Abstract
Two-dimensional polyacrylamide gel electrophoresis (2D PAGE), in combination with matrix-assisted laser desorption ionization-time of flight analysis, and the recently revealed genome sequence of Ralstonia eutropha H16 were employed to detect and identify proteins that are differentially expressed during different phases of poly(3-hydroxybutyric acid) (PHB) metabolism. For this, a modified protein extraction protocol applicable to PHB-harboring cells was developed to enable 2D PAGE-based proteome analysis of such cells. Subsequently, samples from (i) the exponential growth phase, (ii) the stationary growth phase permissive for PHB biosynthesis, and (iii) a phase permissive for PHB mobilization were analyzed. Among several proteins exhibiting quantitative changes during the time course of a cultivation experiment, flagellin, which is the main protein of bacterial flagella, was identified. Initial investigations that report on changes of flagellation for R. eutropha were done, but 2D PAGE and electron microscopic examinations of cells revealed clear evidence that R. eutropha exhibited further significant changes in flagellation depending on the life cycle, nutritional supply, and, in particular, PHB metabolism. The results of our study suggest that R. eutropha is strongly flagellated in the exponential growth phase and loses a certain number of flagella in transition to the stationary phase. In the stationary phase under conditions permissive for PHB biosynthesis, flagellation of cells admittedly stagnated. However, under conditions permissive for intracellular PHB mobilization after a nitrogen source was added to cells that are carbon deprived but with full PHB accumulation, flagella are lost. This might be due to a degradation of flagella; at least, the cells stopped flagellin synthesis while normal degradation continued. In contrast, under nutrient limitation or the loss of phasins, cells retained their flagella.
Ralstonia eutropha H16 is a gram-negative, rod-shaped, and facultatively chemolithoautotrophic hydrogen-oxidizing bacterium that serves as a model organism for polyhydroxyalkanoate (PHA) metabolism. PHAs serve as storage compounds for carbon and energy and are synthesized under unbalanced growth conditions if a carbon source is present in excess and if another macroelement (N, O, P, or S) is depleted at the same time. In addition to the interest of academia, the bacterium has been used in industry for large-scale production of PHAs. These biopolyesters reveal thermoplastic and/or elastomeric properties similar to those of synthetic polymers produced from petrochemicals, like polypropylene (26, 32, 54). Due to their biodegradability and origin from renewable resources, PHAs have attracted much interest for technical and medical applications (3, 20, 62). PHAs are synthesized and accumulated by a large variety of prokaryotes and may represent the major cell constituent, contributing up to about 90% of the cell dry weight (4). Although R. eutropha H16 is able to synthesize different PHAs with short carbon chain lengths (55), poly(3-hydroxybutyric acid) (PHB) is usually the predominant PHA in R. eutropha H16 (12, 23; P. A. Holmes, L. F. Wright, and S. H. Collins, 1981, European patent application 0052459). The synthesis of PHB proceeds in three steps involving the enzymes β-ketothiolase (PhaA), acetoacetyl-coenzyme A reductase (PhaB), and PHA synthase (PhaC) (17, 18, 33). The genes for these three enzymes are located in the PHA operon (phaCAB). Furthermore, several additional genes coding for proteins participating in PHA metabolism are known. Whereas PhaC is essential for PHA biosynthesis in R. eutropha H16, PhaA and PhaB can be replaced by isoenzymes. PHAs are degraded by PHA depolymerases (PhaZ) through hydrolytic or thiolytic cleavage (57). In contrast to extracellular degradation, intracellular degradation of PHAs is far less understood. In R. eutropha H16, seven genes putatively encoding intracellular PHA depolymerases have been identified. Of these enzymes, five depolymerases (encoded by phaZ1 to phaZ5) contain a DepA domain (7, 45, 50, 65) and two different PHA depolymerases (encoded by phaZ6 and phaZ7) share an LpqC domain (1). In addition, two hydroxybutyrate oligomer hydrolase genes were detected (7, 22, 46). At least three of these genes contribute to the intracellular degradation of PHB in R. eutropha H16 (62). PHB synthesis enzymes are constitutively expressed in the bacterium. Therefore, a strict regulation of intracellular PHA depolymerases is required to avoid a futile cycle with simultaneous synthesis and degradation of the polymer. The mechanism of this regulation is still unclear. When the limiting macroelement that caused PHB accumulation is supplied again, degradation (mobilization) of PHB is induced, and the storage compound is used as a carbon and energy source.
