Abstract
Resistance to ciprofloxacin was detected in 111 (48.1%) isolates of Klebsiella pneumoniae from China. GyrA alterations were identified in the ciprofloxacin-resistant and ciprofloxacin-susceptible isolates. The results, including previously published data, indicate that the single substitution Ser83→Ile and three types of double mutations at Ser83 and Asp87 were required for ciprofloxacin resistance (P < 0.05).
Resistance to fluoroquinolones is increasing in Klebsiella pneumoniae strains. Mechanisms of resistance to fluoroquinolones in the Enterobacteriaceae have been shown to be due primarily to alterations in gyrA, which encodes DNA gyrase, a type II topoisomerase (1, 4). The mutations are localized in an area named as the quinolone resistance-determining region (QRDR) (24). DNA sequencing of the GyrA QRDR in clinical isolates showed some alterations associated with fluoroquinolone resistance in K. pneumoniae (8, 11, 24).
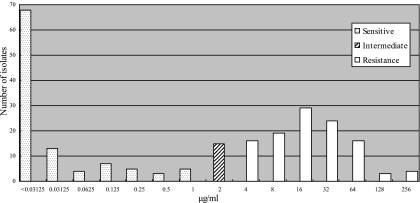
In this paper, 231 consecutive, nonrepetitive isolates of K. pneumoniae were collected from inpatients in three tertiary hospitals in Harbin, the capital city of Heilongjiang Province, between May 2005 and March 2006. The strains were identified with the API 20E system (bioMérieux, Marcy l'Etoile, France) and confirmed as being nonduplicated by randomly amplified polymorphism DNA analysis. MICs of ciprofloxacin and nalidixic acid (Sigma-Aldrich, Inc., St. Louis, MO) were determined by the agar dilution method with Mueller-Hinton agar (BBL Microbiology Systems, Cockeysville, MD) as recommended by the CLSI (formerly NCCLS) (15). The MIC50 and MIC90 of ciprofloxacin were 2 μg/ml and 32 μg/ml, respectively. According to CLSI criteria, 105 (45.5%) and 15 (6.5%) isolates were susceptible (MIC ≤ 1 μg/ml) and intermediately resistant (MIC, 1 to 4 μg/ml) to ciprofloxacin, respectively; 111 isolates (48.1%) had an MIC greater than the breakpoint (MIC ≥ 4 μg/ml) (Fig. 1). Rates of isolation of ciprofloxacin-resistant K. pneumoniae strains in the United States increased from 12.9% in 1991 to 35.6% in 2005 (21). Decreased susceptibility was also found in Europe (1). In China, it was reported that the percentage of ciprofloxacin resistance rose from 2% in 1994 to 18% in 2000 (25). However, a higher percentage of ciprofloxacin resistance was found in this study.
FIG. 1.
Distribution of MICs of ciprofloxacin (n = 231). MICs were determined by the agar dilution method. One hundred eleven isolates (48.1%) were resistant to ciprofloxacin, among which 16, 19, 29, 24, 16, 3, and 4 isolates had MICs of 4 μg/ml, 8 μg/ml, 16 μg/ml, 32 μg/ml, 64 μg/ml, 128 μg/ml, and 256 μg/ml, respectively; 105 isolates (45.5%) were susceptible to ciprofloxacin, with MICs ranging from <0.03125 μg/ml to 1 μg/ml; and 15 (6.5%) were intermediately resistant (MIC, 1 to 4 μg/ml).
To investigate the characteristics of GyrA alterations, gyrA gene fragments were amplified and sequenced in 33 randomly selected isolates representing a range of ciprofloxacin MICs. Primers gyrA-F (5′-TGCGAGAGAAATTACACC), corresponding to positions 299 to 316, and gyrA-R (5′-AATATGTTCCATCAGCCC), complementary to nucleotides 906 to 923 of the K. pneumoniae sequence (GenBank accession number X16817), were used to amplify the gyrA gene fragments with bacterial lysate as a template as described previously (24). PCR products were then sequenced in both directions by use of an ABI 373 automated DNA sequencer (Applied Biosystems, Foster City, CA) with the same primers used for PCR amplification. The nucleotide sequences and the deduced amino acid were compared with that of K. pneumoniae ATCC 13883 (GenBank accession number DQ673325) using the online ClustalW2 multiple sequence alignment program.
