Abstract
The first outbreak of carbapenem-resistant Klebsiella pneumoniae isolates producing the plasmid-encoded carbapenem-hydrolyzing oxacillinase OXA-48 is reported. The 39 isolates belonged to two different clones and were collected at the University Hospital of Istanbul, Turkey, from May 2006 to February 2007, and they coproduced various β-lactamases (SHV-12, OXA-9, and TEM-1 for clone A and CTX-M-15, TEM-1, and OXA-1 for clone B).
Outbreaks of extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae have been extensively reported worldwide (9, 17, 18) and carbapenems often remain the treatment of last resort. However, carbapenem resistance in K. pneumoniae has occasionally been reported and may be due to plasmid-mediated AmpC cephalosporinases associated with porin modifications (3, 14) or carbapenemases (23). Ambler class A β-lactamases of the GES type and the KPC type have been reported for K. pneumoniae isolates from several parts of the world, including for GES Greece (26) and Korea (13) and for KPC the United States (4), Israel (24), and China (6). K. pneumoniae isolates producing metallo-β-lactamase VIM-1 have been identified in Greece (11) and recently in Spain (25) and in Italy (5). The class D β-lactamase OXA-48, possessing significant carbapenemase activity, was identified from sporadic carbapenem-resistant K. pneumoniae isolates from Turkey (1, 10, 22). The blaOXA-48 gene was part of the Tn1999 composite transposon made of two copies of the insertion sequence IS1999 (2).
Here we describe a nosocomial outbreak of carbapenem-resistant K. pneumoniae strains expressing OXA-48.
Thirty-nine nonrepetitive carbapenem-resistant K. pneumoniae isolates were collected from patients hospitalized at the University Hospital of Istanbul, Turkey, from May 2006 to January 2007. Among them, 27 were isolated from clinical samples (4 endotracheal aspirate, 8 pus, 1 ascite, 7 blood culture, 2 wound, 1 urine, and 4 catheter samples). The remaining 12 isolates were grown from routine rectal screening swabs. Most of the patients were hospitalized in intensive care units (ICUs) or for emergency surgery (Table 1). Sixteen patients were infected, and 13 of them were being treated with carbapenems prior to and also after the determination of isolate susceptibility (Table 1). The three untreated patients were suffering from infections which did not require systemic administration of antibiotic. Despite treatment, 9 out of the 16 infected patients died.
TABLE 1.
Clinical features of the 39 isolates of carbapenem-resistant K. pneumoniae isolates
| K. pneumoniae isolate | Clone | Date (mo/day/yr) of:
|
Hospitalization unit | Source | Infected or colonized statusb | Treatment | Outcome | |
|---|---|---|---|---|---|---|---|---|
| Isolation | Hospitalization | |||||||
| 1 | B | 05/24/06 | 05/19/06 | ICU | Cathetera | C | None | Cured |
| 2 | A | 05/26/06 | 05/03/06 | ICU | Catheter | C | None | Cured |
| 3 | B | 05/27/06 | 05/08/06 | Urology | Wound | C | None | Cured |
| 4 | A | 05/28/06 | 04/09/06 | ICU | Blood | I | Imipenem | Dead |
