Abstract
Several different species of clostridia reduced U(VI) to U(IV) to various degrees. The optimal pH for U(VI) reduction is 5 to 6 in most cases; a Clostridium sp. showed the highest rate at pH 4. Nitrate did not affect U(VI) reduction, indicating that this process in clostridia is nitrate independent.
Previously, we showed that a Clostridium sp. (ATCC 53464) and Clostridium sphenoides (ATCC 19403) can reduce uranyl-nitrate, -acetate, and -citrate complexes as well as complexes of other metals, implying that the reduction of uranyl compounds is a general property of this genus (4-6). Apparently, Clostridium is one of major players in uranium reduction in situ in acidic (pH 4) uranium mine pit water (11, 13), at a military facility near Chesapeake Bay, MD (1), and at the ORFRC at Oak Ridge, TN (8). In this study, we investigated the abilities of several strains of clostridia to reduce uranium(VI) under fermentative conditions, viz., the Clostridium sp. (ATCC 53464) that we isolated in our laboratory and C. sphenoides (ATCC 19403), Clostridium acetobutylicum (ATCC 824), and Clostridium pasteurianum (ATCC 7040).
All strains were grown anaerobically in mineral salts medium (4) containing glucose, but only C. sphenoides also metabolized citric acid. The fermentation products, including organic acids and gases (hydrogen and carbon dioxide), were analyzed by high-performance liquid chromatography and gas chromatography. Analysis of culture medium showed that all the strains produced acetic acid and butyric acid. To stabilize the pH of the culture as needed, we added 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.8] or 50 mM MES (morpholineethanesulfonic acid; pH 6.2) to the medium. We removed 4-milliliter aliquots of the culture and measured change in pH and turbidity at 600 nm. Total protein was determined by using a Micro Lowry total protein kit (Sigma) (10).
Ten milliliters of the culture at the late log phase was transferred to an anaerobic, preautoclaved 20-ml serum bottle, and then 0.1 ml of a 10 mM U(VI)-nitrate solution prepared by dissolving UO2(NO3)2 in prereduced deionized water was added via a 1-ml syringe. To determine the U(VI) concentration over time, aliquots of 0.1 to 0.2 ml of the bacterial culture were taken from the serum bottle, and 5 μl of the culture was diluted in 2 ml deionized water and immediately analyzed for U(VI) by a kinetic phosphorescence analyzer (KPA-11A; Chemchek Instruments, Inc.). To assess the effect of pH on U(VI) reduction by clostridia, 10-milliliter aliquots of the bacterial culture at the late log growth phase were adjusted to the required pH values by using 1 N NaOH or HCl.
The culture sample remaining in the serum bottle after sampling for the uranium reduction assay was transferred to a centrifuge tube in the glove box. After its pH was adjusted to 11 with 1 N NaOH to precipitate the uranium, the culture was centrifuged at 10,000 rpm for 10 min, the supernatant discarded, and the pellet resuspended in 2 ml of a 10 mM citric acid solution to extract the uranium species. The solution was filtered through a 0.45-μm membrane filter, and the filtrate was analyzed by UV-visible light (UV-vis) spectrophotometry to determine the absorption spectra of uranium(VI) and -(IV).
