Abstract
Rifaximin, a poorly absorbed rifamycin derivative, is a promising alternative for the treatment of Clostridium difficile infections. Resistance to this agent has been reported, but no commercial test for rifaximin resistance exists and the molecular basis of this resistance has not been previously studied in C. difficile. To evaluate whether the rifampin Etest would be a suitable substitute for rifaximin susceptibility testing in the clinical setting, we analyzed the in vitro rifaximin susceptibilities of 80 clinical isolates from our collection by agar dilution and compared these results to rifampin susceptibility results obtained by agar dilution and Etest. We found rifaximin susceptibility data to agree with rifampin susceptibility; the MICs of both antimicrobials for all isolates were either very low or very high. Fourteen rifaximin-resistant (MIC, ≥32 μg/ml) unique isolates from patients at diverse locations in three countries were identified. Molecular typing analysis showed that nine (64%) of these isolates belonged to the epidemic BI/NAP1/027 group that is responsible for multiple outbreaks and increased disease severity in the United Kingdom, Europe, and North America. The molecular basis of rifaximin and rifampin resistance in these isolates was investigated by sequence analysis of rpoB, which encodes the β subunit of RNA polymerase, the target of rifamycins. Resistance-associated rpoB sequence differences that resulted in specific amino acid substitutions in an otherwise conserved region of RpoB were found in all resistant isolates. Seven different RpoB amino acid substitutions were identified in the resistant isolates, which were divided into five distinct groups by restriction endonuclease analysis typing. These results suggest that the amino acid substitutions associated with rifamycin resistance were independently derived rather than disseminated from specific rifamycin-resistant clones. We propose that rifaximin resistance in C. difficile results from mutations in RpoB and that rifampin resistance predicts rifaximin resistance for this organism.
In recent years, the incidence and severity of Clostridium difficile infection (CDI) has increased significantly. Numerous C. difficile outbreaks in North America, the United Kingdom, and Europe have been caused by the BI/NAP1/027 epidemic group. This clonal group of isolates, which has been characterized by restriction endonuclease analysis (REA), pulsed-field gel electrophoresis, and PCR ribotyping, is described as hypervirulent due to the associated increase in disease severity compared to nonepidemic isolates (12, 13, 21, 24). Treatment failures and CDI recurrences appear to be more frequent as well, which highlights the need for the development of new therapeutic strategies (20). Rifaximin, a poorly absorbed rifamycin derivative with broad efficacy across the gram-positive and gram-negative spectra, has been approved in the United States for the treatment of traveler's diarrhea and is being evaluated for the treatment of CDI (8, 11). Several studies have investigated the efficacy of rifaximin against C. difficile in vitro and in vivo, and although rifaximin appears to have good activity against most C. difficile isolates, some strains for which the MICs are high have been identified (7, 11, 15).
Antimicrobials belonging to the rifamycin group, including rifampin and rifaximin, inhibit protein synthesis in bacteria by binding to RpoB, the β subunit of RNA polymerase (6). Studies of a variety of bacterial genera have shown that exposure to rifamycins in vitro and in vivo can lead to the selection of resistant organisms, which carry specific single amino acid mutations within RpoB (5, 10, 17, 23). These mutations have been systematically mapped in Staphylococcus aureus and Mycobacterium tuberculosis RpoB and occur within a defined region of the protein (17, 19). X-ray crystallography analysis of Thermus aquaticus RpoB complexed with rifampin provided an explanation for the locations of the specific mutations found in rifamycin-resistant organisms (2). In three-dimensional space, the RpoB amino acids that confer rifamycin resistance either directly interact with rifampin or are in close proximity to those that are involved in rifampin interactions (2). However, it is not known whether C. difficile strains for which the rifaximin MICs are high carry RpoB amino acid substitutions characteristic of other rifamycin-resistant bacteria.
We hypothesized that C. difficile isolates for which the MICs of rifaximin are high would be differentiated from isolates for which the MICs are low by specific RpoB amino acid substitutions. In addition, we hypothesized that the rifampin susceptibility and rifaximin susceptibility results for these isolates would be in agreement. Therefore, commercially available rifampin susceptibility test kits could be used to predict the rifaximin susceptibility of C. difficile isolates in the clinical setting.
MATERIALS AND METHODS
C. difficile isolates.
A total of 80 unique patient isolates were selected from our C. difficile collection as follows. Sixteen isolates (4 rifaximin resistant [MICs, ≥32 μg/ml] and 12 nonresistant [MICs, <32 μg/ml]) were selected from previous analyses in which rifaximin susceptibility testing was performed (7, 11). Two rifaximin-resistant isolates were obtained from the clinical practice of one us (S. Johnson, unpublished data).
