Abstract
The susceptibility to 14 antimicrobial agents and the mechanisms of aminopenicillin resistance were studied in 197 clinical isolates of Haemophilus influenzae—109 isolated in 2007 (study group) and 88 isolated in 1997 (control group). Community antibiotic consumption trends were also examined. H. influenzae strains were consecutively isolated from the same geographic area, mostly from respiratory specimens from children and adults. Overall, amoxicillin resistance decreased by 8.4% (from 38.6 to 30.2%). β-Lactamase production decreased by 15.6% (from 33 to 17.4%, P = 0.01), but amoxicillin resistance without β-lactamase production increased by 7.1% (from 5.7 to 12.8%). All β-lactamase-positive isolates were TEM-1, but five different promoter regions were identified, with Pdel being the most prevalent in both years, and Prpt being associated with the highest amoxicillin resistance. A new promoter consisting of a double repeat of 54 bp was detected. Community consumption of most antibiotics decreased, as did the geometric means of their MICs, but amoxicillin-clavulanic acid and azithromycin consumption increased by ca. 60%. For amoxicillin-clavulanic acid, a 14.2% increase in the population with an MIC of 2 to 4 μg/ml (P = 0.02) was observed; for azithromycin, a 21.2% increase in the population with an MIC of 2 to 8 μg/ml (P = 0.0005) was observed. In both periods, the most common gBLNAR (i.e., H. influenzae isolates with mutations in the ftsI gene as previously defined) patterns were IIc and IIb. Community consumption of trimethoprim-sulfamethoxazole decreased by 54%, while resistance decreased from 50 to 34.9% (P = 0.04). Antibiotic resistance in H. influenzae decreased in Spain from 1997 to 2007, but surveillance should be maintained since new forms of resistances may be developing.
Haemophilus influenzae is an important human pathogen that causes community-acquired respiratory tract and invasive infections. The increasing prevalence of antibiotic resistant phenotypes reported in this pathogen limits the choice of a suitable antimicrobial treatment. Antibiotic surveillance studies are necessary to determine trends in national, regional, and local susceptibility patterns and to effectively guide empirical antimicrobial therapy.
The principal resistance mechanism to aminopenicillins described in H. influenzae is β-lactamase production, mainly of the TEM-1 type and sometimes of the ROB-1 type (16). The TEM β-lactamase gene can be associated with different promoter regions: P3, overlapping Pa/Pb, Pdel, which has a 135-bp deletion, and the newly described Prpt that includes a 54-bp insertion in the promoter region (20, 24, 25). These promoters may have different affinities for RNA polymerase, altering β-lactamase expression and giving rise to different levels of resistance to β-lactam antibiotics (25).
The use of oral cephalosporins and amoxicillin-clavulanate has contributed to the spread of β-lactam resistance due to amino acid substitutions in penicillin-binding proteins (PBPs), resulting in β-lactamase-non-producing ampicillin resistance (BLNAR) (7, 11, 27). In addition, the combination of β-lactamase production and alterations in the PBP3 has given rise to a β-lactamase-producing amoxicillin-clavulanic acid-resistant (BLPACR) phenotype (13, 19, 26).
Over the years, the number of antibiotics available for treating respiratory infections has changed considerably. For instance, the use of trimethoprim-sulfamethoxazole, tetracyclines, and amoxicillin has declined in some countries, while the use of new macrolides, quinolones, and amoxicillin-clavulanic acid has increased. However, the impact of these important changes on the antibiotic susceptibility of respiratory H. influenzae has received little attention.
In Spain, high levels of antibiotic resistance have been reported, particularly in capsulated isolates of H. influenzae types b, e, and f (1, 3, 4). However, widespread vaccination in children against H. influenzae type b was established in this country in 1998 and, in recent years, the health authorities have implemented nationwide campaigns for the prudent use of antibiotics, especially in children.
Accordingly, the aims of the present study were (i) to determine the current antimicrobial susceptibility of clinical isolates of H. influenzae in Spain compared to a control group of isolates collected 10 years ago, before the vaccination against H. influenzae type b was introduced; (ii) to compare trends of antibiotic susceptibility in H. influenzae in relation to the evolution of antibiotic consumption by the community; and (iii) to study the prevalence and evolution of the most important mechanisms of resistance to amoxicillin, paying special attention to modifications in PBP3 and β-lactamase genes and their promoter regions.
MATERIALS AND METHODS
Bacterial isolates.
The antibiotic susceptibility of 197 clinical isolates of H. influenzae was studied: 109 of them were isolated in 2007 and constituted the study group, while 88, isolated in 1997, made up the control group. All were consecutively isolated from January to March from the same geographic area covered by the university Hospital Gregorio Marañón, Madrid, Spain. This hospital was chosen for the present study because it is one of the largest hospitals in Spain (1,700 beds), covering a large administrative health area (approximately 756,000 inhabitants), and it systematically submits all its H. influenzae clinical isolates to our Haemophilus Reference lab located 20 km away. All isolates came from individual patients, and no outbreaks or nosocomial infections due to H. influenzae were detected either in 2007 or in 1997. We have previously documented that in the Madrid area nontypeable H. influenzae strains, as in other studies, are largely genetically unrelated (2, 10, 13).
Isolates were identified according to standard microbiological methods (5) and were serotyped by a coagglutination test (Phadebact Haemophilus test; Boule Diagnostics AB, Huddinge, Sweden) that was confirmed by a molecular capsular typing method (8).
Antimicrobial susceptibility testing.
