Abstract
Beta-lactam antibiotics (BLAs) are the first-line agents used against group B streptococci (GBS) infection. A clonal set of four independent, invasive GBS isolates with elevated MICs to BLAs were identified that shared a pbp2x mutation (Q557E) corresponding to a resistance-conferring pneumococcal mutation. BLA sensitivity was restored through allelic replacement or complementation with the wild-type pbp2x.
Group B Streptococcus (GBS) is associated with serious infections in newborn infants, pregnant women, and the elderly. Beta-lactam antibiotics (BLAs) are the primary agents for GBS infection therapy for all age groups and for intrapartum prophylaxis. GBS resistance to BLA, including penicillin (PEN), has not been observed, but the same could once have been said for Streptococcus pneumoniae. PEN-resistant pneumococcal strains were initially detected during the 1970s and subsequently spread world wide. Pneumococcal PEN resistance involves the stepwise accumulation of alterations in three penicillin binding proteins (PBPs), PBP1a, PBP2b, and PBP2x (7, 8, 11). The generation of an altered pbp2x gene is thought to be the first event leading to full PEN resistance in pneumococci (7, 11).
We performed broth dilution susceptibility testing with 5,631 GBS isolates recovered from 1999 to 2005 through Active Bacterial Core Surveillance (http://www.cdc.gov/ncidod/dbmd/abcs/survreports.htm), using PEN and several other BLAs (ampicillin, cefazolin, cefoxitin, cefuroxime, and cefotaxime), as well as clindamycin, erythromycin, telithromycin, levofloxacin, ciprofloxacin, and tetracycline. We red flagged isolates that met any of the following MIC criteria for BLAs: PEN, ≥0.12 μg/ml; cefotaxime, ≥0.12 μg/ml; cefoxitin, ≥8 μg/ml; cefazolin, ≥1 μg/ml; cefuroxime, ≥0.25 μg/ml; ampicillin, ≥ 0.25 μg/ml. Under current guidelines (2), beta-hemolytic streptococcal isolates with MICs to PEN, ampicillin, and cefotaxime of ≤0.12 μg/ml, ≤0.25 μg/ml, and ≤0.5 μg/ml, respectively, are considered sensitive to these antibiotics. There are no guidelines for cefoxitin, cefazolin, and cefuroxime. Bacterial strains were propagated in Todd-Hewitt broth or on THB agar plates.
We red flagged 99 GBS isolates recovered during the period 1999 to 2005, with elevated MICs to one or more BLAs; 41 isolates were immediately available for further study. Upon repeated susceptibility testing, 19 isolates exhibited MICs that were reduced below our cutoff points. The 22 remaining isolates consistently demonstrated elevated MICs for one or more BLAs (Table 1), including 11 with elevated MICs for multiple BLAs, including PEN and/or the extended spectrum cephalosporin cefotaxime.
TABLE 1.
Invasive GBS with elevated MICs to one or more BLAsa
| Laboratory identification | State | Year | Pt age (yr) | Site of isolation | Serotype | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | PEN | CTX | CFX | CFZ | CFR | ||||||
| 3047-05 | MN | 2004 | 49 | Blood | IA | 0.12 | 0.06 | 0.06 | 8 | 0.25 | 0.06 |
| 5662-05 | MN | 2004 | 0 | Blood | IA | 0.12 | 0.06 | 0.06 | 8 | 0.25 | 0.03 |
| 6649-05 | CO | 2005 | 58 | Blood | IA | 0.25 | 0.12 | 0.12 | 4 | 0.5 | ND |
| 7490-03 | MD | 2003 | 64 | Blood | IA | 0.12 | 0.06 | 0.12 | 2 | 0.25 | 0.06 |