Among 33 isolates selected, 27 were revealed to have amino acid alterations in GyrA (Table 1). Isolates that were resistant to ciprofloxacin were also resistant to nalidixic acid, while 3 out of 12 ciprofloxacin-susceptible isolates displayed resistance to nalidixic acid. Twenty-one isolates presented Ser83 changes. The most common mutation was Ser83→Leu, which was present in 13 isolates. A Ser83→Ile substitution was found in six isolates; also, one Tyr substitution and one Thr substitution were found in ciprofloxacin-susceptible isolates. It is notable that almost all the ciprofloxacin-resistant isolates had substitutions at Ser83 by Leu or Ile, and all of the Ser83→Leu changes were combined with Asp87→Asn, which is consistent with data from previous reports (1, 2, 20).
TABLE 1.
Alterations in GyrA and MICs of quinolones in 33 Chinese clinical isolates of K. pneumoniae
| Strain | MIC (μg/ml)
|
Amino acid change at position:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Ciprofloxacin | Nalidixic acid | Ser83 | Asp87 | Arg154 | Ala171 | Gly177 | Leu187 | Val198 | |
| Resistant to ciprofloxacin | |||||||||
| 95 | 128 | >512 | Leu | Asn | Ser | Ile | |||
| 863 | 128 | >512 | Leu | Asn | Ser | Ile | |||
| 149 | 64 | >512 | Leu | Asn | Ser | Ile | |||
| 103 | 64 | >512 | Leu | Asn | Ser | Ile | |||
| 663 | 64 | >512 | Leu | Asn | Ser | Ile | |||
| 94 | 32 | >512 | Leu | Asn | Ser | Ile | |||
| 779 | 32 | >512 | Leu | Asn | Ser | Ile | |||
| 234 | 16 | >512 | Leu | Asn | Ser | Ile | |||
| 3 | 16 | >512 | Leu | Asn | Ser | Ile | |||
| 27 | 8 | >512 | Leu | Asn | Ser | Ile | |||
| 719 | 8 | >512 | Leu | Asn | Ser | Ile | |||
| 205 | 4 | >512 | Leu | Asn | Ser | Ile | |||
| 769 | 32 | >512 | Ile | Arg | |||||
| 828 | 32 | >512 | Ile | Arg | |||||
| 827 | 32 | >512 | Ile | ||||||
| 772 | 32 | >512 | Ile | ||||||
| 721 | 8 | >512 | Ser | Ile | Ile | ||||
| 836 | 4 | >512 | |||||||
| Intermediate resistant to ciprofloxacin | |||||||||
| 753 | 2 | >512 | Ile | ||||||
| 21 | 2 | 16 | |||||||
| 715 | 2 | 16 | |||||||
| Susceptible to ciprofloxacin | |||||||||
| 840 | 1 | 16 | Ser | Ile | Ile | ||||
| 685 | 0.125 | >512 | Ser | Ile | Ile | ||||
| 212 | <0.03125 | 16 | Ser | Ile | Ile | ||||
| 517 | <0.03125 | 8 | Ser | Ile | Ile | ||||
| 838 | 1 | 256 | Tyr | Ser | Ile | Ile | |||
| 49 | 0.5 | >512 | Asn | Ser | Ser | ||||
| 182 | <0.03125 | 4 | Thr | Ser | |||||
| 737 | <0.03125 | 8 | Leu | Ile | |||||
| 724 | <0.03125 | 8 | Ile | ||||||
| 738 | 1 | 8 | |||||||
| 577 | 0.125 | 4 | |||||||
| 760 | <0.03125 | 4 | |||||||
However, comparable with the mutations involving substitutions of Ser83 with Phe, Tyr, or Ile and Asp87 alterations reported in Japanese (4), American (24), and European (11) isolates, a large proportion (12 out of 18 [66.7%]) of Chinese fluoroquinolone-resistant K. pneumoniae isolates demonstrated Ser83→Leu together with Asp87→Asn (Table 1). Although Ser83→Leu is frequently displayed in Escherichia coli (17), the results from China (this study) and Singapore (20) suggested the existence of this alteration in K. pneumoniae. Moreover, most of the isolates with this predominant alteration were highly resistant to ciprofloxacin (MIC ≥ 8 μg/ml), which may be related to the higher prevalence of ciprofloxacin resistance in China. Besides, changes outside the QRDR, such as Ala171→Ser and Val198→Ile, were found in both ciprofloxacin-susceptible and -resistant isolates (Table 1).