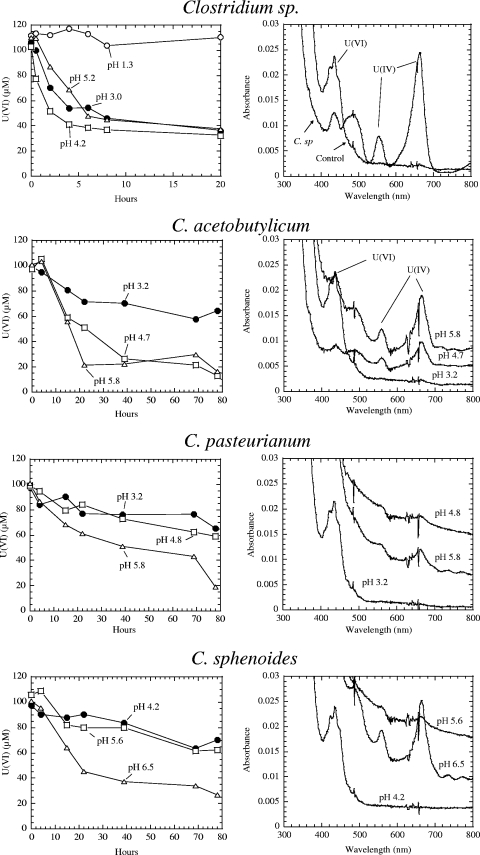
Figure 1 shows the reductions of U(VI) by clostridia at the various pHs and the respective UV-vis spectra for U(VI) (major absorption peak at 438 nm) and U(IV) (major and minor peaks at 662 nm and 560 nm, respectively). Reduction of U(VI) by the Clostridium sp. was observed at pH 3.0, 4.2, and 5.2. The rate of reduction was much faster at pH 4.2 than at pH 3 or 5.2. No reduction of U was observed at pH 1.3. In contrast, C. acetobutylicum showed a marked reduction of uranium(VI) at pH 5.8 and 4.7; at both pHs, the U(IV) peaks at both 662 and 560 nm became very pronounced. At pH 3.2, little U(VI) reduction was observed and only a U(VI) peak was visible. C. pasteurianum exhibited a moderate reduction of U(VI) to U(IV) at pH 5.8, and even at this pH, only the 662-nm peak of U(IV) was observed. Maximum reduction of U(VI) by C. sphenoides occurred at pH 6.5, and lower pHs affected the reduction. These results suggest that by and large that uranium reduction at higher pHs is more favorable for strains other than the Clostridium sp. strain (Fig. 1).
FIG. 1.
Reduction of U(VI) by clostridia and UV-vis spectra of U(VI) and U(IV) at different pHs. The initial pHs of cultures subjected to the reduction assay are given. All strains were cultured in mineral salts medium.
We determined the effect of U(VI) concentration on U reduction by the Clostridium sp. The bacterium reduced 0.6 mM U(VI) at the maximum concentration tested (Fig. 2A). The reduction of U(VI) proceeded at the same rate for about 6 h, followed by lower rates at all U(VI) concentrations (Fig. 2A). We monitored the reduction of U(VI) up to 72 h and found that not all uranium, especially at higher concentrations, was completely reduced to U(IV) (Fig. 2B). Furthermore, the increased concentration of nitrate derived from the added U(VI) source did not affect U(VI) reduction, indicating that this process in the Clostridium sp. is nitrate independent, a characteristic applicable to clostridia and not found in many other anaerobic respiratory bacteria, such as those of the genera Desulfovibrio, Geobacter, and Shewanella.
FIG. 2.
Reduction of U(VI) by the Clostridium sp. at different initial U(VI) concentrations. Uranium was added as UO2(NO3)2. The initial optical density at 600 nm of bacteria was 0.5. The U(VI) concentration is noted.
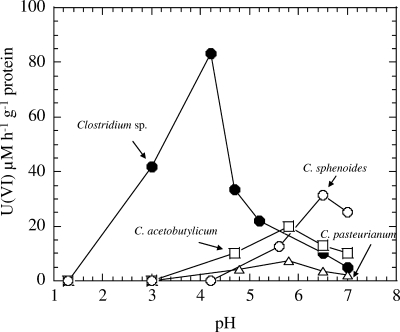
The rates of reduction of 0.1 mM of U(VI) at different pHs by clostridia are shown in Fig. 3. The optimum rates of reduction of U(VI) (μM h−1 g−1 protein) were as follows: Clostridium sp., 83.4 at pH 4.2; C. acetobutylicum, 12.4 at pH 5.8; C. pasteurianum, 7.2 at pH 5.8; and C. sphenoides, 31.3 at pH 6.5. Thus, the Clostridium sp. showed the highest U(VI) reduction, at a rate of 83.4 μM U(VI) h−1 g−1 protein at pH 4, a value that is about 200%, 400%, and 800% higher than those for C. sphenoides, C. acetobutylicum, and C. pasteurianum, respectively, at their optimal pHs of 5.8, 5.8, and 6.5.
FIG. 3.