| 9012-04 | GA | 2004 | 48 | PTL fluid | IA | 0.06 | 0.06 | 0.06 | 16 | 0.25 | 0.06 |
| 5436-03 | OR | 2001 | 53 | Blood | IB | 0.06 | 0.03 | 0.25 | 16 | 1 | 0.25 |
| 3776-04 | MN | 2003 | 71 | Joint | II | 0.25 | 0.12 | 0.12 | 8 | 0.25 | 0.12 |
| 9051-04 | MN | 2004 | 90 | Blood | II | 0.12 | 0.06 | 0.06 | 8 | 0.25 | 0.06 |
| 3444-99 | GA | 1999 | 0 | Blood | III | 0.12 | 0.06 | 0.06 | 8 | 0.25 | 0.06 |
| 7037-99 | GA | 1999 | 0 | Blood | III | 0.12 | 0.06 | 0.12 | 8 | 0.25 | 0.06 |
| 3789-04 | MN | 2003 | 78 | Blood | III | 0.12 | 0.12 | 0.25 | 16 | 1 | 0.12 |
| 6138-03 | NY | 2003 | 90 | Blood | III | 0.06 | 0.12 | 0.25 | 16 | 0.5 | 0.25 |
| 7507-03 | MD | 2003 | 82 | Blood | III | 0.12 | 0.12 | 0.12 | 16 | 0.5 | 0.12 |
| 8607-03 | NY | 2003 | 81 | Blood | III | 0.06 | 0.12 | 0.25 | 16 | 0.5 | 0.12 |
| 3454-03 | MN | 2002 | 73 | PTL fluid | IV | 0.25 | 0.12 | 0.25 | 8 | 0.5 | 0.06 |
| 4214-03 | NY | 2002 | 32 | Bone | NT | 0.25 | 0.12 | 0.12 | 4 | 0.5 | 0.12 |
| 6149-03 | NY | 2003 | 76 | Blood | NT | 0.12 | 0.06 | 0.06 | 8 | 0.25 | 0.06 |
| 7368-04 | MD | 2004 | 45 | Blood | NT | 0.12 | 0.12 | 0.06 | 4 | 0.5 | 0.06 |
| 3386-05 | MD | 2004 | 69 | Blood | V | 0.06 | 0.03 | 0.06 | 16 | 0.12 | ND |
| 4857-03 | NY | 2002 | 84 | Joint | V | 0.25 | 0.12 | 0.06 | 4 | 0.5 | 0.06 |
| 5677-04 | MD | 2003 | 74 | Joint | V | 0.25 | 0.12 | 0.12 | 16 | 1 | 0.12 |
| 9171-03 | GA | 2003 | 49 | Blood | V | 0.12 | 0.06 | 0.12 | 4 | 0.25 | 0.06 |
PTL, preterm labor fluid; AMP, ampicillin; PEN, penicillin; CTX, cefotaxime; CFX, cefoxitin; CFZ, cefazolin; CFR, cefuroxime. MICs that are considered unusually high are underlined. The four isolates that shared the Q557E pbp2x mutation and multilocus sequence type ST19 are in bold.
Four GBS isolates (3789-04, 6138-03, 7507-03, and 8607-03) were analyzed immediately, since they shared the same capsular serotype III and MIC profile, features consistent with the successful expansion of a single mutant. These isolates were recovered from three different states during 2003 from elderly bacteremic patients (Table 1) and exhibited elevated MICs to PEN (0.12 μg/ml), cefotaxime (0.12 to 0.25 μg/ml), and cefoxitin (16 μg/ml); one isolate also displayed a relatively high MIC to cefazolin (1.0 μg/ml) and another to cefuroxime (0.25 μg/ml). These four isolates remained sensitive to the antibiotics of other drug classes, with the exception of three isolates that were tetracycline resistant.
We amplified and sequenced the deduced PEN binding region-encoding segments (PBRs) of pbp2x, pbp2b, and pbp1a from the four serotype III isolates. For pbp1a, the primers GBS1aF (5′-GACATCTACAACAGTGACACTTACA-3′) and GBS1aR (5′-GTAGGACATCATTGCACGATAAAC-3′) were used to amplify and sequence a 960-bp amplicon. For pbp2b (1,091-bp amplicon), the primers GBS2bF (5′-GCAAGAAGGTAAGTCAGGAAGAAA-3′) and GBS2bR (5′-GATCAATCATATCTCGTGCAACTAATT-3′) were used. For pbp2x (1,040-bp amplicon), the primers GBS2xF (5′-CTGGTTCAACAATGAAGGTAATGA-3′) and GBS2xR (5′-TAGTAGCTGTTCATTCTCAGCAAG-3′) were used. We found sequence identity between the 960-to-1,091-bp amplicon of pbp2b and that of pbp1a with the corresponding sequences in the three published GBS genomes (GenBank accession numbers CP000114, AE014204, and AL766844).