The antibiotic susceptibility of all H. influenzae clinical isolates was determined at the same time by the broth microdilution method according to the CLSI guidelines (6) on Haemophilus test medium. Microtiter plates (Sensititre Enzyme 13; Trek Diagnostics, Inc., Westlake, OH) were inoculated to produce a final density of approximately 5 × 105 CFU/ml, which was regularly controlled by colony counting on chocolate agar after overnight incubation. The inoculated plates were incubated at 35°C for 20 to 24 h in 5% CO2. The MIC was defined as the lowest concentration of antibiotic that inhibited growth. Amoxicillin, amoxicillin-clavulanic acid (2:1 ratio), cefaclor, cefuroxime, cefotaxime, cefixime, cefpodoxime, chloramphenicol, tetracycline, ciprofloxacin, nalidixic acid, telithromycin, clarithromycin, azithromycin, and trimethoprim-sulfamethoxazole were evaluated.
β-Lactamase production was determined by the chromogenic cephalosporin test with nitrocephin as substrate (21).
Antibiotic quality control results for H. influenzae ATCC 49247 and ATCC 49766 were within recommended values according to guidelines for MIC tests (6).
β-Lactamase-negative isolates, which were nonsusceptible to amoxicillin (MIC of ≥2 μg/ml), were categorized as intermediate (amoxicillin MIC of 2 μg/ml) and resistant (amoxicillin MIC of ≥4 μg/ml), as previously described (10).
PCR and DNA sequence determinations.
Amplification of the ftsI gene between nucleotide positions bp 936 and 1640, corresponding to amino acid positions 327 to 540, was carried out using the primers and PCR conditions described previously (7).
The blaTEM gene and its promoter region were assessed by PCR using the following olignucleotide primer pairs (20): forward (5′-AAT TCT TGA AGA CGA AAG GG-3′) and reverse (5′-GTG TTA TCA CAC ATG GTT ATG-3′) (from nucleotides 3740 to 4341 of the Ampr gene transposed on plasmid pBR322, GenBank accession no. V00613) and forward (5′-CAT AAC CAT GAG TGA TAA CAC-3′) and reverse (5′-ACG CTC AGT GGA ACG AAA AC-3′) (from nucleotides 4321 to 4949). Sequencing was performed in forward and reverse directions, and the sequences were compared to the nucleotide sequence of the blaTEM-1 gene and its promoter region in public databases (GenBank accession no. AB194682).
The PCR program for ftsI and tem genes was as follows: denaturation at 94°C for 5 min, 25 amplification cycles of denaturation at 94°C for 30 s, annealing of primers at 55°C for 30 s, and primer extension at 72°C for 30 s were carried out, followed by a final primer extension step at 72°C for 7 min.
Reactions were performed in a 25-μl final volume, with the PureTaq Ready-To-Go PCR bead method (Amersham Biosciences), containing 5 μl of DNA template, 10 mM Tris-HCl (pH 9), 50 mM KCl, 1.5 mM MgCl2, 1 μM concentrations of each primer, 200 μM concentrations of each deoxynucleoside triphosphate, and 2.5 U of Taq polymerase. PCR products were visualized by 1% agarose gel electrophoresis and ethidium bromide staining, and all PCR products were purified with a PCR purification kit (Qiagen, Inc.) and were subjected to automated DNA sequencing.
The purified products were sequenced by using an Applied Biosystems BigDye Terminator v3.1 cycle sequencing kit (Perkin-Elmer, Warrington, United Kingdom) according to the instructions of the manufacturer. The products were resolved and analyzed with an ABI Prism 377 DNA sequencer. Nucleotide sequences were analyzed with DNAstar (Madison, WI) software; the Rd (ATCC 51907) strain (9) was included in sequence alignments as a reference.
Genotype definition.
We classified our H. influenzae strains into four main genotypes: gBLNAS, strains without a detectable resistance mechanism; gBLNAR, strains with mutations in the ftsI gene belonging to previously defined gBLNAR groups (7, 27); gBLPAR, strains producing β-lactamase and without ftsI amino acid substitutions; and gBLPACR, strains that both produce β-lactamase and have amino acid substitutions in the ftsI gene (15, 18).
Statistical analyses.
Differences in the prevalence of antibiotic resistance were assessed by the Fisher exact test. The MIC values data based on double dilution of the antibiotics were converted into log2 values; the log2 values were compared by using the unpaired t test. Associations were determined by calculation of odds ratios (OR) with 95% confidence intervals (CI). A P value of <0.05 was considered statistically significant. Statistical analyses were performed by using the GraphPad Prism version 3.02 (GraphPad Prism Software, Inc.) computer program.
Antibiotic consumption.
Antibiotic consumption in Spain was determined according to the ATC/defined daily dose(s) (DDD) methodology as previously described (11, 28) and expressed as DDD per 1,000 inhabitants per day.
Nucleotide sequence accession number.
The nucleotide sequence data of the 2Prpt promoter reported here is listed in the GenBank nucleotide database under accession no. EU531509.
RESULTS
Clinical isolates.
The clinical and microbiological features of the collected isolates from 2007 and 1997 were similar ( Table 1).
TABLE 1.