| 5 | B | 05/28/06 | 04/24/06 | ICU | Blood | I | Imipenem + ciprofloxacin | Dead |
| 6 | A | 06/13/06 | 05/27/06 | ICU | Catheter | C | None | Cured |
| 7 | B | 06/13/06 | 05/31/06 | ICU | Ascites | I | Imipenem + netilmicin | Dead |
| 8 | A | 06/14/06 | 05/23/06 | ICU | Catheter | C | None | Cured |
| 9 | A | 06/14/06 | 05/31/06 | ICU/Emergency Surgery | Pus | I | None | Cured |
| 10 | A | 06/25/06 | 05/05/06 | Transplantation unit | Pus | I | Imipenem | Dead |
| 11 | A | 07/14/06 | 10/30/06 | General Surgery | Rectal swab | C | None | Dead |
| 12 | A | 08/12/06 | 11/17/06 | ICU/Emergency Surgery | Rectal swab | C | None | Cured |
| 13 | A | 09/30/06 | 08/15/06 | ICU/Emergency Surgery | Rectal swab | C | None | Cured |
| 14 | B | 10/23/06 | 10/17/06 | ICU | Endotracheal | C | None | Cured |
| 15 | A | 11/13/06 | 11/05/06 | ICU/Emergency Surgery | Blood | I | Imipenem | Dead |
| 16 | B | 11/16/06 | 10/02/06 | ICU | Endotracheal | I | Imipenem | Dead |
| 17 | B | 11/18/06 | 10/27/06 | ICU/Neurosurgery | Urine | C | None | Cured |
| 18 | A | 11/22/06 | 10/03/06 | ICU/Emergency Surgery | Pus | I | None | Dead |
| 19 | B | 11/25/06 | 10/22/06 | Chest | Endotracheal | C | None | Dead |
| 20 | A | 12/01/06 | 11/22/06 | ICU/Emergency Surgery | Pus | I | Imipenem + amikacin | Cured |
| 21 | A | 12/01/06 | 11/13/06 | ICU/Emergency Surgery | Pus | C | None | Cured |
| 22 | B | 12/02/06 | 11/10/06 | ICU/Emergency Surgery | Rectal swab | C | None | Cured |
| 23 | A | 12/06/06 | 10/23/06 | ICU/Emergency Surgery | Rectal swab | C | None | Cured |
| 24 | B | 12/07/06 | 12/01/06 | ICU/Emergency Surgery | Blood | I | Imipenem + ciprofloxacin | Cured |
| 25 | A | 12/07/06 | 12/04/06 | ICU | Pus | I | Imipenem + netilmicin | Dead |
| 26 | A | 12/08/06 | 10/04/06 | ICU | Rectal swab | C | None | Cured |
| 27 | A | 12/12/06 | 11/22/06 | General Surgery | Blood | I | Piperacillin + tazobactam + ciprofloxacin | Cured |
| 28 | A | 12/17/06 | 11/13/06 | Urology | Pus | I | None | Cured |
| 29 | B | 12/19/06 | 12/01/06 | ICU | Endotracheal | C | None | Cured |
| 30 | A | 12/19/06 | 12/30/06 | General Surgery | Rectal swab | C | None | Dead |
| 31 | A | 12/30/06 | 12/26/06 | ICU/Emergency Surgery | Rectal swab | C | None | Dead |
| 32 | A | 12/30/06 | 12/22/06 | ICU | Rectal swab | C | None | Cured |
| 33 | A | 01/04/07 | 01/31/07 | ICU/Emergency Surgery | Wound | C | None | Cured |
| 34 | B | 01/05/07 | 12/31/06 | ICU/Emergency Surgery | Rectal swab | C | None | Cured |
| 35 | A | 01/13/07 | 12/20/06 | ICU/Emergency Surgery | Rectal swab | C | None | Cured |
| 36 | B | 01/16/07 | 01/04/06 | ICU/Emergency Surgery | Rectal swab | C | None | Cured |
| 37 | A | 01/29/07 | 07/09/06 | ICU | Blood | I | Imipenem + ciprofloxacin | Dead |
| 38 | A | 02/02/07 | 01/31/07 | ICU/Emergency Surgery | Pus | I | Ciprofloxacin + metronidazole | Cured |
| 39 | B | 02/10/07 | 01/15/07 | ICU/Emergency Surgery | Blood | I | Imipenem | Dead |
All catheters corresponded to central venous catheters.
C, colonized; I, infected.