DNA sequencing of GyrA in clinical strains has revealed some mutations in the QRDR associated with fluoroquinolone resistance. However, QRDR alterations were also found in isolates susceptible to ciprofloxacin in this study and others (8, 11, 13, 19, 24). In order to explore the role of individual alteration types found in K. pneumoniae in ciprofloxacin resistance, alterations in Ser83 and Asp87 of GyrA were reviewed, based on articles found in the PubMed database, and the association between ciprofloxacin resistance and the individual alteration was analyzed by means of the SPSS 13.0 statistical package using Fisher's exact test or Pearson chi-square test. In total, types of GyrA alterations carried by 138 strains were found among 185 isolates of K. pneumoniae with an exact MIC, which included 152 strains in 11 published articles and 33 isolates in this study (Table 2).
TABLE 2.
Amino acid changes in the GyrA QRDR of K. pneumoniae
| Susceptibility to ciprofloxacin | Amino acid change at position:
|
No. of isolates | Reference(s) and/or source | |
|---|---|---|---|---|
| 83 | 87 | |||
| Resistant | 9 | 2, 18; this study | ||
| Tyr | 15 | 1, 8, 11, 14, 20, 24 | ||
| Phe | 10 | 1, 2, 24 | ||
| Ile | 7 | 1, 2, 20; this study | ||
| Phe | Asn | 17 | 1, 2, 4, 5, 6, 11 | |
| Leu | Asn | 13 | 20; this study | |
| Tyr | Asn | 10 | 20, 24 | |
| Phe | Tyr | 4 | 1, 11 | |
| Phe | Ala | 3 | 4, 5, 6 | |
| Phe | Gly | 3 | 4, 5, 24 | |
| Tyr | Tyr | 2 | 1, 2 | |
| Ile | Asn | 1 | 20 | |
| Intermediate resistant | 1 | 18 | ||
| Tyr | 14 | 1, 4, 5, 8, 20 | ||
| Phe | 5 | 1, 4, 5 | ||
| Ile | 1 | This study | ||
| Gly | 3 | 4, 5; this study | ||
| Phe | Gly | 1 | 4, 5 | |
| Susceptible | 37 | 4, 5,1, 4, 5, 11, 14, 18, 25; this study | ||
| Tyr | 17 | 4, 5,4, 5, 8, 11, 20; this study | ||
| Phe | 7 | 4, 5,4, 5, 11, 18, 24 | ||
| Leu | 1 | This study | ||
| Ile | 1 | This study | ||
| Thr | 1 | This study | ||
| Gly | 1 | 4, 5 | ||
| Asn | 1 | This study | ||
| Total | 185 | |||
Although seven types of single alterations were detected in 84 strains, only Ser83→Ile was distributed differently between the ciprofloxacin-resistant and ciprofloxacin-susceptible isolates (P < 0.005), with the Ile substitution occurring more frequently in the former group. The distribution of other single substitutions such as Ser83→Tyr, Ser83→Phe, and Ser83→Leu showed no statistical differences between the two groups. Eight types of double mutations involving both Ser83 and Asp87 were found exclusively in 54 ciprofloxacin-resistant isolates; however, only three types of double mutations, Ser83→Phe plus Asp87→Asn, Ser83→Leu plus Asp87→Asn, and Ser83→Tyr plus Asp87→Asn, were associated with ciprofloxacin resistance (P < 0.05). Thus, the three types of double mutations and the single mutation Ser83→Ile are required for ciprofloxacin resistance in K. pneumoniae. Also, most of the isolates carrying such mutations had MICs exceeding 16 μg/ml, which indicates that these alterations in GyrA are prone to conferring high-level resistance to ciprofloxacin. The resistance phenotype of isolates with the “silent” alterations (mutations having no statistical association with ciprofloxacin resistance) may be attributed to other factors affecting antibiotic susceptibility, such as a change in the penetration of agents resulting from energy-dependent efflux and porin loss (11, 12), differential expression of a resistant gene (7), and activities of regulatory loci like mar and sox, which induce decreased porin expression and increased efflux (9, 10, 16).