Comparison between the H. influenzae study group isolated in 2007 and the control group isolated in 1997
| Parameter | No. of isolates (%) for:
|
Fisher exact test
|
||
|---|---|---|---|---|
| 2007 (n = 109) | 1997 (n = 88) | OR (95% CI) | P | |
| Age | ||||
| Adults | 35 (32.1) | 32 (36.4) | 0.83 (0.46-1.50) | 0.55 |
| Children | 61 (56) | 49 (55.7) | 1.01 (0.57-1.78) | 1.00 |
| Unknown | 13 (11.9) | 7 (7.9) | 1.57 (0.60-4.11) | 0.48 |
| Clinical source | ||||
| Respiratory | 102 (93.6) | 76 (86.4) | 2.30 (0.86-6.12) | 0.10 |
| Conjunctivitis | 43 (39.4) | 37 (42) | 0.90 (0.50-1.59) | 0.77 |
| Otitis | 16 (14.7) | 9 (10.2) | 1.51 (0.63-3.60) | 0.39 |
| Sputum | 5 (4.6) | 3 (3.4) | 1.36 (0.31-5.86) | 0.73 |
| Other respiratorya | 37 (34) | 24 (27.3) | 1.37 (0.74-2.53) | 0.35 |
| Invasive | 2 (1.8) | 5 (5.7) | 0.31 (0.06-1.60) | 0.25 |
| Other | 5 (4.6) | 7 (7.9) | 0.56 (0.17-1.82) | 0.38 |
| Microbiological data | ||||
| Biotype I | 13 (11.9) | 13 (14.8) | 0.78 (0.34-1.79) | 0.67 |
| Biotype II | 53 (48.6) | 45 (51.1) | 0.90 (0.51-1.59) | 0.77 |
| Biotype III | 32 (29.3) | 23 (26.1) | 1.17 (0.62-2.20) | 0.63 |
| Biotypes IV, V, and VI | 11 (10.1) | 7 (7.9) | 1.30 (0.48-3.50) | 0.80 |
| Capsulated strains | 1, type f (0.9) | 2, type b (2.3) | 0.40 (0.04-4.50) | 0.59 |
This value includes bronchial aspirate, bronchoalveolar lavage, and respiratory secretion samples.
Antibiotic susceptibility. (i) Overall susceptibility.
The antibiotic MIC90s for H. influenzae in 2007 and 1997 were similar, although some differences were observed in the geometric means (Table 2). Statistical comparisons of the MICs using the unpaired t test revealed significant differences between the two populations for five antibiotics: amoxicillin (P = 0.05), cefaclor (P = 0.003), chloramphenicol (P = 0.008), azithromycin (P = 0.0001), and trimethoprim-sulfamethoxazole (P = 0.03) (Table 2); no differences were observed for the remaining nine antibiotics. For the azithromycin MICs, the 25th percentiles were 1 μg/ml (for 1997) and 2 μg/ml (for 2007), while the 75th percentiles were 2 μg/ml (for 1997) and 4 μg/ml (for 2007). For amoxicillin MICs, the 75th percentiles were ≥64 μg/ml (for 1997) and 4 μg/ml (for 2007); for amoxicillin-clavulanic acid, the 75th percentiles were 1 μg/ml (for 1997) and 2 μg/ml (for 2007). Most importantly, the geometric mean of azithromycin increased 36.88% between 1997 and 2007, while it decreased for amoxicillin (21.23%), cefaclor (15.30%), chloramphenicol (14.29%), and trimethoprim-sulfamethoxazole (42.60%) (Table 2). A variation in the geometric mean of ≥14% was statistically significant as determined by the unpaired t test.
TABLE 2.
Antibiotic susceptibility distributions of 14 antimicrobials against 197 isolates of H. influenzaea
| Antibiotic | MIC (μg/ml)
|
Pb | Geometric mean
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC50
|
MIC90
|
Range
|
||||||||
| 1997 | 2007 | 1997 | 2007 | 1997 | 2007 | 1997 | 2007 | % Variation | ||
| Amoxicillin | 1 | 0.5 | ≥64 | 32 | ≤0.25-≥64 | ≤0.25-≥64 | 0.05 | 2.83 | 1.67 | -41.0 |
| Amoxicillin-clavulanic acid | 0.5 | 0.5 | 2 | 2 | 0.25-4 | 0.25-4 | 0.49 | 0.78 | 0.84 | +7.69 |
| Cefaclor | 4 | 4 | 16 | 16 | 1-32 | ≤0.5-32 | 0.003 | 5.23 | 4.43 | -15.30 |
| Cefuroxime | 1 | 1 | 4 | 4 | 0.25-8 | 0.25-8 | 0.07 | 1.16 | 1.07 | -7.76 |
| Cefotaxime | ≤0.01 | ≤0.01 | 0.06 | 0.06 | ≤0.01-0.12 | ≤0.01-0.25 | 0.94 | 0.18 | 0.18 | 0.00 |
| Cefixime | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06-0.5 | ≤0.06-0.25 | 0.66 | 0.063 | 0.062 | -1.59 |
| Cefpodoxime | ≤0.06 | ≤0.06 | 0.25 | 0.25 | ≤0.06-0.25 | ≤0.06-0.5 | 0.85 | 0.084 | 0.086 | +2.38 |
| Chloramphenicol | ≤0.5 | ≤0.5 | 1 | ≤0.5 | ≤0.5-8 | ≤0.5-8 | 0.008 | 0.63 | 0.54 | -14.29 |
| Tetracycline | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5-≥8 | ≤0.5-4 | 0.14 | 0.55 | 0.51 | -7.27 |
| Telithromycin | ≥2 | ≥2 | ≥2 | ≥2 | 0.5-≥2 | 0.5-≥2 | 0.19 | 1.72 | 1.73 | +0.58 |
| Clarithromycin | 8 | 8 | 16 | 16 | 2-32 | 2-16 | 0.65 | 9.15 | 8.91 | -2.62 |
| Azithromycin | 2 | 2 | 4 | 4 | 0.25-8 | 0.5-8 | 0.0001 | 1.60 | 2.19 | +36.88 |
| Trimethoprim-Sulfamethoxazole | 1 | ≤0.25 | 16 | 16 | ≤0.25-≥64 | ≤0.25-32 | 0.03 | 1.69 | 0.97 | -42.60 |
| Ciprofloxacin | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ||||||
Significant values are indicated in boldface.
As determined by an unpaired t test.
(ii) Amoxicillin susceptibility.