PHB is accumulated as granules in the cytoplasm of cells. At the surfaces of PHB granules, four different types of proteins are bound: (i) PHA synthases (PhaC), (ii) intracellular PHA depolymerases (PhaZ), (iii) phasins (PhaP), and (iv) a regulator of phasin expression. Phasins are considered a class of structural proteins and consist of at least one hydrophobic domain, which binds to the surfaces of PHB granules, and hydrophilic or amphiphilic domains, which are exposed to the cytoplasm. This layer of phasins stabilizes the granules, thus preventing the coalescence of granules and also binding of cytosolic proteins to the hydrophobic granule surface (60). In R. eutropha H16, four genes for phasin homologues occur, all of which are transcribed under conditions permissive for PHB synthesis. PhaP1 is the predominant phasin (37). Biosynthesis of at least PhaP1 is controlled by the autoregulative transcriptional repressor PhaR (36).
R. eutropha H16 exhibits peritrichous flagellation. While most cells appear to be strongly flagellated in the early exponential growth phase, the number of flagellated cells decreases during the course of the exponential phase (4). However, flagellation in the stationary growth phase and a putative linkage to PHB metabolism have not been investigated. Flagellin is encoded by the fliC gene and represents the main structural protein of bacterial flagella (28). Changes in flagellation have been reported for only a few microorganisms, for example, the peritrichously flagellated Escherichia coli (39, 40) and the polarly flagellated Pseudomonas aeruginosa (13, 14). These changes occur in response to environmental changes, such as variations in the availability of nutrients and macroelements. The polarly flagellated Caulobacter crescentus is another example of an organism that shows changes in flagellation; during its unique cell division cycle, it divides asymmetrically, producing daughter cells with different polar structures and differing cell fates, where initiation of flagellar assembly is closely tied to cell division (30).
Flagellar and chemotaxis genes are usually organized as clusters, but nonflagellar genes tend to be inserted within these clusters, as well. The numbers and organization of clusters vary in different bacteria. The transcription of all flagellar genes proceeds hierarchically to ensure that each protein is synthesized when needed in the assembly process of flagella (42). Although different mechanisms of regulation of flagella and motility genes have been found in bacteria, three general hierarchical schemes of flagellar-gene expression have been identified, each possessing a master regulator (class I) controlling the expression of all other flagellar genes: the Enterobacteriaceae system (master regulator, FlhD/FlhC) (2, 10), the C. crescentus system (master regulator, CtrA) (30), and the scheme of the Vibrio/pseudomonad group (FlrA) (41). In addition to these master regulator proteins, sigma factors are required for activation of class II genes. Enteric bacteria use the housekeeping sigma factor σ70, and similarly, C. crescentus needs σ73. In contrast, the Vibrio/pseudomonad group requires the alternative sigma factor σ54 for transcription of class II genes. RNA polymerases containing σ54 are able to bind to promoters, but activation of transcription is dependent on the availability of an upstream enhancer. Therefore, σ54-dependent promoters are strictly regulated. At least in the close relatives of R. eutropha, the pseudomonads, the activities of a subset of these enhancers are controlled by phosphorylation in response to environmental signals.
In addition to flagellar genes, several regulatory genes belong to class II. In enteric bacteria, fliA encodes the flagellum-specific sigma factor σ28, which mediates the expression of late flagellar genes. For expression of genes of one subset of class III (class IIIa), FlhD/FlhC are also necessary, whereas the class IIIb subset does not require these proteins. Interestingly, some of the class II genes are activated by σ28 themselves. Class II of C. crescentus possesses a two-component signal transduction pathway (FlbED). The phosphorylated form of the response regulator FlbD is essential to activate σ54 for transcription of class III and class IV genes in this organism. Class II of the Vibrio/pseudomonad group comprises both fliA and a two-component signal transduction pathway (flrBC or homologues). The transcription of class III genes is regulated by the phosphorylated form of the response regulator FlrC, which activates σ54. In contrast, expression of class IV requires σ28 (42).
