tables 3 and 4). Because overexpression of the GyrB wild-type subunit in a wild-type background did not result in resistance, it is possible that the MOA of the compound involves the entire GyrAB complex (and not just the GyrB subunit itself). In addition, the D437N gyrB mutation, which confers resistance to QPT-1, did not result in a significantly increased level of resistance to fluoroquinolones and actually increased the susceptibility to novobiocin, reinforcing the notion that QPT-1, like the fluoroquinolones, may involve the formation of a lethal intermediate (DNA-protein complex or breaks). Although there has been one mention of a GyrB D437N mutation in association with fluoroquinolone resistance (12), this mutation was seen after only three rounds of selection and, as we have shown here, does not by itself confer resistance to fluoroquinolones. We therefore conclude that the MOA of QPT-1 is related to but distinct from that of the fluoroquinolone class.
The salient structural feature of QPT-1 is its fused morpholinylquinoline core bearing an appended spirocyclic barbituric acid moiety. This compound is the first example of a structurally novel class of bacterial TopoII inhibitors. Studies with the fluoroquinolones and novobiocin have provided ample precedent that these targets are essential in bacteria and that the development of clinically relevant agents is possible.
Without any structural modification, QPT-1 was found to be selectively active against bacteria, with no measurable inhibitory activity either against purified human TopoII or in a eukaryotic cell proliferation assay. Although spontaneous resistance to the active enantiomer of this compound was found to be moderate for H. influenzae (1 × 10−8 at 4× the MIC), encouragingly, it was low for gram-positive bacteria (<1 × 10−10 at 4× the MIC). In addition, the compound demonstrated efficacy when it was administered orally, attractive pharmacokinetic parameters, and no observed adverse effects when it was used at high doses. Finding such a lead compound with so many desirable attributes straight out of a screening campaign, prior to any modification by medicinal chemistry, gives new hope that alternative antibacterial discovery efforts can lead to novel agents which are desperately needed in the clinic.
Supplementary Material
Acknowledgments
We acknowledge Jana Jensen for formulation work and Steve Dunham and Phoebe Roberts for critical reading of the manuscript.
Footnotes
Published ahead of print on 2 June 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Andries, K., P. Verhasselt, J. Guillemont, H. W. Gohlmann, J. M. Neefs, H. Winkler, J. Van Gestel, P. Timmerman, M. Zhu, E. Lee, P. Williams, D. de Chaffoy, E. Huitric, S. Hoffner, E. Cambay, C. Truffot-Pernot, N. Lounis, and V. Jarlier. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223-227. [DOI] [PubMed] [Google Scholar]
- 2.Artsimovitch, I., C. Clement, A. S. Lynch, and R. A. Landick. 2003. New class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science 24:650-654. [DOI] [PubMed] [Google Scholar]
- 3.Barbachyn, M. R., G. L. Bundy, P. J. Dobrowolski, A. R. Hurd, A. R. Romero, D. J. McNamara, J. R. Palmer, L. M. Thomasco, P. L. Toogood, J. C. Ruble, D. A. Sherry, D. L. Romero, and G. E. Martin. April 2007. Tricyclic tetrahydroquinoline antibacterial agents. U.S. patent 7,208,490 B2.
- 4.Belanger, A. E., A. Lai, M. A. Brackman, and D. J. Leblanc. 2002. PCR-based ordered genomic libraries: a new approach to drug target identification for Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2507-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, E. D., and G. D. Wright. 2005. New targets and screening approaches in antimicrobial drug discovery. Chem. Rev. 105:759-774. [DOI] [PubMed] [Google Scholar]
- 6.Ehmann, D. E., J. E. Demeritt, K. G. Hull, and S. L. Fisher. 2004. Biochemical characterization of an inhibitor of Escherichia coli UDP-N-acetylmuramyl-l-alanine ligase. Biochim. Biophys. Acta 1698:167-174. [DOI] [PubMed] [Google Scholar]
- 7.Ejim, L. J., J. E. Blanchard, K. P. Koteva, R. Sumerfield, N. H. Elowe, J. D. Chechetto, E. D. Brown, M. S. Junop, and G. D. Wright. 2007. Inhibitors of bacterial cystathionine beta-lyase: leads for new antimicrobial agents and probes of enzyme structure and function. J. Med. Chem. 50:755-764. [DOI] [PubMed] [Google Scholar]
- 8.Fujimoto-Nakamura, M., H. Ito, Y. Oyamada, T. Nishino, and J. Yamagishi. 2005. Accumulation of mutations in both gyrB and parE genes is associated with high-level resistance to novobiocin in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3810-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, X., A. Alian, R. Stroud, and P. R. Ortiz de Montellano. 2006. Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from M. tuberculosis. J. Med. Chem. 49:6308-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper, D. C. 1998. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin. Infect. Dis. 27:S54-S63. [DOI] [PubMed] [Google Scholar]
- 11.ILAR, Commission of Life Sciences, and National Research Council. 1996. Guide for the Care and Use of Laboratory Animals. National Academy Press, Washington, DC.