A total of 30.2% of the 2007 isolates had an amoxicillin MIC ≥ 4 μg/ml, compared to 38.6% in 1997. In addition, a higher proportion of β-lactamase positive isolates from 1997 had an amoxicillin MIC of ≥32 μg/ml (P = 0.04) (Table 3). Nevertheless, while β-lactamase production decreased by 15.5% from 1997 to 2007 (P = 0.01) (Table 3), non-β-lactamase-mediated amoxicillin resistance increased by 7.1% (from 5.7% in 1997 to 12.8% in 2007). In fact, the percentage of β-lactamase-negative isolates with an amoxicillin MIC of ≥2 μg/ml increased from 13.6% in 1997 to 28.9% in 2007 (P = 0.04) (Table 3).
TABLE 3.
Antimicrobial susceptibility comparison between the H. influenzae study group isolated in 2007 and the control group isolated in 1997
| Parameter | No. of isolates (%) for:
|
% Change | Fisher exact test
|
||
|---|---|---|---|---|---|
| 2007 (109) | 1997 (88) | OR (95% CI) | Pd | ||
| Amoxicillin resistance (MIC ≥ 4 μg/ml) | 33 (30.2) | 34 (38.6) | -8.4 | 0.69 (0.38-1.25) | 0.23 |
| β-Lactamase-positive isolates | 19 (17.4) | 29 (32.9) | -15.5 | 0.43 (0.22-0.83) | 0.01 |
| Amoxicillin resistance in β-lactamase-negative isolates | 14 (12.8) | 5 (5.7) | +7.1 | 1.99 (0.68-5.85) | 0.31 |
| Amoxicillin, MIC ≥ 2 μg/mla | 26 (28.9) | 8 (13.6) | +15.3 | 2.60 (1.08-6.20) | 0.04 |
| Amoxicillin, MIC ≥ 32 μg/mlb | 15 (78.9) | 29 (100) | -21.1 | 2.45 (1.02-5.90) | 0.04 |
| Amoxicillin-clavulanic acid, MIC = 2 to 4 μg/mlc | 25 (27.8) | 8 (13.6) | +14.2 | 0.05 (0.003-1.16) | 0.02 |
| gBLNAS | 60 (55) | 43 (48.9) | +6.1 | 1.30 (0.73-2.25) | 0.40 |
| gBLPAR | 16 (14.7) | 17 (19.3) | -4-.6 | 0.72 (0.34-1.52) | 0.44 |
| gBLNAR | 30 (27.5) | 16 (18.2) | +9.3 | 1.71 (0.86-3.40) | 0.13 |
| gBLPACR | 3 (2.8) | 12 (13.6) | -15.4 | 0.13 (0.036-0.45) | 0.0004 |
| Azithromycin, MIC = 2 to 8 μg/ml | 95 (87.2) | 58 (66) | +21.2 | 3.50 (1.72-7.20) | 0.0005 |
| Trimethoprim-sulfamethoxazole, MIC ≥ 4 μg/ml | 38 (34.9) | 44 (50) | -15.1 | 0.53 (0.30-0.95) | 0.04 |
| Trimethoprim-sulfamethoxazole and amoxicillin simultaneous resistance | 16 (14.7) | 23 (26.1) | -11.4 | 0.49 (0.24-1.00) | 0.04 |
This values includes intermediate and resistant isolates (9). Total β-lactamase-negative isolates: 90 in 2007 and 59 in 1997.
Total β-lactamase-positive isolates: 19 in 2007 and 29 in 1997.
Total β-lactamase-negative isolates: 90 in 2007 and 59 in 1997.
Significant values are indicated in boldface.
(iii) Amoxicillin-clavulanic acid susceptibility.
Resistance to amoxicillin-clavulanic acid was not detected according to current breakpoints (6). However, the number of β-lactamase-negative isolates with an amoxicillin-clavulanic acid MIC of ≥2 μg/ml was higher in 2007 (27.8%) than in 1997 (13.6%) (P = 0.02) (Table 3).
(iv) Cephalosporin susceptibility.
The proportion of cefaclor-nonsusceptible isolates (resistant and intermediate) decreased 12.2% from 17% in 1997 to 4.8% in 2007. No resistance to the other cephalosporins was found.
(v) Macrolide susceptibility.
Clarithromycin resistance (MIC of 32 μg/ml) was 0% in 2007 and 2.3% in 1997. The proportion of isolates with an azithromycin MIC of 8 μg/ml (the susceptibility breakpoint is currently defined as ≤4 μg/ml) (6) was 0.9% in 2007 and 1.1% in 1997. However, the percentage of isolates with an azithromycin MIC from 2 to 8 μg/ml significantly increased in isolates from 2007 in relation to isolates from 1997 (P = 0.0005) (Table 3).
(vi) Trimethoprim-sulfamethoxazole susceptibility.
The prevalence of trimethoprim-sulfamethoxazole resistance decreased from 50% in 1997 to 34.9% in 2007, a decrease of 15.1% (P = 0.04) (Table 3). Also, resistance to trimethoprim-sulfamethoxazole was more prevalent in amoxicillin-resistant (57.4%) strains than in amoxicillin-susceptible strains (28.7%) (P = 0.0002, OR = 3.34, 95% CI = 1.78 to 6.26). Simultaneous resistance to trimethoprim-sulfamethoxazole and amoxicillin decreased from 26.1% in 1997 to 14.7% in 2007 (P = 0.04) (Table 3).
Amoxicillin susceptibility in relation to resistance genotype.
Of the 2007 H. influenzae isolates, 60 (55.0%) had no detectable mechanism of resistance (gBLNAS genotype) compared to 43 (48.9%) in the control group from 1997 (P = 0.40). Sixteen isolates (14.7%) were gBLPAR in 2007 compared to 17 (19.3%) in 1997; thirty isolates (27.5%) were gBLNAR in 2007 compared to sixteen isolates (18.2%) in 1997. The number of gBLPACR strains decreased from 12 isolates (13.6%) in 1997 to 3 isolates (2.8%) in 2007 (P = 0.0004) (Table 3).