Antibiotic susceptibility was determined by the disk diffusion method according to Clinical and Laboratory Standards Institute guidelines (7). Additionally, the MICs of several antibiotics were determined by using E-test strips (AB Biodisk, Solna, Sweden). Thirty-seven isolates were resistant to all penicillins and expanded cephalosporins. They exhibited heterogeneous decreased susceptibility to carbapenems, with meropenem and ertapenem being resistant, whereas MICs of imipenem ranged from 4 to 32 μg/ml (Table 2). Isolates 11 and 21 were resistant to carbapenems but remained susceptible to cefotaxime, ceftazidime, and aztreonam. Taking resistance to β-lactam and non-β-lactam antibiotics into account, the following three distinct resistance patterns were defined: group 1 included 23 isolates susceptible to ciprofloxacin but resistant to aminoglycosides; group 2 included 14 isolates resistant to ciprofloxacin but susceptible to gentamicin; and group 3 included isolates 11 and 21, susceptible to ceftazidime and ciprofloxacin but resistant to aminoglycosides. Double-disk synergy tests were performed for ESBL detection as described previously (12) and positive results were obtained for isolates of groups 1 and 2 but not for the two isolates of group 3. These various resistance patterns may explain the treatment failure observed for most of the infected patients.
TABLE 2.
MICs of β-lactams and non-β-lactamsa
| Drug | MIC (μg/ml) against indicated isolate (genotype)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| K. pneumoniae clone A (blaOXA-48blaTEM-1blaSHV-12blaOXA-9) | K. pneumoniae clone B (blaOXA-48blaTEM-1blaCTX-M-15blaOXA-1) | E. coli J53(pAa) (blaCTX-M-15blaTEM-1blaOXA-1) | E. coli J53(pAb) or -(pBb) (blaOXA-48) | E. coli J53(pBa) (blaSHV-12blaTEM-1blaOXA-9) | E. coli J53 | K. pneumoniae isolate 11 (blaOXA-48blaTEM-1blaOXA-1) | K. pneumoniae isolate 21 (blaOXA-48blaTEM-1blaOXA-9) | |
| Amoxicillin | >512 | >512 | >512 | >512 | >512 | 1 | >512 | >512 |
| Amoxicillin + clavulanic acid | >512 | >512 | >513 | >512 | >512 | 1 | >512 | >512 |
| Ticarcillin | >512 | >512 | >514 | >512 | >512 | 4 | >512 | >512 |
| Ticarcillin + clavulanic acid | >512 | >512 | >515 | >512 | >512 | 4 | >512 | >512 |
| Piperacillin | >512 | >512 | >512 | >512 | >512 | 4 | >512 | >512 |
| Piperacillin + tazobactam | >512 | >512 | >4-8 | 64 | >4-8 | 4 | >512 | >512 |
| Cefotaxime | >512 | >512 | >32 | 0,5 | 4 | 0.5 | 4 | 4 |
| Ceftazidime | >512 | >512 | 128 | 1 | 64 | 1 | 2 | 2 |
| Cefepime | >512 | >512 | 4 | <0.5 | 0.5 | 0.125 | 4 | 4 |
| Aztreonam | >512 | >512 | 8 | <0.125 | 64 | 0.5 | 0.5 | 0.5 |
| Cefoxitin | 16-512 | 32->512 | 2-4 | 4-8 | 2-4 | 4 | 16 | >32 |
| Moxalactam | 16->512 | 16->512 | <0.5 | 2 | <0.5 | 1 | ||
| Imipenem | 4-32 | 4-16 | <1 | 0.5 | <1 | 2 | 8-16 | 8-16 |
| Meropenem | >32 | 6-32 | <1 | 0.12 | <1 | <1 | 16 | 16 |
| Ertapenem | >32 | >32 | <1 | 0.25 | <1 | <1 | >32 | >32 |
| Kanamycin | >256 | 128 | ||||||
| Tobramycin | >64 | >64 | ||||||
| Netilmicin | >128 | 3 | ||||||
| Amikacin | 16 | 2 | ||||||
| Gentamicin | >64 | 1 | ||||||
| Sulfamides | >256 | >256 | ||||||
| Trimethoprim | >32 | >32 | ||||||
| Tetracycline | >256 | >256 | ||||||
| Nalidixic acid | >64 | >64 | ||||||
| Norfloxacin | 8 | >256 | ||||||
| Ofloxacin | 2 | >64 | ||||||
| Ciprofloxacin | 1 | >128 | ||||||
MICs are for K. pneumoniae isolates of clones A and B, E. coli J53 harboring natural plasmids pAa or pAb from clone A, E. coli J53 harboring natural plasmids pBa or pBb from clone B, K. pneumoniae isolates 11 and 21, and E. coli J53, as indicated.