Within pbp2x, we identified a single base change that predicted a glutamine (Q)-to-glutamate (E) substitution at residue 557 of PBP2x (GenBank accession number EU570239). The GBS PBP2x Q557E substitution corresponds to a well-described resistance-conferring mutation, Q552E, in pneumococci (14, 15) (Fig. 1).
FIG. 1.
Alignment of PBP2x segments encompassing the critical residue 552 (pneumococci), corresponding to residue 557 (GBS) from PEN-resistant S. pneumoniae (top, penR Spn, strain 5259 [15]), the penicillin-sensitive S. pneumoniae TIGR4 (penS Spn) strain, the GBS strain 860703 from this study with elevated beta-lactam MICs, and the corresponding segments from wild type (wt) GBS strains with basal sensitivities to beta-lactam antibiotics (GenBank accession numbers CP000114, AE014204, and AL766844). Vertical lines indicate identical residues, colons indicate conserved substitutions, and nonconserved substitutions have no marking.
To verify the functional significance of the Q557E mutation in PBP2x for GBS BLA sensitivity, we used previously described methods (4, 6, 9) using the GBS serotype III clinical isolate 8607-03 as the test organism (Table 2). All strains were constructed with the absence of BLA selection. We amplified the normal or “wild-type” (WT) GBS pbp2x allele (identical in overlap with the three published GBS genome sequences), along with its upstream ribosome binding site and promoter region by using the primers PBP2xF (5′-GTGACTTTTTTTAAAAAGCTAAAAAAAATCTTC-3′) and PBP2xR (5′-CTGGCAGTCCATACCACATCCCCTAATAAAG-3′). This amplicon was cloned into the expression vector pDCerm to create the pPBP2xWT plasmid. The pPBP2xWT plasmid and the empty vector were introduced by electroporation into GBS clinical isolate 8607-03. We found that reintroducing the WT GBS pbp2x allele conferred an 8- to 16-fold decrease in MICs for PEN, cefotaxime, cefoxitin, and cefazolin, essentially restoring sensitivity to these BLAs to that of values commonly associated with GBS.
TABLE 2.
MICs due following complementation or allelic replacement of the GBS serotype III clinical isolate 8607-03 pbp2x(Q557E) mutation with the normal allelea
| Laboratory identification | PBP2x derivative | MIC(μg/ml)
|
||||
|---|---|---|---|---|---|---|
| AMP | PEN | CTX | CFX | CFZ | ||
| 8607-03 | Q557E | 0.06 | 0.12 | 0.25 | 16 | 0.5 |
| 8607-03 plus vector | Q557E | 0.06 | 0.12 | 0.25 | 16 | 0.5 |
| 8607-03 plus pPBP2xWT | Q557E (chromosome) | 0.06 | 0.015 | 0.015 | 2 | 0.06 |
| 8607(PBP2xWT) | Multicopy WT pbp2x | |||||
| Allelic exchange | WT | 0.06 | 0.015 | 0.03 | 2 | 0.06 |
AMP, ampicillin; PEN, penicillin; CTX, cefotaxime; CFX, cefoxitin; CFZ, cefazolin. MICs that are considered unusually high are in bold.
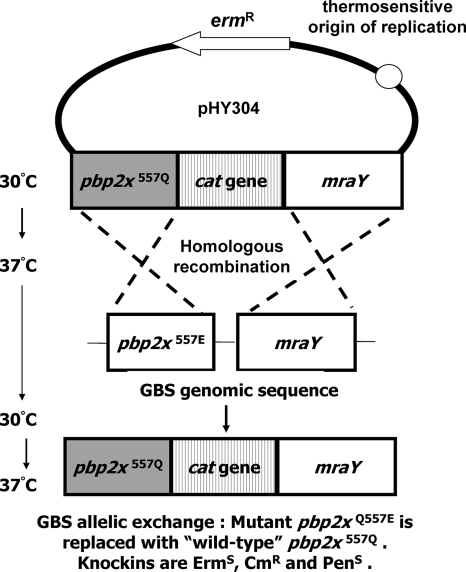
To exclude the potential confounding factors of PBP2x overexpression from the pPBP2xWT multicopy plasmid, we replaced the implicated chromosomal pbp2x(Q577E) allele of strain 8607-03 with the prevalent pbp2x(WT) allele exactly according to previously described methods (4, 6, 9) (Fig. 2). Precise in-frame chromosomal replacement of the pbp2x(Q577E) allele in strain 8607-03 with the pbp2x(WT) allele-cat sequence was verified by PCR and sequencing. Replacement of the Q557E allele with the WT allele restored sensitivity to PEN, cefotaxime, cefoxitin, and cefazolin (Table 2).