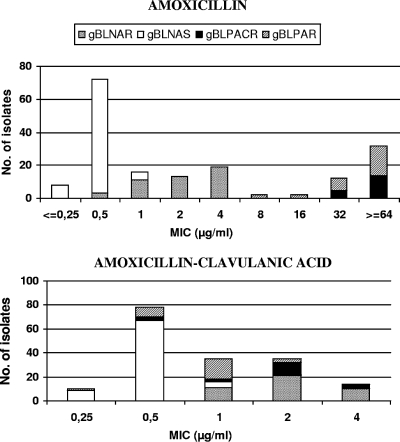
The distribution of MICs is depicted in Fig. 1 according to the mechanisms of β-lactam resistance. Those isolates with an amoxicillin MIC of 2 to 4 μg/ml were gBLNAR, while the highest levels of amoxicillin resistance (32 to ≥64 μg/ml) were attained in isolates that produce β-lactamase, gBLPAR and gBLPACR (Fig. 1). In the case of amoxicillin-clavulanic acid, isolates with MICs of 2 μg/ml were mostly gBLNAR but also included gBLPACR and gBLPAR, while only gBLNAR and gBLPACR had MICs of 4 μg/ml (Fig. 1).
FIG. 1.
Amoxicillin and amoxicillin-clavulanic acid MIC distribution for H. influenzae in relation to mechanisms of amoxicillin resistance. Abbreviations: gBLNAR, H. influenzae isolates with mutations in the ftsI gene as previously defined; gBLNAS, H. influenzae isolates without a resistance mechanism; gBLPACR, H. influenzae isolates with both mechanisms (β-lactamase production and amino acid substitutions in the ftsI gene); and gBLPAR, H. influenzae β-lactamase-producing isolates.
The effectiveness of β-lactam antibiotics against the 197 H. influenzae isolates according to their resistance genotype showed that the MIC90s for amoxicillin, amoxicillin-clavulanic acid, and cefuroxime of the gBLNAR isolates were about eightfold higher than the gBLNAS isolates, fourfold higher for cefaclor, and threefold higher for cefotaxime (data not shown); no differences were found between the two study periods.
Trends in the mutation patterns of the ftsI gene.
Amino acid substitutions in the ftsI gene of gBLNAR and gBLPACR isolates and their susceptibility to amoxicillin are listed in Table 4. Groups I and II had similar frequencies in H. influenzae isolates from 2007 and 1997: group I, 6.1% (in 2007) and 7.1% (in 1997), and group II, 93.9% (in 2007) and 92.8% (in 1997). However, some rearrangement was found within group II, as group IIa increased from 0.0% (in 1997) to 15.1% (in 2007), while groups IIb, IIc, and IId decreased by 3.8, 4.0, and 5.2%, respectively, between 1997 and 2007.
TABLE 4.
Amino acid substitutions in the ftsI gene in H. influenzae
| Group | Amino acid substitutions
|
MIC AMX range (μg/ml) | Overall geometric meanb
|
No. of isolates
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Near STVK motif
|
Thr-352 | Lys-355 | Leu-356 | Met-377 | Met-391 | Ala-437 | Ile-449 | Ile-475 | Gly-490 | Surrounding KTG motif
|
||||||||||||||
| Lys-344 | Ile-348 | Asp-350 | Ala-502 | Arg-517 | Ile-519 | Asn-526 | Ala-530 | Thr-532 | AMX | AMC | 1997 | 2007 | Total | |||||||||||
| I | His | Ser | 1-≥64 | 1.59 | 2.00 | 2 | 2 | 4 | ||||||||||||||||
| IIa | Lys | 0.5-4 | 2.00 | 1.74 | 3 | 3 | ||||||||||||||||||
| Lys | Ser | 1-4 | 2 | 2 | ||||||||||||||||||||
| IIb | Val | Lys | 2-4 | 2.00 | 2.00 | 2 | 2 | |||||||||||||||||
| Glu | Val | Lys | 1 | 1 | 1 | |||||||||||||||||||
| Asn | Ile | Val | Lys | 1-≥64 | 4 | 6 | 10 | |||||||||||||||||
| Asn | Ile | Glu | Val | Lys | 2-16 | 1 | 2 | 3 | ||||||||||||||||
| Asn | Ser | Val | Leu | Lys | 1 | 2 | 2 | |||||||||||||||||
| IIc | Thr | Lys | 0.5-32 | 2.18 | 1.80 | 7 | 6 | 13 | ||||||||||||||||
| Asn | Thr | Lys | 1-≥64 | 6 | 8 | 14 | ||||||||||||||||||
| IId | Val | Lys | 0.5-≥64 | 2.00 | 2.00 | 4 | 3 | 7 | ||||||||||||||||
| Ma | Ile | ≤0.25 | 0.52 | 0.53 | 1 | 1 | ||||||||||||||||||
| Leu | 0.5 | 2 | 2 | |||||||||||||||||||||
| Thr | ≥64 | 1 | 1 | |||||||||||||||||||||
| Asn | ≤0.25-≥64 | 8 | 3 | 11 | ||||||||||||||||||||
| Asn | Ser | 0.5 | 2 | 2 | 4 | |||||||||||||||||||
| Val | 0.5 | 1 | 1 | |||||||||||||||||||||
| Val | 0.5 | 1 | 1 | |||||||||||||||||||||
| Ser | 0.5 | 2 | 2 | |||||||||||||||||||||
| Arg | Asn | Gly | Thr | Val | Ile | 0.5 | 1 | 1 | ||||||||||||||||
| Arg | Asn | Gly | Thr | Val | Ile | Ser | ≤0.25 | 1 | 1 | |||||||||||||||
M, miscellaneous.
The amoxicillin (AMX) value was calculated for the β-lactamase-negative isolates only (n = 149); the amoxicillin-clavulanic acid (AMC) value was calculated for the entire collection (n = 197).