In summary, we found the single mutation Ser83→Ile and the double mutation Ser83→Leu plus Asp87→Asn to be associated with ciprofloxacin resistance in China. By reviewing all the alterations in the GyrA QRDR, we demonstrated that a single change, Ser83→Ile, and three types of double alterations, Ser83→Phe plus Asp87→Asn, Ser83→Leu plus Asp87→Asn, and Ser83→Ile plus Asp87→Asn, were required for ciprofloxacin resistance. These results suggest that effectivity of a certain mutation should be considered when studying the alterations of GyrA associated with ciprofloxacin resistance.
Nucleotide sequence accession numbers.
The partial sequences of the variant gyrA genes in clinical isolates of K. pneumoniae have been submitted to the GenBank database under accession numbers EU430280 through EU430289.
Acknowledgments
This work was supported in part by grants from the NSFC (J0730858 and 30700032), the China Postdoctoral Science Foundation (20070410913), Key Technologies R&D Program of China (2004BA720A09-02), the Outstanding Youth Foundation of Heilongjiang Province (JC04-05), and the Heilongjiang Provincial Postdoctoral Science Foundation.
Footnotes
Published ahead of print on 27 May 2008.
REFERENCES
- 1.Brisse, S., D. Milatovic, A. C. Fluit, J. Verhoef, N. Martin, S. Scheuring, K. Köhrer, and F. L. Schmitz. 1999. Comparative in vitro activities of ciprofloxacin, clinafloxacin, gatifloxacin, levofloxacin, moxifloxacin, and trovafloxacin against Klebsiella pneumoniae, Klebsiella oxytoca, Enterobacter cloacae, and Enterobacter aerogenes clinical isolates with alterations in GyrA and ParC proteins. Antimicrob. Agents Chemother. 43:2051-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brisse, S., and J. Verhoef. 2001. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 51:915-924. [DOI] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Deguchi, T., A. Fukuoka, M. Yasuda, M. Nakano, S. Ozeki, E. Kanematsu, Y. Nishino, S. Ishihara, Y. Ban, and Y. Kawada. 1997. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 41:699-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deguchi, T., M. Yasuda, T. Kawamura, M. Nakano, S. Ozeki, E. Kanematsu, Y. Nishino, and Y. Kawada. 1997. Improved antimicrobial activity of DU-6859a, a new fluoroquinolone, against quinolone-resistant Klebsiella pneumoniae and Enterobacter cloacae isolates with alterations in GyrA and ParC proteins. Antimicrob. Agents Chemother. 11:2544-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deguchi, T., T. Kawamura, M. Yasuda, M. Nakano, H. Fukuda, H. Kato, N. Kato, Y. Okano, and Y. Kawada. 1997. In vivo selection of Klebsiella pneumoniae strains with enhanced quinolone resistance during fluoroquinolone treatment of urinary tract infections. Antimicrob. Agents Chemother. 41:1609-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu, Y., F. Zhang, W. Zhang, X. Chen, Y. Zhao, J. Ma, L. Bao, W. Song, T. Ohsugi, T. Urano, and S. Liu. 2007. Differential expression of blaSHV related to susceptibility to ampicillin in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 29:344-347. [DOI] [PubMed] [Google Scholar]
- 8.Gruteke, P., W. Goessens, J. Van Gils, P. Peerbooms, N. Lemmens-Den Toom, M. Van Santen-Verheuvel, A. Van Belkum, and H. Verbrugh. 2003. Patterns of resistance associated with integrons, the extended-spectrum β-lactamase SHV-5 gene, and a multidrug efflux pump of Klebsiella pneumoniae causing a nosocomial outbreak. J. Clin. Microbiol. 41:1161-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kern, W. V., M. Oethinger, A. S. Jellen-Ritter, and S. B. Levy. 2000. Nontarget gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 44:814-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linde, H. J., F. Notka, C. Irtenkauf, J. Decker, J. Wild, H. H. Niller, P. Heisig, and N. Lehn. 2002. Increase in MICs of ciprofloxacin in vivo in two closely related clinical isolates of Enterobacter cloacae. J. Antimicrob. Chemother. 49:625-630. [DOI] [PubMed] [Google Scholar]