The aim of this study was to analyze changes in the proteome of R. eutropha H16 from the growth phase to the PHB accumulation phase and the PHB mobilization phase. Proteome analysis is a direct measurement of proteins in terms of their presence and relative abundance, providing an accurate picture of the state of a living cell (61). Two-dimensional polyacrylamide gel electrophoresis (2D PAGE) is the method of choice to analyze the proteome for differentially expressed proteins (15). Since the R. eutropha H16 database is available (35), protein spots from 2D gels were analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) and allocated to their coding genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
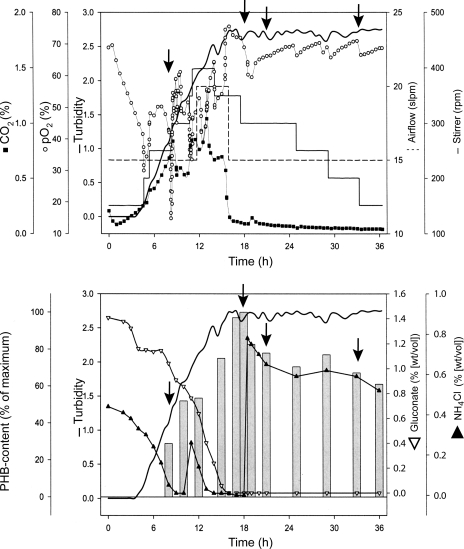
The bacterial strains used in this study are listed in Table 1. In the first cultivation (36-h) experiment, cells of R. eutropha were grown in a fed-batch mode (for detailed cultivation parameters, see Fig. 2) in a Biostat D650 fermentor. The culture volume was 400 liters. The mineral salts medium (MM) used contained 1.5% (wt/vol) sodium gluconate as a carbon source. NH4Cl served as a nitrogen source at an initial concentration of 0.05% (wt/vol) (see Fig. 2). The oxygen concentration was measured to allow reaction to a decrease in the partial O2 pressure by an increase in the airflow and stirring rate to avoid growth limitation by the macroelement. Adjustments of the airflow and the stirring rate were documented, as well (see Fig. 2). After 11 h, a small amount of NH4Cl was fed to increase the cell concentration. Within the late exponential and early stationary phases, large amounts of PHB were synthesized. After 18 h of cultivation, ammonium was added to the medium to induce the degradation of stored PHB granules to mobilize this intracellular carbon source. For long-term cultivation (72 h), cells were grown in 2.5-liter Erlenmeyer flasks equipped with baffles at 30°C in 500 ml MM supplemented with 1% (wt/vol) sodium gluconate (47). To promote extensive accumulation of PHB, the concentration of NH4Cl in MM was 0.1% (wt/vol) for these long-term cultivations. Alternatively, cells were grown for 72 h in tryptic soy broth (TSB) (Becton Dickinson; 2.7% [wt/vol]), and additionally with 0.5% (wt/vol) sodium gluconate, to suppress PHB accumulation in the stationary phase.
TABLE 1.
Strains of Ralstonia eutropha used in this study
| R. eutropha strain | Relevant phenotype or genotype | Reference or source |
|---|---|---|
| H16 | Wild type | DSMa 428 |
| Re 1052 | ΔphaP1 | 64 |
| ΔphaP2 ΩKm | ΔphaP2 | M. Pötter |
| ΔphaP3 | ΔphaP3 | 38 |
| ΔphaP4 | ΔphaP4 | 38 |
| ΔphaP1 ΔphaP2 ΩKm | ΔphaP1 ΔphaP2 | 38 |
| ΔphaP1 ΔphaP2 ΔphaP3 | ΔphaP1 ΔphaP2 ΔphaP3 | 38 |
| ΔphaP1 ΔphaP2 ΔphaP3 ΔphaP4 | ΔphaP1 ΔphaP2 ΔphaP3 ΔphaP4 | 38 |
| PHB−4 | Mutant of H16; PHB− | DSM 541; 48 |
| HF09 | ΔrpoN | 44 |
DSM, Deutsche Sammlung für Mikroorganismen und Zellkulturen GmbH.
FIG. 2.
Fed-batch fermentation of R. eutropha H16. Fermentation was performed in a Braun Biostat DU 30 bioreactor at 30°C. The culture medium (30 liters MM), with starting concentrations of 1.5% (wt/vol) sodium gluconate and 0.05% (wt/vol) NH4Cl, was inoculated with a preculture (5% [vol/vol]). The upper diagram shows different physical fermentation parameters. In the lower diagram, the turbidity (optical density at 600 nm), the concentration of the carbon and nitrogen sources, and the relative PHB content (percent of maximum value) are shown. The arrows mark the withdrawal of samples that were subjected to protein extraction and analysis (8 h, 18 h, 21 h, and 33 h).