Rates of reduction of U(VI) added as uranyl nitrate by clostridia at different pHs.
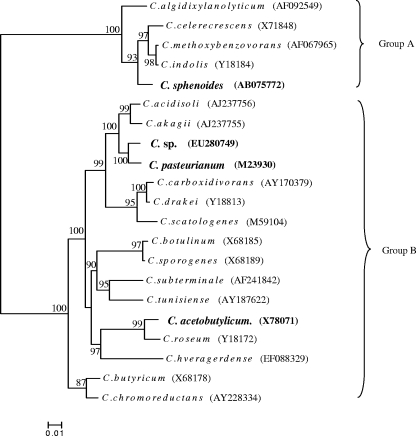
We identified that the Clostridium sp. was the closest phenotypically to C. pasteurianum (unpublished data). Like C. pasteurianum, the Clostridium sp. can fix nitrogen. By a partial 16S rRNA gene sequencing, we obtained further evidence verifying their phylogeny affiliations. The total DNAs of strains were isolated as described previously (20). The 16S rRNA genes were amplified from isolates with the PCR primers FD1 (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1540R (5′-AAG GAG GTG ATC CAG CC-3′) (17, 19). The DNA sequences were determined with a BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems), using a 3700 DNA analyzer (Perkin-Elmer) and the primer 529R (5′-CGC GGC TGC TGG CAC-3′) (19). We compared the obtained sequences against those in GenBank through the NCBI Internet web service and aligned them with ClustalW (15). Phylogenetic analyses of 16S rRNA gene sequences were undertaken using MEGA version 4.0 (14). A phylogeny tree was constructed by a neighbor-joining algorithm under a ρ distance model, with the bootstrap computed 500 times. Based on a DNA sequence of about 400 bp, we constructed a phylogeny tree containing all the strains used in this study and some other clostridia (Fig. 4). This phylogeny tree shows that only C. sphenoides is classified into group A, while all other three strains that we explored fall into group B.
FIG. 4.
Phylogenetic relationships between strains used in this study (in bold) and other clostridia. Bootstrap (n = 500) values above 50 are shown. GenBank accession numbers are in parentheses.
The phylogeny analysis agreed with a previous phenotypic analysis showing that the Clostridium sp. is closest to C. pasteurianum. The partial 16S rRNA sequence of the Clostridium sp. is 98% identical to that of C. pasteurianum. Nevertheless, the difference between the two is evident in terms of uranium reduction. Similarly, although Clostridium butyricum is phylogenetically related to a novel electrochemically active and Fe(III)-reducing bacterium, it does not itself necessarily possess the same properties (9). Given the plasticity of the bacterial genome, which is subject to frequent recombination and reconstruction (12), it is not surprising to observe differences in some characteristics of the Clostridium sp. and C. pasteurianum.
With their widespread occurrence in soils, sediments, and low-level radioactive wastes, clostridia could play a significant role in the in situ reduction of uranium and other metals, particularly at acidic pHs and in nitrate-rich environments (1, 2, 7, 8, 11, 13). We found that all of the clostridia tested not only reduced uranium but differed in extent of reduction and that the pH of the medium strongly influences the reduction. The Clostridium sp., which was isolated from an acidic metal-contaminated site (3), has higher tolerance to harsh environmental conditions, such as low pH, and a stronger ability for fermentation and reduction of U(VI) and other metals in acidic environments.
Although the mechanisms of U(VI) reduction by Desulfovibrio, Geobacter, and Shewanella species are known (16, 18), the mechanisms of uranium reduction by clostridia remain unclear. Our previous study showed that it is an enzymatic process, since it occurred only in the presence of growing or resting cells; neither the organic acid metabolites generated, the extracellular components of the culture, nor heat-killed cells could reduce uranium anaerobically (4). Other conjectures were that the reducing power generated from glucose fermentation caused uranium reduction. Thus, Petrie at al. (11) demonstrated that glucose amendment to the growth medium enriched the numbers of gram-positive, spore-forming bacteria and, since some of the highest rates of U(VI) reduction in situ occurred upon amendment, that fermentative processes were involved (7). Our results herein show that optimizing the conditions for fermentation resulted in better U(VI) reduction, supporting this hypothesis.