The remaining 62 isolates were obtained from patients enrolled in a multicenter clinical treatment trial, which compared the efficacy of the toxin-binding polymer tolevamer to vancomycin and metronidazole for the treatment of CDI (14). The study was conducted in the United States and Canada with patients enrolled from 2005 to 2007 (14). This sample of 62 isolates had not previously been tested for susceptibility. If two or more isolates were obtained from the same patient, had the same susceptibility profile, and could not be differentiated at the molecular level, only one representative was included in our study. All of the C. difficile isolates in this study were characterized by HindIII REA as previously described (3).
Susceptibility testing.
Agar dilution susceptibility testing was performed as previously described, by using the Clinical and Laboratory and Standards Institute-recommended reference agar dilution method for anaerobes (4, 7). Rifaximin (Salix Pharmaceuticals, Inc., Morrisville, NC) was dissolved in methanol and then diluted in 0.1 M phosphate buffer (pH 7.4) plus 0.45% sodium dodecyl sulfate. Rifampin (Sigma-Aldrich, St. Louis, MO) was dissolved in methanol and diluted in water. The range of rifaximin concentrations tested was 0.0009 to 256 μg/ml. The range of rifampin concentrations tested was 0.00001 to 256 μg/ml. The interpretation of endpoints was conducted according to Clinical and Laboratory and Standards Institute guideline M11-A7, and C. difficile ATCC 70005 was used as a control (4). Rifampin Etests were performed according to the manufacturer's instructions (AB Biodisk, Solna, Sweden), with brucella blood agar (Anaerobe Systems, San Jose, CA). The range of rifampin MICs detected by Etest was 0.002 to 32 μg/ml. Although no susceptibility or resistance breakpoints have been determined for rifaximin or rifampin against anaerobes, we designated isolates for which the MICs were ≥32 μg/ml resistant.
Identification of sequence differences in the rpoB gene.
The rpoB gene of C. difficile QCD-32g58 (accession no. NZ_AAML04000015) was identified by a BLASTn search of the database for that sequence (http://www.ncbi.nlm.nih.gov/genomes/geblast.cgi?gi=5410) by using the rpoB gene from C. difficile strain 630 as the query sequence (accession no. NC_009089.1) (1). C. difficile QCD-32g58 rpoB was then used as the template for designing oligonucleotide primers. PCR amplification and sequencing of rpoB were carried out on 19 C. difficile isolates by using primers listed in Table 1 and the FailSafe PCR system with reaction buffer E, according to the manufacturer's recommendations (Epicenter Biotechnologies, Madison, WI). Amplicons were either directly purified from the PCR or gel purified, as required (Qiagen Inc., Valencia CA). The entire rpoB gene was sequenced from eight strains (four resistant and four nonresistant isolates). The genomic DNA templates for these experiments were prepared as previously described (18).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) | Use |
|---|---|---|
| DGP22 | AATATTGATGATATAGATCACCTAGGAAACA | Forward primer, amplification/sequencing of rpoB |
| DGP23 | AAATTGTGCTTTACCACCTAGTGGCT | Reverse primer, amplification/sequencing of rpoB |
| DGP26 | GATGCGTTAGAATGAACTATTG | Forward primer, amplification of rpoB |
| DGP27 | AAGTGCATCATCTTTATTAAATGC | Forward primer, amplification/sequencing of rpoB |
| DGP28 | GAAAGAGCTGGATTCGAAGTG | Forward primer, amplification/sequencing of rpoB |
| DGP29 | GAAGGTTATAACTACGAGGATGCT | Forward primer, amplification/sequencing of rpoB |
| DGP30 | GAATAACTGTTGGATATATGTATTACTTG | Forward primer, amplification/sequencing of rpoB |
| DGP31 | TCAAACAAGGAGTTCTCTCC | Reverse primer, amplification/sequencing of rpoB |
| DGP32 | ACTTCTAGAACCTGACCGATGT | Reverse primer, amplification/sequencing of rpoB |
| DGP33 | TATATCTATCTCTGTTTCCATCTTCTC | Reverse primer, amplification/sequencing of rpoB |