Preparation of protein samples for proteome analysis.
To obtain crude extracts from cells, which were cultivated in MM, 1 volume of crack solution (8 M urea, 2% [vol/vol] Triton X-114) was added to 1 volume of culture broth (usually 25 ml). The mixture was then incubated for 1.5 h at room temperature on a gyratory shaker to break the cells. Cells grown in TSB medium were harvested by centrifugation (15 min; 3,500 × g; 4°C), and the resulting cell pellets were then treated with crack solution as described above. In these cases, the volume of crack solution corresponded to the volume of the centrifuged cell suspension.
PHB and cell debris were separated from the crude extract by centrifugation (1 h; 60,000 to 70,000 × g; 4°C), and proteins were subsequently extracted from the supernatant. For this, an aliquot of 15 ml supernatant for each sample was transferred into a 50-ml plastic tube, and 10 ml Tris-equilibrated phenol and ice-cold H2Odest were added to 40 ml. The samples were stirred, incubated for 10 min at 72°C, and then chilled for 10 min on ice. After centrifugation (15 min; 3,500 × g; 4°C), 20 ml of the aqueous supernatant was removed and discarded, and 10 ml ice-cold deionized water was added to the solution remaining in the centrifugation tube. The samples were stirred and again heated for 10 min at 72°C, chilled for 10 min on ice, and centrifuged (15 min; 3,500 × g; 4°C) as described above. This washing procedure was then repeated for a second time. Usually, two washing steps were enough to clear the supernatant sufficiently to precipitate the proteins. Therefore, as much of the supernatant as possible was removed and discarded, thereby avoiding a loss of proteins located in the interphase. The proteins were subsequently precipitated from the remaining solution by adding 2 volumes of chilled acetone to the tubes. After 10 min of incubation on ice, the samples were centrifuged (15 min; 3,500 × g; 4°C). Whereas the supernatant was carefully removed and discarded, the protein pellet was resuspended in ice-cold acetone (5 ml) and again centrifuged (15 min; 3,500 × g; 4°C). The washed protein pellet was air dried by incubation at room temperature to evaporate the acetone and was then stored at −20°C.
SDS-PAGE.
Protein samples were mixed with denaturation buffer (0.225 M Tris-HCl [pH 6.8], 50% [wt/vol] glycerol, 5% [wt/vol] sodium dodecyl sulfate [SDS], 0.05% [wt/vol] bromophenol blue, 0.25 mM dithiothreitol [DTT]) and were separated in SDS-12% (wt/vol) polyacrylamide gels as described by Laemmli (25). The proteins were stained with Coomassie brilliant blue (59).
2D PAGE. (i) First dimension (IEF).
To the dried protein pellets, 250 μl rehydration buffer A (9 M urea, 4% [wt/vol] CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 100 mM DTT, dissolved in H2O to a volume of 10 ml) was added, and the mixture was incubated at room temperature for 2 h. To ensure effective rehydration, the samples were stirred several times during this period. The protein solutions were then transferred to Eppendorf tubes and centrifuged (5 min; 11,000 × g). The supernatants were mixed with 150 μl rehydration buffer B (2.5 ml rehydration buffer A, 125 μl ampholyte solution, pH 3 to 10 [Serva], 125 μl Triton X-100, a trace amount of bromophenol blue). The isoelectric focusing (IEF) strips (pH 5 to 8; 11 cm; Bio-Rad) were passively rehydrated overnight at room temperature with prepared protein solutions (maximum, 400 μl each) while overlaid with mineral oil. After rehydration, the strips were focused in a focusing tray (likewise overlaid with mineral oil) by a series of voltage increases, 250 V (1 h), 500 V (1 h), 1,000 V (1 h), 6,000 V (18 h), and 500 V (up to 99 h), at 20°C to a total value of at least 100 kVh.
(ii) Second dimension and gel staining.