Specific primers were used for the detection of β-lactamase-encoding genes that had been previously identified in K. pneumoniae 11978, namely, blaTEM, blaSHV, blaOXA-1, blaOXA-9, and blaOXA-48 (22). In addition, PCR experiments were performed to identify other ESBL-encoding genes by use of specific primers for blaCTX-M, blaVEB, blaPER, and blaGES, which had been designed previously (9, 15, 19). All isolates from group 1 were positive for the blaOXA-48, blaTEM, blaSHV, and blaOXA-9 genes; all isolates from group 2 were positive for the blaOXA-48, blaTEM, blaCTX-M, and blaOXA-1 genes; and isolates from group 3 were positive for the blaOXA-48, blaOXA-9, and blaTEM genes only. Sequence analysis of the entire genes revealed perfect identity with the blaCTX-M-15, blaOXA-1,blaOXA-9, blaOXA-48, blaSHV-12, and blaTEM-1 genes.
Isoelectrofocusing analysis performed as described elsewhere (20) confirmed gene identification for each group, with pI values of 7.4, 7.2, 8.2, 8.9, and 5.4, corresponding to OXA-48, OXA-1, SHV-12, CTX-M-15, and TEM-1, respectively (data not shown). As shown for K. pneumoniae 11978, OXA-9 was not expressed, and no isoelectric focusing band at the corresponding pI was found. No additional β-lactamase signal was observed for isolates from group 3.
The genetic relationship between the different isolates studied by pulsed-field gel electrophoresis (8) revealed only two distinct profiles, defining clone A as grouping 25 isolates from groups 1 and 3 and clone B as corresponding to the 14 isolates of group 2 (Fig. 1 and data not shown). It is noteworthy that clones A and B were genetically distinct from K. pneumoniae 11978, which is known to express another ESBL clavulanic acid-inhibited determinant, SHV-2a (22) (data not shown).
FIG. 1.
Pulsed-field gel electrophoresis patterns of OXA-48-producing K. pneumoniae isolates. Lanes: 1 to 3, three isolates of clone A harboring the blaOXA-48, blaOXA-9, blaSHV-12, and blaTEM-1 genes; 4 to 6, three isolates of clone B harboring the blaOXA-48, blaOXA-1, blaCTX-M-15, and blaTEM-1 genes; M, molecular mass markers.
Conjugation experiments were performed with one isolate belonging to clone A and with one isolate belonging to clone B by use of amoxicillin, ceftazidime, or cefotaxime as the selective agent as described previously (22). Plasmid DNA extraction according to the Kieser technique (16) showed that the blaOXA-48 gene was carried by a 70-kb plasmid present in each clone (named pAb and pBb for clones A and B, respectively), while the other β-lactamases genes were carried on plasmids of more than 150 kb (pAa and pBa) and were identified by PCR (Table 2 and data not shown).