| DGP34 | GCTGAAACTGGTCTTATGTTAACT | Reverse primer, amplification/sequencing of rpoB |
| DGP35 | CCTAAAAGGTCAATTATCTCTGCATC | Reverse primer, amplification/sequencing of rpoB |
| DGP36 | TGGAAAATTGTACATAATACATAATATATGC | Forward primer for amplification/sequencing of rpoB |
| DGP37 | ATGGAGCAGAAAGAGTTATAGTAAGTCA | Forward primer, amplification/sequencing of rpoB conserved region |
| DGP38 | TCATCAGTAACTGTTGATGTTTCTTTATCA | Reverse primer, amplification/sequencing of rpoB conserved region |
PCR amplification and sequencing of the genomic region corresponding to amino acids 136 to 550 of RpoB were conducted for 10 rifaximin-resistant isolates and 1 nonresistant isolate with the primers listed in Table 1. This amino acid sequence corresponds to the defined region of RpoB in S. aureus and M. tuberculosis where mutations leading to rifamycin resistance commonly occur (17). DNAs were extracted from overnight agar cultures of these 11 isolates by resuspending C. difficile cells in 50 μl sterile deionized water until the suspension was visibly turbid. The suspension was boiled for 10 min, incubated on ice for 5 min, and then centrifuged at 22,000 × g at 4°C for 10 min, and the supernatant was used in a PCR. Ten microliters of the crude DNA preparation was used in each reaction mixture, with a final volume of 50 μl. DNA sequencing was carried out at the DNA Services Facility, Research Resources Center, University of Illinois at Chicago.
RESULTS
Rifaximin and rifampin MICs for C. difficile isolates are comparable.
Rifampin and rifaximin MICs determined by agar dilution and rifampin MICs obtained by Etest were consistent. The MICs of both rifamycin derivatives were either high (>32 μg/ml) or low MICs (≤0.002 μg/ml) by both susceptibility testing methods. None of the MICs fell within the range of 0.002 to 32 μg/ml. The rifampin and rifaximin MICs were >32 μg/ml for 14 of the 80 unique patient isolates.
Most rifaximin-resistant isolates were from epidemic REA group BI.
Nine of the 14 isolates that were resistant to rifampin and rifaximin (MICs, >32 μg/ml) were typed as epidemic REA group BI. This REA group is responsible for the large North American and European outbreaks reported in previous studies (13, 16, 24). These 9 resistant BI isolates accounted for all of the resistant isolates identified within the group of 62 from the tolevamer study (14). The overall proportion of BI group isolates in this 62-isolate sample was 51.6%. The remaining five resistant isolates were typed as REA group R (n = 2), K (n = 1), N (n = 1), and CF (n = 1) isolates (Table 2).
TABLE 2.
Rifaximin and rifampin MICs for C. difficile clinical isolatesa
| Antimicrobial agent (method) | MIC (μg/ml)
|
||
|---|---|---|---|
| Range | 50% of strains | 90% of strains | |
| Rifaximin (AD)b | 0.0019->256 | 0.0078 | >256 |
| Rifampin (AD) | 0.00006->256 | 0.0009 | >256 |
| Rifampin (Etest) | <0.002->32 | <0.002 | >32 |
See Table S1 in the supplemental material.
AD, agar dilution.
Amino acid sequence substitutions in C. difficile RpoB are associated with rifamycin resistance.
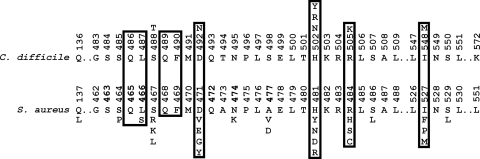
RpoB of C. difficile QCD-32g58 (accession no. NZ_AAML04000015) was identified and aligned with RpoB of S. aureus (accession no. CAG39568), M. tuberculosis (accession no. ABR05024), and T. aquaticus (accession no. CAB65465) (data not shown). Figure 1 depicts the conserved region of C. difficile RpoB aligned with that of S. aureus, which was the most similar protein of the three comparison sequences (66% amino acid identity with C. difficile RpoB). Twenty-five of the 26 S. aureus RpoB single amino acid substitutions that are known to be associated with rifamycin resistance occur between residues 137 and 529, which are equivalent to amino acids 136 to 550 of C. difficile RpoB (Fig. 1) (17). This region was analyzed in all 14 rifamycin-resistant and 5 nonresistant C. difficile isolates.
FIG. 1.