IEF strips were equilibrated in equilibration buffer (6.70 ml 1.5 M Tris-HCl [pH 8.8], 72 g urea, 10 g SDS, 87 g glycerol, a trace amount of bromophenol blue, 200 ml H2Odest) for 20 min with slight agitation and then transferred for alkylation to equilibration buffer containing 150 mM iodacetamide. After 20 min of incubation, likewise with slight agitation, the strips were briefly rinsed with water, placed on top of 12% (wt/vol) SDS-polyacrylamide 2D gels, and fixed by overlaying them with sealing solution (25 ml SDS running buffer containing 125 mg agarose and a trace amount of bromophenol blue). The gels were run for 1.5 h at 20°C and 50 V with constant current and then for 7 h at 20°C and 200 V with constant current in a Dodeca cell (Bio-Rad) filled with running buffer (192 mM glycerol, 0.1% SDS, 25 mM Tris-HCl [pH 8.8]). The gels were then stained with Coomassie brilliant blue (4 g Serva Blue G, 450 ml methanol, 90 ml acetic acid, 1,000 ml H2Odest [60]); the solution was filtered, the staining solution was removed, and the gels were briefly washed with water. Destaining was done in destaining solution (330 ml methanol, 100 ml acetic acid, 1,000 ml H2Odest) for about 5 h and subsequently for about 12 h in 10% (vol/vol) acetic acid. All staining and destaining procedures were done under slight agitation.
Software-based analysis of 2D gel images.
Images from 2D PAGE were analyzed using Decodon Delta 2D software (Decodon GmbH, Greifswald, Germany). From replicates comprising various samples from identical stages of cultivation (8 h, six replicates; 18 h, seven replicates; 21 h, eight replicates; 33 h, two replicates) an average fusion image was created for each group (after the necessary warping steps were performed). The spots were color coded according to their expression profiles in the dual-channel images of these fusions. For further comparison of the protein patterns during the different stages, spot quantities were likewise determined wuth Delta 2D software. For this, a proteome map comprising all gel images of each of the four groups was created using the union fusion approach of the software. Spot boundaries on the proteome map were detected and transferred to the original images, and the spots were automatically quantified by the software. The spot quantities given represent the relative portion (percent volume) in an individual spot of the total protein present on the respective average fusion image. For normalization, the 10 quantitatively most predominant spots were excluded from the normalization set. Normalization aims to mitigate systematic differences between gel images, which can occur by variation in protein loading, imaging exposure times, and dye/stain efficiencies. It is a common procedure to exclude some of the quantitatively predominant spots from normalization, because these spots disturb the quantification process of weaker spots (for further details, see reference 6).
Protein preparation, mass spectrometry and data analysis.
Spots were cut from the 2D gels and transferred to Eppendorf tubes. MALDI-TOF analysis was performed, employing the method of Shevchenko et al. (51), at the Institut für Integrierte Funktionelle Genomik of the Faculty of Medicine of the University of Münster and at the Institut für Mikrobiologie, Ernst-Moritz-Arndt Universität, Greifswald, Germany. For this, proteins were tryptically digested, and the mass spectra of the protein fragments were revealed by MALDI-TOF (Institut für Integrierte Funktionelle Genomik, TofSpec-2E [Micromass, Manchester, England]; Ernst-Moritz-Arndt Universität, MALDI-TOF Proteome Analyzer 4800 [Applied Biosystems, Foster City, CA]). The parameters for the measurements were set as described by Voigt et al. (58), except that the signal-to-noise ratio for the TOF-TOF measurements was raised to 10). These spectra were compared with hypothetical spectra from the R. eutropha H16 data bank, and proteins were identified by using the computer software GPMAW (Lighthouse Data, Denmark), as well as Mascot, a program associated with MALDI-TOF (search parameters were as in reference 58).
Electron microscopy.
For electron microscopic investigations, 200 μl of culture broth was mixed with 200 μl of 2.5% (wt/vol) glutardialdehyde in Sörensen's phosphate buffer, pH 7.3 (5). This aliquot was then diluted at a ratio of 1:1 with Sörensen's phosphate buffer, pH 7.3. From these suspensions, 3 μl was placed on pioloform-coated copper mesh grids for 1 min; subsequently, the excess liquid was removed using filter paper. Then, the samples were stained with 0.5% phosphate tungsten acid, pH 7.0. The samples were investigated in a transmission electron microscope (H-500; Hitachi Ltd., Tokyo, Japan) operated at 75 kV. Photographs were taken in the bright-field mode with Agfa-Gevaert 23D56 film.
PHA quantification.