Within group II, the most common mutations in 1997 and 2007 were groups IIb, 32.1% (1997) and 27.3% (2007), and IIc, 46.4% (1997) and 42.4% (2007). All of the ftsI mutation groups had geometric means of about 2 μg/ml for amoxicillin (β-lactamase-negative isolates) and amoxicillin-clavulanic acid (Table 4).
The miscellaneous group (group M in Table 4) had overall prevalences of 18.2% in 1997 and 8.2% in 2007. Group M presented geometric means of 0.5 μg/ml for amoxicillin and amoxicillin-clavulanic acid (Table 4). Geometric means of 3.4, 0.84, 0.01, 0.06, and 0.06 μg/ml were found for cefaclor, cefuroxime, cefotaxime, cefixime, and cefpodoxime, respectively, similar to those obtained in isolates without any mechanism of resistance, suggesting that these mutations may not be associated with decreased susceptibility to these antibiotics (Table 4). Within group M, the most important mutation pattern (eight isolates in 1997 and three isolates in 2007) was characterized by the Asp350-to-Asn amino acid substitution (Table 4).
Characterization of the promoter regions of β-lactamases.
Overall, the proportion of the different promoter regions found in the β-lactamase-positive isolates was as follows: 39.6% (42.1% in 2007 and 37.9% in 1997) of the isolates had the Pdel promoter, 33.3% (31.6% in 2007 and 34.5% in 1997) had the Pa/Pb promoter, 22.9% (15.8% in 2007 and 27.6% in 1997) had the Prpt promoter, and 2.1% (5.3% in 2007 and 0% in 1997) had the P3 promoter.
A new variant of the Prpt promoter (here designated as 2Prpt) was detected in a nontypeable clinical isolate taken in 2007 from a child with otitis media. 2Prpt consisted of a double repeat of bp 145 to 198 inserted after bp 198 (Fig. 2). The double insertion provides additional −10 and −35 sequences equal to the Prpt promoter, in such a way that blaTEM-2Prpt seems to have three promoters. The new promoter and the promoter corresponding to Prpt are likely stronger than the coexisting P3 promoter as the −35 regions are a closer match to the consensus sequence (Fig. 2).
FIG. 2.
Promoter region (bp 1 to 208) of blaTEM (23). Underlining represents region in P3 which is replicated once in Prpt and twice in 2Prpt. *, Point of insertion in P3 to create 2Prpt (text in boldface represents inserted sequences). Boxed regions represent −10 and −35 sequence of promoter P3, circled regions represent −10 and −35 sequence of promoter Prpt, and dotted circles represent new −10 and −35 regions in 2Prpt.
The highest geometric mean for amoxicillin was observed for isolates harboring the Prpt promoter (56.42 μg/ml), followed by Pdel promoter (44.44 μg/ml) and Pa/Pb promoter (41.50 μg/ml) (data not shown); for amoxicillin-clavulanic acid, the geometric means were 1.13, 1.29, and 0.96 μg/ml for the Prpt, Pdel, and P3 promoters, respectively (data not shown).
Antibiotic consumption.
A decrease in antibiotic consumption at the community level was observed for amoxicillin, cefaclor, cefuroxime, cefixime, cotrimoxazole, and clarithromycin (Table 5); conversely, amoxicillin plus β-lactamase inhibitor and azithromycin experienced an important increase of ca. 60% (Table 5).
TABLE 5.
Community antimicrobial consumption in Spain
| WHO ATC code | Antibiotic | Consumption (DDD/1,000 inhabitants/day) in:
|
Variation (%) | |
|---|---|---|---|---|
| 1997 | 2006 | |||
| J01C | β-Lactam antibacterials, penicillins | |||
| J01CA04 | Amoxicillin | 6.5 | 4.1 | -36.9 |
| J01CR02 | Amoxicillin plus β-lactamase inhibitor | 4.4 | 7.2 | +63.6 |
| J01D | Cephalosporins | |||
| J01DC04 | Cefaclor | 0.4 | 0.1 | -75.0 |
| J01DC02 | Cefuroxime | 1.2 | 1.0 | -16.7 |
| J01DD08 | Cefixime | 0.5 | 0.3 | -40.0 |
| J01EE01 | Trimethoprim-sulfamethoxazole | 0.7 | 0.3 | -54.1 |
| J01FA | Macrolides | |||
| J01FA09 | Clarithromycin | 1.2 | 0.9 | -25.0 |
| J01FA10 | Azithromycin | 0.5 | 0.8 | +60.0 |
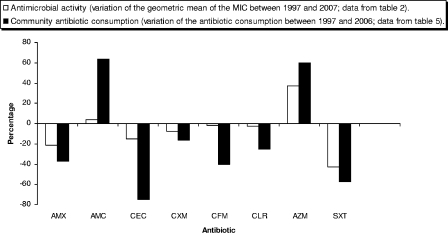
Simultaneous variation on antibiotic consumption and antibiotic susceptibility is presented in Fig. 3. For a number of antibiotics (amoxicillin, cefaclor, cefuroxime, cefixime, clarithromycin, and cotrimoxazole), both antibiotic consumption and antimicrobial resistance decreased over the study period. In contrast, amoxicillin-clavulanic acid and azithromycin experienced parallel increases in antibiotic consumption and antibiotic geometric means (Fig. 3).
FIG. 3.
Comparison of percentages of variation between community antibiotic consumption and antibiotic activity in H. influenzae (Spain, 1997 to 2007). Drug abbreviations: AMX, amoxicillin; AMC, amoxicillin-clavulanic acid; CEC, cefaclor; CXM, cefuroxime; CFM, cefixime; CLR, clarithromycin; AZM, azithromycin; and SXT, trimethoprim-sulfamethoxazole.