Locations of amino acid substitutions within the conserved region of RpoB in rifamycin-resistant C. difficile relative to those which are associated with resistance in S. aureus. Amino acid numbering for S. aureus is based on strain MRSA252 (accession no. CAG39568), and for C. difficile, amino acid numbering is based on strain QCD-32g58 (accession no. NZ_AAML04000015). Amino acid substitutions associated with rifamycin resistance are shown above (C. difficile) or below (S. aureus) the amino acid sequence (17). Boxed residues are equivalent to T. aquaticus RpoB amino acids that directly interact with rifampin (2).
The predicted amino acid sequence of the analyzed region of RpoB (amino acids 136 to 550) from 19 C. difficile isolates was conserved and could clearly be divided into two categories according to phenotype, i.e., rifamycin resistant and nonresistant. All 14 resistant isolates had sequence differences within this region compared to the 5 nonresistant isolates; a total of seven amino acid substitutions were identified at five locations (Table 3; Fig. 1). Four of these amino acids were equivalent to amino acids of T. aquaticus RpoB that interact with rifampin (Fig. 1) (2). The locations of all of the amino acid substitutions corresponded to locations where substitutions were associated with rifamycin resistance in S. aureus RpoB (Fig. 1) (17). All of the resistant C. difficile isolates were obtained from separate patients and originated from at least 10 different hospitals in Canada (Quebec and Nova Scotia), the United States (Illinois, Delaware, California, Vermont, and Maryland), and Argentina (Table 3).
TABLE 3.
RpoB sequence substitutions, REA groups, and geographic origins of 14 rifaximin-resistant isolates
| RpoB sequence substitution(s) | REA group (no. of isolates) | Geographic origin(s) (no. of isolates) |
|---|---|---|
| S488T, R505K | BI (1) | Quebec |
| D492N, R505K | K (1) | Illinois location aa |
| H502N, R505K | BI (4) | Delaware (1), California locations ca (2) and da (1) |
| H502N, R505K | R (2) | Argentina (2) |
| H502Y | CF (1) | Illinois location ba |
| H502R | N (1) | Illinois location ba |
| R505K, I548M | BI (1) | Vermont |
| R505Kb | BI (3) | Maryland (1), Nova Scotia (2) |
Rifamycin-resistant isolates were obtained from two geographically separate hospitals or clinics in Illinois (locations a and b) and two in California (locations c and d).
Agar dilution reproducibly yielded rifampin MICs of 128 μg/ml and rifaximin MICs of >256 μg/ml for the three resistant isolates with just the R505K amino acid substitution. The MICs of both rifampin and rifaximin for the other 11 resistant isolates were >256 μg/ml by agar dilution.
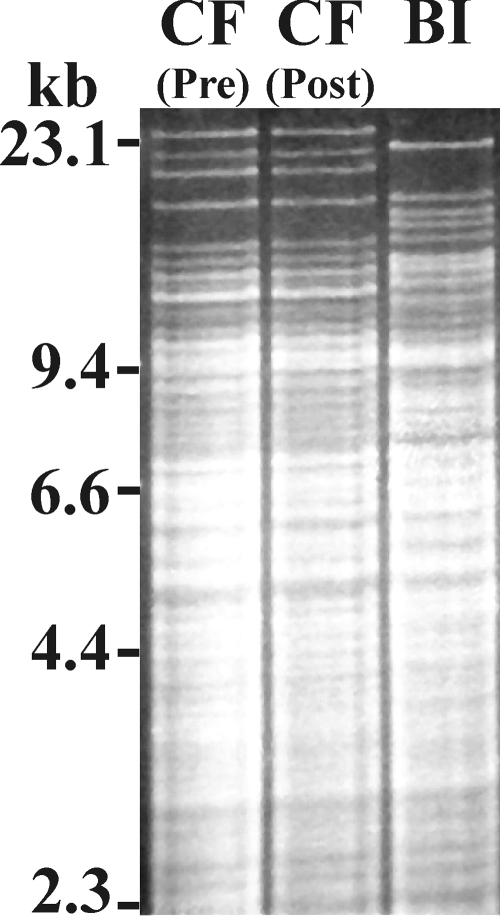
One set of paired patient specimens came from a patient before and after the completion of two courses of rifaximin, which was used in an attempt to interrupt multiple recurrences of C. difficile disease (11). These two isolates had identical REA patterns (group CF strains, Fig. 2), but the rifaximin MICs for the pre- and posttreatment isolates were 0.0039 and >256 μg/ml, respectively. Remarkably, comparison of the entire rpoB sequences of these two isolates revealed that they differed by only a single nucleotide, which resulted in an amino acid substitution within the conserved region of RpoB of the resistant isolate (H502Y) (Table 3).