- 12.Ito, H., Y. Hiroaki, M. Bogaki-Shonai, N. Toshiyuki, H. Hattori, and S. Nakamura. 1994. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2014-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, C. L., and S. A. Kahn. 1986. Nucleotide sequence of the enterotoxin B gene from Staphylococcus aureus. J. Bacteriol. 166:29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy, S. B. 2005. Antibiotic resistance—the problem intensifies. Adv. Drug Deliv. Rev. 57:1443-1450. [DOI] [PubMed] [Google Scholar]
- 15.Liu, S., J. S. Chang, J. T. Herberg, M. M. Horng, P. K. Tomich, A. H. Lin, and K. R. Marotti. 2006. Allosteric inhibition of Staphylococcus aureus d-alanine:d-alanine ligase revealed by crystallographic studies. Proc. Natl. Acad. Sci. USA 103:15178-15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Mwangi, M. M., S. W. Wu, Y. Zhou, K. Sieradzki, H. de Lencastre, P. Richardson, D. Bruce, E. Rubin, E. Myers, E. D. Siggia, and A. Tomasz. 2007. Tracking the in vivo evolution of multi-drug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 104:9451-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2000. Approved standard M7-A5: methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 19.Overby, K. M., and J. F. Barrett. 2005. Antibiotics: where did we go wrong? Drug Discov. Today 10:45-52. [DOI] [PubMed] [Google Scholar]
- 20.Payne, D. J., M. N. Gwynn, D. J. Holmes, and D. L. Pompliano. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 21.Peterson, L. R. 2005. Squeezing the antibiotic balloon: the impact of antimicrobial classes on emerging resistance. Clin. Microbiol. Infect. 11(Suppl. 5):4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Projan, S. 2003. Why is big pharma getting out of antibacterial drug discovery? Curr. Opin. Microbiol. 6:427-430. [DOI] [PubMed] [Google Scholar]
- 23.Schenk, S., and R. Ladagga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 24.Shlaes, D. M. 2003. The abandonment of antibacterials: why and wherefore? Curr. Opin. Pharmacol. 3:470-473. [DOI] [PubMed] [Google Scholar]
- 25.Silver, L. L., and K. A. Bostian. 1993. Discovery and development of new antibiotics: the problem of antibiotic resistance. Antimicrob. Agents Chemother. 37:377-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamper, G. F., K. L. Longenecker, E. H. Fry, C. G. Jakob, A. S. Florjancic, Y. G. Gu, D. D. Anderson, C. S. Cooper, T. Zhang, R. F. Clark, Y. Cia, C. L. Black-Schaefer, J. Owen McCall, C. G. Lerner, P. J. Hajduk, B. A. Beutel, and V. S. Stoll. 2006. Structure-based Optimization of MurF inhibitors. Chem. Biol. Drug Des. 67:58-65. [DOI] [PubMed] [Google Scholar]
- 27.Talbot, G. H., J. Bradley, J. E. Edwards, Jr., D. Gilbert, M. Scheld, and G. J. Bartlett. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42:657-668. [DOI] [PubMed] [Google Scholar]
- 28.Verboom, W., D. N. Reinhoudt, R. Visser, and S. Harkema. 1984. “tert-Amino effect” in heterocyclic synthesis. Formation of N-heterocycles by ring-closure reactions of substituted 2-vinyl-N,N-dialkylanilines. J. Org. Chem. 49:269-276. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.