DISCUSSION
Several observations of clinical and epidemiological interest can be made from the data presented here. First, the proportion of H. influenzae isolates producing β-lactamase decreased by 15.5% between two sets of isolates collected 10 years apart. Second, because the proportion of resistant isolates without β-lactamase production increased by 7.1%, overall amoxicillin resistance decreased by only 8.4% (from 38.6 to 30.2%). Finally, while all β-lactamase-positive isolates were of the classical TEM 1 type, up to five different promoter regions were detected. The Pdel promoter was the most prevalent in both years, followed by the Prpt promoter, but the prevalence of Prpt decreased by 11.8% from 1997 to 2007, while the proportion of Pdel increased by 4.2%; these changes may be important because the Prpt promoter showed the greatest resistance to amoxicillin (data not shown).
One possible limitation of the present study was that susceptibility data from one hospital were compared to national average consumption data. The data from Table 2 suggest that over long periods of time altering the antibiotic consumption by the community has important consequences on the antibiotic susceptibility of H. influenzae. A decrease in antibiotic resistance is only occasionally observed in the scientific literature, but it is well documented here for amoxicillin (a 36.9% decrease in consumption, a 8.4% decrease in resistance, and a 15.5% decrease in β-lactamase production) and trimethoprim-sulfamethoxazole (a 54.1% decrease in consumption and a 15.1% decrease in resistance). In addition, simultaneous resistance to amoxicillin and trimethoprim-sulfamethoxazole decreased by 11.4%.
Furthermore, in addition to the decreasing prevalence of β-lactamase, the H. influenzae population with an amoxicillin MIC of ≥32 μg/ml decreased. These results are in accordance with data from a recent surveillance study in the United States and with European studies, which confirmed a decrease in the incidence of β-lactamase production in this pathogen (14, 17).
Two antibiotics, amoxicillin-clavulanic acid and azithromycin, demonstrated an increase in consumption over the study period of ca. 60%. The increases were recent, and we noticed little impact on true antibiotic resistance as measured by current breakpoints. However, we have documented some changes in the H. influenzae population that may indicate ongoing adaptations to this increase in antibiotic pressure. For amoxicillin-clavulanic acid, we observed a 14.2% increase in the H. influenzae population presenting an MIC of 2 to 4 μg/ml (P = 0.02), and for azithromycin we found a 21.2% increase in the population presenting an MIC of 2 to 8 μg/ml (P = 0.0005).
In our study, the percentage of isolates with an amoxicillin MIC of ≥2 μg/ml (intermediate and resistant) (10) without β-lactamase production increased from 13.6% in 1997 to 28.9% in 2007, while the gBLNAR group increased from 18.2% in 1997 to 27.5% in 2007. However, in Europe and the United States the number of ampicillin-resistant non-β-lactamase-producing strains remained relatively constant and low in recent years (14, 17). This difference may be the consequence of the large amoxicillin-clavulanic acid consumption detected in Spain that is among the highest in Europe (12). In accordance with data previously reported by us (11), the vast majority of the gBLNAR isolates in the present study had amino acid substitutions at the Lys-Thr-Gly motif, the most common being Asn-526 to Lys. However, in the present study no amino acid substitutions were found at positions 385 and 389, previously found to be related to a decreased susceptibility to cefotaxime and cefixime (11, 22).
As in other reports (20, 24), in our β-lactamase-positive isolates the most prevalent promoter was Pdel. The geometric mean for amoxicillin was higher for the Pdel promoter than for the Pa/Pb promoter, as reported by Tristram et al. (24). In our study, the Prpt promoter presented the highest geometric means for amoxicillin and cefaclor, suggesting that it may be associated with increased levels of resistance to these two antibiotics, but a number of additional factors such as plasmid location and plasmid copy numbers may also determine the level of susceptibility.
In summary, we report here data that are of clinical and epidemiological interest that may be important to other countries as well. Overall, H. influenzae antibiotic resistance decreased in Spain from 1997 to 2007. In this period, nationwide campaigns for the prudent use of antibiotics have been implemented by the health authorities and conjugate vaccines against H. influenzae type b (a microorganism that was highly antibiotic resistant in Spain and constituted a reservoir for antibiotic resistance genes) were introduced. However, antibiotic surveillance studies on this pathogen should be maintained since we noticed that amoxicillin-clavulanic acid consumption, as well as amoxicillin resistance, in β-lactamase negative isolates experienced a relevant increase. Also, the azithromycin MICs in the study population increased in parallel with the important increase observed in the community consumption of azithromycin.
Acknowledgments
This study was supported by research grants from the Instituto de Salud Carlos III, Fondo de Investigaciones Sanitarias (reference 04/0899), the REIPI Network (reference RD 06/0008/0023), and the Network of Excellence GRACE (PL 518226). S.G.-C. is a recipient of a fellowship from the Instituto de Salud Carlos III (reference 05/0033).
Footnotes
Published ahead of print on 27 May 2008.
REFERENCES
- 1.Campos, J., S. García-Tornel, and I. Sanfeliu. 1984. Susceptibility studies of multiply resistant Haemophilus influenzae isolated from pediatric patients and contacts. Antimicrob. Agents Chemother. 25:706-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos, J., M. Hernando, F. Román, M. Pérez-Vázquez, B. Aracil, J. Oteo, F. de Abajo, et al. 2004. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J. Clin. Microbiol. 42:524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos, J., F. Román, M. Pérez-Vázquez, B. Aracil, J. Oteo, E. Cercenado, et al. 2003. Antibiotic resistance and clinical significance of Haemophilus influenzae type f. J. Antimicrob. Chemother. 52:961-966. [DOI] [PubMed] [Google Scholar]
- 4.Campos, J., F. Román, M. Pérez-Vázquez, J. Oteo, B. Aracil, E. Cercenado, et al. 2003. Infections due to Haemophilus influenzae serotype E: microbiological, clinical, and epidemiological features. Clin. Infect. Dis. 37:841-845. [DOI] [PubMed] [Google Scholar]
- 5.Campos, J., and J. A. Sáez-Nieto. 2001. gram-negative infections: Haemophilus and other clinically relevant gram-negative coccobacilli, p. 557 to 580. In N. Cimolai (ed.), Laboratory diagnosis of bacterial infections. Marcel Dekker, Inc., New York, NY.