FIG. 2.
REA patterns of HindIII-digested genomic DNAs purified from two group CF C. difficile isolates obtained from a patient prior to rifaximin therapy [CF (Pre)] and after treatment failure [CF (Post)] (11). The REA pattern of a group BI strain is shown for comparison.
DISCUSSION
In this study, 80 C. difficile unique patient isolates were analyzed for susceptibility to rifaximin and rifampin. Comparison of the agar dilution rifaximin and rifampin MICs to rifampin Etest results showed that the rifampin Etest can reliably predict rifaximin resistance in C. difficile (Table 2). Rifamycin susceptibility testing of these isolates yielded a bimodal distribution pattern. The MICs for the isolates were either very high or very low, which agrees with previous findings (7, 15). In addition, rifamycin resistance was detected among the members of epidemic REA group BI. These isolates accounted for more than half of the 14 resistant unique patient isolates included in this study (Table 3). However, determination of the clinical implications of our findings will require the correlation of C. difficile rifaximin resistance with treatment response in patients infected with resistant strains. Clinical correlation studies will be particularly important, considering that patients undergoing rifaximin therapy can have fecal rifaximin concentrations as high as 8,000 μg/g (9).
Fourteen of the 80 isolates we analyzed were resistant to rifaximin and rifampin (MICs, >32 μg/ml). Compared to nonresistant isolates, sequence differences in the rpoB gene of the resistant isolates resulted in amino acid substitutions that most likely explain this phenotype. No other rifamycin resistance mechanisms have been characterized in C. difficile. RpoB amino acid substitutions associated with rifamycin resistance were not clonal and were most likely selected for due to exposure to these antimicrobials (Table 3). Analysis of the REA grouping of the rifaximin-resistant isolates showed that the same sequence substitutions (H502N and R505K) could be detected in more than one REA group (Table 3). This result is not unexpected, since the equivalent amino acids of the T. aquaticus RpoB ortholog directly interact with rifampin (Fig. 1) (2). In addition, previous studies with S. aureus and M. tuberculosis have shown that independently derived mutations in these residues are frequently detected (17, 19, 22).
In our study, the nine rifamycin-resistant isolates within the BI epidemic group had four RpoB sequence differences compared to nonresistant isolates. The R505K amino acid substitution was found in all of the resistant BI group isolates, either as the only substitution or in addition to S488T, H502N, or I548M (Table 3). Each of these isolates was sampled from a separate patient and originated from four different states within the United States and two Canadian provinces (Table 2). These data suggest that the BI isolates were unlikely to have resulted from the clonal dissemination of a single resistant strain. This information further demonstrates that rifamycin therapy could be responsible for the selection of these resistant isolates in individual patients. This conclusion is also supported by the results from the paired patient isolates, which were sampled before rifaximin therapy and after treatment failure (Fig. 2) (11). The RpoB sequence of the rifaximin-resistant posttreatment isolate differed by only one amino acid from that of the nonresistant pretreatment strain (H502Y) (Table 3 and Fig. 1).
We have shown that the rifampin Etest is a suitable surrogate for rifaximin susceptibility testing by agar dilution. Rifaximin-resistant C. difficile isolates all carried sequence substitutions within a conserved RpoB region compared to nonresistant isolates. Molecular typing and analysis of geographic origins determined that these isolates were likely to be independently derived and that rifamycin therapy may have selected for them. We have shown that rifamycin resistance in clinical isolates of C. difficile may be more common than initially suspected, particularly among epidemic BI isolates (7). Further studies are warranted to address the clinical implications of these findings.
Supplementary Material
Acknowledgments
Kristin Nagaro, Adam Cheknis, and Walter Zukowski are thanked for conducting REA typing. James Osmolski is thanked for assistance with MIC experiments.
This study was supported by the U.S. Department of Veterans Affairs Research Service (D.N.G., S.J., and D.W.H.), NIH grant AI 050122 (D.W.H.), and Genzyme Corporation (D.N.G., S.J., and D.W.H.). The rifaximin compound used was graciously supplied by Salix Pharmaceuticals, Inc.