Acknowledgments
We thank C. J. Dodge for assistance and Avril D. Woodhead for editorial help.
This research was funded by the Environmental Remediation Sciences Program, Environmental Remediation Sciences Division, Office of Biological and Environmental Research (OBER), Office of Science, U.S. Department of Energy, under contract no. DE-AC02-98CH10886.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Dong, W., G. Xie, T. R. Mille, M. P. Franklin, T. P. Oxenberg, E. J. Bouwer, W. P. Ball, and R. U. Halden. 2006. Sorption and bioreduction of hexavalent uranium at a military facility by the Chesapeake Bay. Environ. Pollut. 142:132-142. [DOI] [PubMed] [Google Scholar]
- 2.Francis, A. J., S. Dobbs, and B. Nine. 1980. Microbial activity of trench leachates from shallow-land, low-level radioactive waste disposal sites. Appl. Environ. Microbiol. 40:108-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francis, A. J., and C. J. Dodge. 1988. Anaerobic microbial dissolution of transition and heavy metal oxides. Appl. Environ. Microbiol. 54:1009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis, A. J., C. J. Dodge, F. Lu, G. P. Halada, and C. R. Clayton. 1994. XPS and XANES studies of uranium reduction by Clostridium sp. Environ. Sci. Technol. 28:636-639. [DOI] [PubMed] [Google Scholar]
- 5.Francis, A. J., C. J. Dodge, and G. E. Meinken. 2002. Biotransformation of pertechnetate by Clostridia. Radiochim. Acta 90:791-797. [Google Scholar]
- 6.Francis, A. J., G. A. Joshi-Tope, C. J. Dodge, and J. B. Gillow. 2002. Biotransformation of uranium and transition metal citrate complexes by Clostridia. J. Nucl. Sci. Technol. 3(Suppl.):935-938. [Google Scholar]
- 7.Istok, J. D., J. M. Senko, L. R. Krumholz, D. Watson, M. A. Bogle, A. Peacock, Y. J. Chang, and D. C. White. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Tech. 38:468-475. [DOI] [PubMed] [Google Scholar]
- 8.Madden, A. S., A. C. Smith, D. V. Balkwill, L. A. Fagan, and T. J. Phelps. 2007. Microbial uranium immobilization independent of nitrate reduction. Environ. Microbiol. 9:2321-2330. [DOI] [PubMed] [Google Scholar]
- 9.Park, H. S., B. H. Kim, H. S. Kim, H. J. Kim, G. T. Kim, M. Kim, I. S. Chang, Y. K. Park, and H. I. Chang. 2001. A novel electrochemically active and Fe(III)-reducing bacterium phylogenetically related to Clostridium butyricum isolated from a microbial fuel cell. Anaerobe 7:297-306. [Google Scholar]
- 10.Peterson, G. L. 1977. Simplification of protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83:346-356. [DOI] [PubMed] [Google Scholar]
- 11.Petrie, L., N. N. North, S. L. Dollhpf, D. L. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuster, S. C., and G. Gottschalk. 2005. Microbial genomics in its second decade. Curr. Opin. Microbiol. 8:561-563. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl. Environ. Microbiol. 69:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 15.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wall, J. D., and L. R. Krumholz. 2006. Uranium reduction. Annu. Rev. Microbiol. 60:149-166. [DOI] [PubMed] [Google Scholar]
- 17.Weisburg, W. W., S. M. Barns, J. S. Nickels, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkins, M. J., F. R. Livens, D. J. Vaughan, and J. R. Lloyd. 2006. The impact of Fe(III)-reducing bacteria on uranium mobility. Biogeochemistry 78:125-150. [Google Scholar]
- 19.Ye, Q., Y. Roh, S. L. Carroll, B. Blair, J. Zhou, C. L. Zhang, and M. W. Fields. 2004. Alkaline anaerobic respiration: isolation and characterization of a novel alkaliphilic and metal-reducing bacterium. Appl. Environ. Microbiol. 70:5595-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou, J. Z., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-332. [DOI] [PMC free article] [PubMed] [Google Scholar]