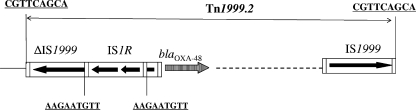
The genetic environment of the blaOXA-48 gene was determined by PCR using specific primers for the insertion sequence IS1999, located upstream and downstream of the blaOXA-48 gene in Tn1999 (2). A structure identical to Tn1999, as found in K. pneumoniae 11978, was identified in clone A isolates, whereas in clone B isolates, the IS1999 element located upstream of the blaOXA-48 gene was truncated by IS1R, which had targeted the transposase gene of IS1999 (Fig. 2). Whereas IS1999 was shown to provide −35 and −10 promoter sequences for the expression of the blaOXA-48 gene in K. pneumoniae 11978 and thus acts similarly in isolates belonging to the clone A (Fig. 3A), it is likely that the promoter driving the expression in clone B isolates was made of a −35 box located inside IS1R together with the −10 box originally located in IS1999 (Fig. 3B). This hybrid promoter possesses the features of a strong promoter for blaOXA-48 gene expression, with a more efficient −35 sequence and an optimal 17-bp spacing between the −35 and −10 boxes. Since the genetic structures identified upstream of the blaOXA-48 gene varied, their role in blaOXA-48 gene expression was studied by measuring hydrolytic activities as described previously (21). Hydrolysis of imipenem (100 μM) obtained from Escherichia coli(pAb) (23 [± 0.002] mU/mg of protein−1) was twofold lower than that obtained from E. coli(pBb) (48 [± 0.013] mU/mg of protein−1). This result indicated a higher expression of the blaOXA-48 gene in clone B.
FIG. 2.
Schematic representation of Tn1999.2 described for clone B. The coding regions are represented as boxes, with arrows indicating their transcription orientations.
FIG. 3.
Nucleotide sequence of the −35 and −10 putative promoter regions within IS1999 described for clone A (A) and clone B (B). The left inverted repeats (IRL) of IS1999 and of IS1R are shaded in gray, and the −35 and −10 putative sequences are underlined. The tnpA transposase gene is indicated.
Our study identified the first large outbreak of OXA-48-positive carbapenem-resistant K. pneumoniae isolates. Most isolates produced multiple β-lactamases, including ESBLs, narrow-spectrum oxacillinases, and penicillinases, therefore leading to resistance to all β-lactams. Several OXA-48-producing isolates also expressed ESBL SHV-12, whereas others expressed ESBL CTX-M-15. This association was not due to the dissemination of single plasmids, since the ESBL and OXA-48 genes were not located on the same plasmids. The OXA-48 producers identified here were not clonally related to the previously identified OXA-48-positive K. pneumoniae 11978 isolate from Turkey, indicating the concomitant spread of several OXA-48-producing clones in Istanbul. This identification of K. pneumoniae isolates harboring the worldwide-spread CTX-M-15 determinant but also the OXA-48 carbapenemase is worrying, since carbapenems are often the last resort for treating infections due to ESBL-producing strains. In addition, we showed here that a higher level of OXA-48 expression was likely due to a particular genetic structure providing a stronger promoter.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France, and mostly by a grant of the European community (LSHM-CT-2005-018705).
Footnotes
Published ahead of print on 2 June 2008.
REFERENCES
- 1.Aktas, Z., C. Bal Kayacan, I. Schneider, B. Can, K. Misilli, and A. Bauernfeind. 2008. Carbapenem-hydrolyzing oxacillinase OXA-48 persists in Klebsiella pneumoniae in Istanbul, Turkey. Chemotherapy 54:101-106. [DOI] [PubMed] [Google Scholar]
- 2.Aubert, D., T. Naas, C. Héritier, L. Poirel, and P. Nordmann. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of β-lactam resistance genes. J. Bacteriol. 188:6506-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratu, S., D. Landman, R. Haag, R. Recco, A. Eramo, M. Alam, and J. Quale. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165:1430-1435. [DOI] [PubMed] [Google Scholar]
- 5.Cagnacci, S., L. Gualco, S. Roveta, S. Mannelli, L. Borgianni, J. D. Docquier, F. Dodi, M. Centanaro, E. Debbia, A. Marchese, and G. M. Rossolini. 2008. Bloodstream infections caused by multidrug-resistant Klebsiella pneumoniae producing the carbapenem-hydrolysing VIM-1 metallo-beta-lactamase: first Italian outbreak. J. Antimicrob. Chemother. 61:296-300. [DOI] [PubMed] [Google Scholar]
- 6.Cai, J. C., H. W. Zhou, R. Zhang, and G.-X. Chen. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement M100-S18. Clinical and Laboratory Standards Institute, Baltimore, MD.