- 11.Martínez-Martínez, L., A. Pascual, C. Conejo Mdel, I. García, P. Joyanes, A. Doménech-Sánchez, and V. J. Benedí. 2002. Energy-dependent accumulation of norfloxacin and porin expression in clinical isolates of Klebsiella pneumoniae and relationship to extended-spectrum β-lactamase production. Antimicrob. Agents Chemother. 46:3926-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Martínez, L., A. Pascual, S. Hernandez-Alles, D. Alvarez-Diaz, A. I. Suarez, J. Tran, V. J. Benedi, and G. A. Jacoby. 1999. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Martínez, L., I. García, S. Ballesta, V. J. Benedí, S. Hernández-Allés, and A. Pascual. 1998. Energy-dependent accumulation of fluoroquinolones in quinolone-resistant Klebsiella pneumoniae strains. Antimicrob. Agents Chemother. 42:1850-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mery De la Fuente, C. M., S. P. Dauros, T. H. Bello, Y. M. Domínguez, M. S. Mella, A. M. Sepúlveda, Z. R. Zemelman, and R. G. González. 2007. Mutations in gyrA and gyrB genes among strains of gram-negative bacilli isolated from Chilean hospitals and their relation with resistance to fluoroquinolones. Rev. Med. Chil. 135:1103-1110. [DOI] [PubMed] [Google Scholar]
- 15.NCCLS. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. NCCLS, Wayne, PA.
- 16.Oethinger, M., I. Podglajen, W. V. Kern, and S. B. Levy. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents Chemother. 42:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piddock, L. J. 1999. Mechanisms of fluoroquinolone resistance: an update 1994-1998. Drugs 58:11-18. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Martínez, J. M., C. Velasco, I. García, M. E. Cano, L. Martínez-Martínez, and A. Pascual. 2007. Mutant prevention concentrations of fluoroquinolones for Enterobacteriaceae expressing the plasmid-carried quinolone resistance determinant qnrA1. Antimicrob. Agents Chemother. 51:2236-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sader, H. S., A. T. Ferreira, I. I. Tosin, A. C. Gales, L. S. Keim, J. M. Carbadillo, S. J. Mello, Jr., and W. Tavares. 1998. Piperacillin/tazobactam: evaluation of its in vitro activity against bacteria isolated in two Brazilian hospitals and an overview of its antibacterial activity, pharmacokinetic properties and therapeutic potential. Braz. J. Infect. Dis. 2:241-255. [PubMed] [Google Scholar]
- 20.Schneiders, T., S. G. Amyes, and S. B. Levy. 2003. Role of AcrR and ramA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob. Agents Chemother. 47:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strahilevitz, J., D. Engelstein, A. Adler, V. Temper, A. E. Moses, C. Block, and A. Robicsek. 2007. Changes in qnr prevalence and fluoroquinolone resistance in clinical isolates of Klebsiella pneumoniae and Enterobacter spp. collected from 1990 to 2005. Antimicrob. Agents Chemother. 51:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Reference deleted.
- 24.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, R., K. Eggleston, V. Rotimi, and R. J. Zeckhauser. 7 April 2006, posting date. Antibiotic resistance as a global threat: evidence from China, Kuwait and the United States. Global Health 2:6. doi: 10.1186/1744-8603-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]