FIG. 2.
Scheme for the targeted “knock-in” allelic replacement mutagenesis of GBS strain 8607-03, possessing an elevated MIC for beta-lactam antibiotics, with the prevalent pbp2x(WT) allele.
Multilocus sequence typing of these four serotype III GBS isolates, performed as described (www.mlst.net), revealed that they shared multilocus sequence type ST19, a genotype common among serotype III isolates recovered from adult and neonatal GBS disease cases (12).
The 22 GBS isolates described here are considered sensitive to PEN, ampicillin, and cefotaxime (2); however, they appear to carry mutations that confer MICs to BLAs that are well beyond baseline levels. Indeed, the observed MIC to PEN of 0.12 μg/ml meets the current clinical definition of PEN intermediate as applied to pneumococci in meningitis cases (2).
In contrast to PEN-nonsusceptible pneumococci, which typically exhibit mosaicism in which the PBRs consist of sections of nonpneumococcal origin (5, 11, 13), the four GBS isolates described in this study shared a single base substitution within the PBP2x PBR. A single-point mutation (Q557E) rather than mosaicism is consistent with an organism that undergoes recombination less frequently than the pneumococcus (12) and also shares lower DNA homology with suspected viridans streptococcal donors.
A recent presentation indicated that GBS clinical isolates recovered in Japan with increased MICs to PEN (0.25 to 1.0 μg/ml) contained the Q557E substitution and, in addition, contained from one to four additional mutations within the PBP2x PBR (10). In view of these isolates with higher reported MICs, it is possible that the Q557E substitution is a common first step in the pathway to BLA resistance, with higher MICs conferred additively through subsequent substitutions. Increases in MICs to BLAs among GBS isolates have also been reported recently in Hong Kong (1), but without PBP sequence analysis.
One major group of BLA-resistant pneumococcal strains contains the Q552E mutation (14, 15), which corresponds to the GBS Q557E substitution. The Q552E substitution is the key change responsible for the majority of reduced BLA susceptibility in these fully resistant pneumococcal strains that characteristically have mosaic PBP2x PBRs containing up to 27 additional substitutions (15). Compared to PBP2x from sensitive pneumococci, a Q552E mutant derivative lowered the active site serine acylation for both PEN and cefotaxime (14).
Detection of the Q557E substitution in four independent GBS invasive disease isolates of the same clonal type indicates that this alteration potentially occurred within one strain, with subsequent expansion and spread to at least two additional geographic areas (Table 1). This possibility is consistent with the lack of a discernible growth disadvantage in standard growth curve comparisons that could indicate a defect in general fitness (data not shown). The emergence of a physiologically fit first-step GBS pbp2x (Q557E) mutant is of concern, since the accumulation of additional mutations could possibly lead to full PEN resistance. GBS strains that can survive even modest BLA concentrations could potentially have a selective advantage during carriage by antibiotic-exposed individuals, increasing the likelihood of further mutations, incrementally increasing resistance. In addition, possibly the original mutant has accumulated resistance to an additional antibiotic class, since three of the four isolates of this MLST type were tetracycline resistant.
We have recently obtained suggestive sequence data indicating additional first-step mutations within the invasive GBS isolates from our surveillance, with the identification of a T394I/T555S double-substitution PBP2x variant and a T546N PBP1a variant (corresponding to isolates 5436-03 and 3454-03, respectively, in Table 1). The PBP2x T555S substitution corresponds to another pneumococcal PBP2x substitution (T550A) that lowers the acylation efficiency of PBP2x by cefotaxime (14). The PBP1a T546N substitution lies within a highly conserved bacterial PBP1a Gly-Thr-Gly motif, where closely neighboring mutations potentially affect accessibility of the transpeptidase active site (3). Future experimentation will reveal whether these additional variant PBP derivatives function as potential first steps in resistance to BLAs. Detection and quantitation of mutations that could continue the progression to BLA resistance in GBS are warranted.