Samples were subjected to methanolysis in the presence of 85% (vol/vol) methanol and 15% (wt/vol) sulfuric acid. The resulting 3-hydroxybutyric acid methyl esters were analyzed by gas chromatography (8, 56).
RESULTS
Development of a protein extraction protocol for PHB-harboring cells of R. eutropha suitable to perform 2D PAGE.
An appropriate sample preparation is crucial for the success of a 2D gel electrophoresis-based proteomics experiment. The specific properties of the studied organism, as well as the solubility, size, charge, and pI of the proteins, must be considered during sample preparation. In the case of PHB-harboring cells of R. eutropha, standard procedures cannot be applied to extract proteins from cell lysates by phenol/acetone extraction because of the significantly disturbing influence of accumulated PHB.
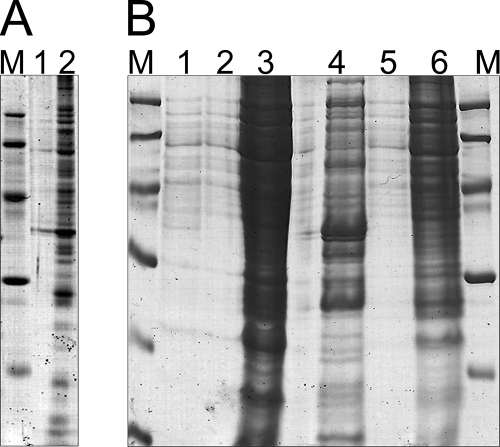
It was therefore necessary to develop an adequately modified protein extraction method for a 2D gel-based proteomic approach applicable to whole cells of R. eutropha. The modified method was partially based on a previous procedure (60) and employed a “crack solution” enabling the removal of PHB granules without simultaneous loss of proteins. This crack solution comprised an aqueous solution of 8 M urea and 2% (vol/vol) Triton X-114 to break the cells and to solubilize cytosolic, membrane-bound, and PHB granule-associated proteins. The ionic detergent SDS is a perfect detergent to disrupt all types of noncovalent interactions of proteins; however, it is not compatible with IEF and would have to be removed before IEF. Therefore, the nonionic chaotrope urea was used for breaking most noncovalent interactions. Because urea is very efficient in breaking hydrogen bonds but is less efficient in disrupting hydrophobic interactions, the nonionic detergent Triton X-114 was also added. After treatment with crack solution, PHB granules were removed from the cell lysates by centrifugation, and proteins were subsequently extracted from the supernatant with a phenol-plus-acetone solution. The negative influence of accumulated PHB on protein extraction is shown in Fig. 1A, in which samples of the wild-type H16 and the PHB-negative mutant PHB−4 were compared after a conventional protein extraction protocol was applied (16) to cells cultivated under conditions permissive for PHB accumulation. Whereas only very small amounts of protein were extracted from wild-type cells, much larger amounts of proteins, and also many more protein species, were extracted from the mutant cells (Fig. 1A). If the modified method was applied, the amount of proteins extracted from cells of the wild type was comparable to that extracted from cells of the PHB-negative mutant (Fig. 1B). The modified method therefore enabled protein extraction from cells of R. eutropha H16 that had accumulated PHA without quantitative and qualitative losses of proteins.
FIG. 1.
Optimization of the protein extraction protocol. (A) Protein extraction was performed by classical treatment with phenol/acetone starting from sedimented cells previously cultivated in MM under conditions promoting PHB accumulation, and it showed disturbance of protein extraction by PHB. Protein pellets were dissolved in rehydration buffer A, and for both samples, the polyacrylamide gel (12% [wt/vol]) was loaded with 15 μl protein solution and 5 μl denaturation buffer (0.225 M Tris-HCl [pH 6.8], 50% [wt/vol] glycerol, 5% [wt/vol] SDS, 0.05% [wt/vol] bromophenol blue, 0.25 mM DTT). Lane 1, R. eutropha H16; lane 2, R. eutropha PHB−4. (B) Application of the modified protein extraction protocol, including ultracentrifugation, which was developed in this study. Lane 1, R. eutropha H16, supernatant after ultracentrifugation (for details, see “Preparation of protein samples for proteome analysis” in the text); lane 2, R. eutropha H16, sedimented proteins from ultracentrifugation after resuspension in crack solution; lane 3, R. eutropha H16 after protein extraction from the supernatant and resuspension of the resulting protein pellet in crack solution; lane 4, R. eutropha PHB−4, supernatant after ultracentrifugation; lane 5, R. eutropha PHB−4, sedimented proteins from ultracentrifugation after resuspension in crack solution; lane 6, R. eutropha PHB−4, after protein extraction from supernatant and resuspension of the resulting protein pellet in crack solution. For each lane, 15 μl of the solution was loaded, together with 5 μl denaturation buffer. Lanes M, molecular mass markers.