Footnotes
Published ahead of print on 16 June 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 3.Clabots, C. R., S. Johnson, K. M. Bettin, P. A. Mathie, M. E. Mulligan, D. R. Schaberg, L. R. Peterson, and D. N. Gerding. 1993. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J. Clin. Microbiol. 31:1870-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Dreses-Werringloer, U., I. Padubrin, L. Kohler, and A. P. Hudson. 2003. Detection of nucleotide variability in rpoB in both rifampin-sensitive and rifampin-resistant strains of Chlamydia trachomatis. Antimicrob. Agents Chemother. 47:2316-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmann, G., K. O. Honikel, F. Knüsel, and J. Nüesch. 1967. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta 145:843-844. [DOI] [PubMed] [Google Scholar]
- 7.Hecht, D. W., M. A. Galang, S. P. Sambol, J. R. Osmolski, S. Johnson, and D. N. Gerding. 2007. In vitro activities of 15 antimicrobial agents against 110 toxigenic Clostridium difficile clinical isolates collected from 1983 to 2004. Antimicrob. Agents Chemother. 51:2716-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoover, W. W., E. H. Gerlach, D. J. Hoban, G. M. Eliopoulos, M. A. Pfaller, and R. N. Jones. 1993. Antimicrobial activity and spectrum of rifaximin, a new topical rifamycin derivative. Diagn. Microbiol. Infect. Dis. 16:111-118. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, Z. D., S. Ke, E. Palazzini, L. Riopel, and H. Dupont. 2000. In vitro activity and fecal concentration of rifaximin after oral administration. Antimicrob. Agents Chemother. 44:2205-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, S., C. Schriever, M. Galang, C. P. Kelly, and D. N. Gerding. 2007. Interruption of recurrent Clostridium difficile-associated diarrhea episodes by serial therapy with vancomycin and rifaximin. Clin. Infect. Dis. 44:846-848. [DOI] [PubMed] [Google Scholar]
- 12.Killgore, G., A. Thompson, S. Johnson, J. Brazier, E. Kuijper, J. Pepin, E. H. Frost, P. Savelkoul, B. Nicholson, R. J. van den Berg, H. Kato, S. P. Sambol, W. Zukowski, C. Woods, B. Limbago, D. N. Gerding, and L. C. McDonald. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442-2449. [DOI] [PubMed] [Google Scholar]
- 14.Louie, T., M. Gerson, D. Grimard, S. Johnson, A. Poirier, K. Weiss, J. Peppe, J. Donovan, and D. Davidson. 2007. Results of a phase III trial comparing tolevamer, vancomycin and metronidazole in patients with Clostridium difficile-associated diarrhea (CDAD), abstr. K-425a, p. 219. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother., American Society for Microbiology, Washington, DC.
- 15.Marchese, A., A. Salerno, A. Pesce, E. A. Debbia, and G. C. Schito. 2000. In vitro activity of rifaximin, metronidazole and vancomycin against Clostridium difficile and the rate of selection of spontaneously resistant mutants against representative anaerobic and aerobic bacteria, including ammonia-producing species. Chemotherapy 46:253-266. [DOI] [PubMed] [Google Scholar]
- 16.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433-2441. [DOI] [PubMed] [Google Scholar]
- 17.Murphy, C. K., S. Mullin, M. S. Osburne, J. van Duzer, J. Siedlecki, X. Yu, K. Kerstein, M. Cynamon, and D. M. Rothstein. 2006. In vitro activity of novel rifamycins against rifamycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 50:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connor, J. R., D. Lyras, K. A. Farrow, V. Adams, D. R. Powell, J. Hinds, J. K. Cheung, and J. I. Rood. 2006. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 61:1335-1351. [DOI] [PubMed] [Google Scholar]
- 19.O'Sullivan, D. M., T. D. McHugh, and S. H. Gillespie. 2005. Analysis of rpoB and pncA mutations in the published literature: an insight into the role of oxidative stress in Mycobacterium tuberculosis evolution? J. Antimicrob. Chemother. 55:674-679. [DOI] [PubMed] [Google Scholar]
- 20.Pepin, J., M. E. Alary, L. Valiquette, E. Raiche, J. Ruel, K. Fulop, D. Godin, and C. Bourassa. 2005. Increasing risk of relapse after treatment of Clostridium difficile colitis in Quebec, Canada. Clin. Infect. Dis. 40:1591-1597. [DOI] [PubMed] [Google Scholar]
- 21.Pepin, J., L. Valiquette, and B. Cossette. 2005. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. Can. Med. Assoc. J. 173:1037-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portugal, I., S. Maia, and J. Moniz-Pereira. 1999. Discrimination of multidrug-resistant Mycobacterium tuberculosis IS6110 fingerprint subclusters by rpoB gene mutation analysis. J. Clin. Microbiol. 37:3022-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 24.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.