Acknowledgments
This work was funded by the CDC's Emerging Infections Program Network and CDC's Antimicrobial Resistance Working Group. Work at the University of California, San Diego, was supported by NIH grant HD051796 and an American Heart Association established investigator award (V.N.).
We thank the hospitals and laboratories that participate in the CDC's Emerging Infections Program Network and Active Bacterial Core surveillance system. We thank the CDC Division of Bacterial Diseases Streptococcus Laboratory for serotyping, antibiotic susceptibility testing, and multilocus sequence typing support. We thank K. Kimura for sharing the presentation from the 46th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy.
Footnotes
Published ahead of print on 9 June 2008.
REFERENCES
- 1.Chu, Y. W., C. Tse, G. K. Tsang, D. K. So, J. T. Fung, and J. Y. Lo. 2007. Invasive group B Streptococcus isolates showing reduced susceptibility to penicillin in Hong Kong. J. Antimicrob. Chemother. 60:1407-1409. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing. CLSI M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Contreras-Martel, C., V. Job, A. M. Di Guilmi, T. Vernet, O. Dideberg, and A. Dessen. 2006. Crystal structure of penicillin-binding protein 1a (PBP1a) reveals a mutational hotspot implicated in beta-lactam resistance in Streptococcus pneumoniae. J. Mol. Biol. 355:684-696. [DOI] [PubMed] [Google Scholar]
- 4.Doran, K. S., E. J. Engelson, A. Khosravi, H. C. Maisey, I. Fedtke, O. Equils, K. S. Michelsen, M. Arditi, A. Peschel, and V. Nizet. 2005. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J. Clin. Investig. 115:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowson, C. G., A. Hutchison, J. A. Brannigan, R. C. George, D. Hansman, J. Liñares, A. Tomasz, J. M. Smith, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakenbeck, R., K. Kaminski, A. König, M. van der Linden, J. Paik, P. Reichmann, and D. Zähner. 1999. Penicillin-binding proteins in beta-lactam-resistant streptococcus pneumoniae. Microb. Drug Resist. 5:91-99. [DOI] [PubMed] [Google Scholar]
- 8.Jamin, M., C. Damblon, S. Millier, R. Hakenbeck, and J. M. Frère. 1993. Penicillin-binding protein 2x of Streptococcus pneumoniae: enzymic activities and interactions with beta-lactams. Biochem. J. 292:735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeng, A., V. Sakota, Z. Li, V. Datta, B. Beall, and V. Nizet. 2003. Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 185:1208-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura, K., J. Wachino, S. Kurokawa, K. Suzuki, N. Yamane, Y. Shibata, and Y. Arakawa. 2006. Emergence of penicillin-resistant group B streptococci. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1286.
- 11.Laible, G., B. G. Spratt, and R. Hakenbeck. 1991. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 5:1993-2002. [DOI] [PubMed] [Google Scholar]
- 12.Luan, S. L., M. Granlund, M. Sellin, T. Lagergård, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin, C., T. Briese, and R. Hakenbeck. 1992. Nucleotide sequences of genes encoding penicillin-binding proteins from Streptococcus pneumoniae and Streptococcus oralis with high homology to Escherichia coli penicillin-binding proteins 1A and 1B. J. Bacteriol. 174:4517-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouz, N., A. M. Di Guilmi, E. Gordon, R. Hakenbeck, O. Dideberg, and T. Vernet. 1999. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae. Role in the specificity for beta-lactam antibiotics. J. Biol. Chem. 274:19175-19180. [DOI] [PubMed] [Google Scholar]
- 15.Pernot, L., L. Chesnel, A. Le Gouellec, J. Croizé, T. Vernet, O. Dideberg, and A. Dessen. 2004. A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics. J. Biol. Chem. 279:16463-16470. [DOI] [PubMed] [Google Scholar]