- 8.Girlich, D., A. Karim, L. Poirel, M. H. Cavin, C. Vernyand, and P. Nordmann. 2000. Molecular epidemiology of an outbreak due to IRT-2 β-lactamase-producing strains of Klebsiella pneumoniae in a geriatric department. J. Antimicrob. Chemother. 45:467-473. [DOI] [PubMed] [Google Scholar]
- 9.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulmez, D., N. Woodford, M. F. I. Palepou, S. Mushtaq, G. Metan, Y. Yakupogullari, S. Kocagoz, O. Uzun, G. Hascelik, and D. M. Livermore. 2008. Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. Int. J. Antimicrob. Agents 31:523-526. [DOI] [PubMed] [Google Scholar]
- 11.Ikonomidis, A., D. Tokatlidou, I. Kristo, D. Sofianou, A. Tsakris, P. Mantzana, S. Pournaras, and A. N. Maniatis. 2005. Outbreaks in distinct regions due to a single Klebsiella pneumoniae clone carrying a blaVIM-1 metallo-β-lactamase gene. J. Clin. Microbiol. 43:5344-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 13.Jeong, S. H., I. K. Bae, D. Kim, S. G. Hong, J. S. Song, J. H. Lee, and S. H. Lee. 2005. First outbreak of Klebsiella pneumoniae clinical isolates producing GES-5 and SHV-12 extended-spectrum β-lactamases in Korea. Antimicrob. Agents. Chemother. 49:4809-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaczmarek, F. M., F. Dib-Hajj, W. Shang, and T. D. Gootz. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob. Agents Chemother. 50:3396-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS. Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 16.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 17.Livermore, D. M., and P. M. Hawkey. 2005. CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56:451-454. [DOI] [PubMed] [Google Scholar]
- 18.Mena, A., V. Plasencia, L. García, O. Hidalgo, J. I. Ayestarán, S. Alberti, N. Borrell, J. L. Pérez, and A. Oliver. 2006. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J. Clin. Microbiol. 44:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., L. Cabanne, H. Vahaboglu, and P. Nordmann. 2005. Genetic environment and expression of the extended-spectrum β-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob. Agents Chemother. 49:1708-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel, L., D. Girlich, T. Naas, and P. Nordmann. 2001. OXA-28, an extended-spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., M. Guibert, S. Bellais, T. Naas, and P. Nordmann. 1999. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob. Agents Chemother. 43:1098-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel, L., C. Héritier, V. Tolün, and P. Nordmann. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents. Chemother. 48:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samra, Z., O. Ofir, Y. Lishtzinsky, L. Madar-Shapiro, and J. Bishara. 2007. Outbreak of carbapenem-resistant Klebsiella pneumoniae producing KPC-3 in a tertiary medical centre in Israel. Int. J. Antimicrob. Agents 30:525-529. [DOI] [PubMed] [Google Scholar]
- 25.Tato, M., T. M. Coque, P. Ruíz-Garbajosa, V. Pintado, J. Cobo, H. S. Sader, R. N. Jones, F. Baquero, and R. Cantón. 2007. Complex clonal and plasmid epidemiology in the first outbreak of Enterobacteriaceae infection involving VIM-1 metallo-β-lactamase in Spain: toward endemicity? Clin. Infect. Dis. 45:1171-1178. [DOI] [PubMed] [Google Scholar]
- 26.Vourli, S., P. Giakkoupi, V. Miriagou, E. Tzelepi, A. C. Vatopoulos, and L. S. Tzouvelekis. 2004. Novel GES/IBC extended-spectrum β-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol. Lett. 234:209-213. [DOI] [PubMed] [Google Scholar]