- 6.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th ed. Approved standard M100-S17. CLSI, Wayne, PA.
- 7.Dabernat, H., C. Delmas, M. Seguy, R. Pelisser, G. Faucon, S. Bennamani, and C. Pasquier. 2002. Diversity of β-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob. Agents Chemother. 46:2208-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 10.Fluit, A. C., A. Florijn, J. Verhoef, and D. Milatovic. 2005. Susceptibility of European β-lactamase-positive and -negative Haemophilus influenzae isolates from the periods 1997/1998 and 2002/2003. J. Antimicrob. Chemother. 56:133-138. [DOI] [PubMed] [Google Scholar]
- 11.García-Cobos, S., J. Campos, E. Lázaro, F. Román, E. Cercenado, C. García-Rey, M. Pérez-Vázquez, J. Oteo, and F. de Abajo. 2007. Ampicillin-resistant non-β-lactamase-producing Haemophilus influenzae in Spain: recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob. Agents Chemother. 51:2564-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goossens, H., M. Ferech, R. Vander Stichele, M. Elseviers, et al. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579-587. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa, K., K. Yamamoto, N. Chiba, R. Kobayashi, K. Nagai, M. R. Jacobs, P. C. Appelbaum, K. Sunakawa, and K. Ubukata. 2003. Diversity of ampicillin-resistance genes in Haemophilus influenzae in Japan and the United States. Microb. Drug Resist. 9:39-46. [DOI] [PubMed] [Google Scholar]
- 14.Heilmann, K. P., C. L. Rice, A. L. Miller, N. J. Miller, S. E. Beekmann, M. A. Pfaller, S. S. Richter, and G. V. Doern. 2005. Decreasing prevalence of beta-lactamase production among respiratory tract isolates of Haemophilus influenzae in the United States. Antimicrob. Agents Chemother. 49:2561-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hotomi, M., K. Fujihara, D. S. Billal, K. Suzuki, T. Nishimura, S. Baba, and N. Yamanaka. 2007. Genetic characteristics and clonal dissemination of β-lactamase non-producing ampicillin-resistant (BLNAR) Haemophilus influenzae isolated from the upper respiratory tract in Japan. Antimicrob. Agents Chemother. 51:3969-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, R. N. Grüneberg, et al. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolates from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 17.Jansen, W. T., A. Verel, M. Beitsma, J. Verhoef, and D. Milatovic. 2006. Longitudinal European surveillance study of antibiotic resistance of Haemophilus influenzae. J. Antimicrob. Chemother. 58:873-877. [DOI] [PubMed] [Google Scholar]
- 18.Kim, I. S., C. S. Ki, S. Kim, W. S. Oh, K. R. Peck, J. H. Song, K. Lee, and N. Y. Lee. 2007. Diversity of ampicillin-resistance genes and antimicrobial susceptibility patterns in Haemophilus influenzae isolated in Korea. Antimicrob. Agents Chemother. 51:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matic, V., B. Bozdogan, M. R. Jacobs, K. Ubukata, and P. C. Appelbaum. 2003. Contribution of β-lactamase and PBP amino acid substitutions to amoxicillin/clavulanate resistance in β-lactamase-positive, amoxicillin/clavulanate-resistant Haemophilus influenzae. J. Antimicrob. Chemother. 52:1018-1021. [DOI] [PubMed] [Google Scholar]
- 20.Molina, J. M., J. Cordoba, A. Monsoliu, N. Diosdado, and M. Gobernado. 2003. Haemophilus influenzae and β-lactam resistance: description of bla TEM gene deletion. Rev. Esp. Quimioter. 16:195-203. Spanish. [PubMed] [Google Scholar]
- 21.O′Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shinger. 1972. Novel method for detection of β-lactamase by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanbongi, Y., T. Suzuki, Y. Osaki, N. Senju, T. Ida, and K. Ubukata. 2006. Molecular evolution of beta-lactam-resistant Haemophilus influenzae: 9-year surveillance of penicillin-binding protein 3 mutations in isolates from Japan. Antimicrob. Agents Chemother. 50:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tristram, S. G., R. Hawes, and J. Souprounov. 2005. Variation in selected regions of blaTEM genes and promoters in Haemophilus influenzae. J. Antimicrob. Chemother. 56:481-484. [DOI] [PubMed] [Google Scholar]
- 25.Tristram, S. G., and S. Nichols. 2006. A multiplex PCR for β-lactamase genes of Haemophilus influenzae and description of a new blaTEM promoter variant. J. Antimicrob. Chemother. 58:183-185. [DOI] [PubMed] [Google Scholar]
- 26.Ubukata, K. 2003. Problems associated with high prevalence of multidrug-resistant bacteria in patients with community-acquired infections. J. Infect. Chemother. 9:285-291. [DOI] [PubMed] [Google Scholar]
- 27.Ubukata, K., Y. Shibasaki, K. Yamamoto, N. Chiba, K. Hasegawa, Y. Takeuchi, K. Sunakawa, M. Inoue, and M. Konno. 2001. Association of amino acid substitutions in penicillin-binding protein 3 with β-lactam resistance in β-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 45:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. 2005. WHO Collaborating Centre for Drug Statistics Methodology: guidelines for ATC classification and DDD assignment. World Health Organization, Oslo, Norway.