Growth protocol of R. eutropha wild-type strain H16.
Cells of R. eutropha H16 were cultivated in MM that contained 1.5% (wt/vol) sodium gluconate as a carbon source. NH4Cl served as a nitrogen source at an initial concentration of 0.05% (wt/vol) (Fig. 2). The oxygen concentration was measured to allow reaction to a decrease in the partial O2 pressure by an increase in the airflow and stirring rate to avoid growth limitation by the macroelement. Employing the modified protocol, the proteome of R. eutropha H16 was characterized at different stages of cultivation. During the first stage of cultivation (8 h), cells were still in the exponential growth phase, where no limitation of nutrients occurred, and the PHB content was therefore relatively low. After 11 h, NH4Cl was fed to increase the cell concentration. While the nitrogen source was depleted immediately, the carbon source was still available. The second stage (18 h) represented cells of the early stationary growth phase with maximum PHB content as a consequence of conditions permissive for PHB accumulation (a surplus of the carbon source and limitation of nitrogen). After 18 h of cultivation, ammonium was added to the medium to induce the degradation of stored PHB granules to mobilize this intracellular carbon source. When the next samples were taken (at 21 and 33 h), the cells were in the early or late phase of PHB mobilization, respectively.
Proteome analysis of R. eutropha wild-type strain H16 during different phases of PHB metabolism.
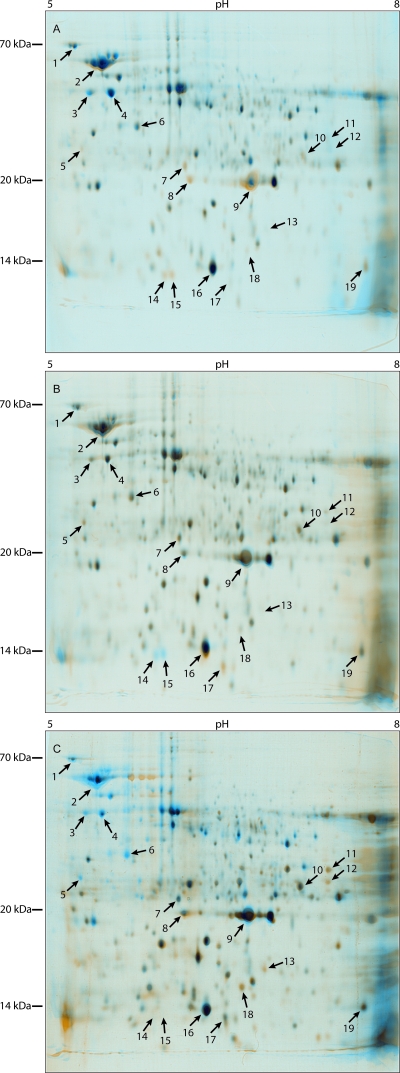
Extracting the proteins directly from the culture broth allowed the detection of intra- and extracellular proteins. Proteins with pIs between 5.0 and 8.0 were well separated in the 2D gels. The gels were scanned, and pictures of replicate groups were fused and color coded by the Delta 2D software. Proteome expression profiles are shown as dual-channel images in Fig. 3. Proteins that showed different expression levels during the time course of the experiment are marked in Fig. 3 and were quantified with Delta 2D software (Fig. 4). Values are given as percent relative volume. The data were normalized as described in Materials and Methods. Proteins showing altered expression levels in these four samples were excised and subjected to MALDI-TOF mass spectrometry analysis, and the corresponding genes were identified in the R. eutropha H16 database. In addition, some protein spots were assigned according to the results of previous studies (37). The proteins detected are listed in Table 2.
FIG. 3.
Changes in the R. eutropha H16 proteome during the time course of the experiment as revealed by 2D PAGE. Samples were taken from the cultivation experiment shown in Fig. 2. Proteins were focused using pH 5 to 8 nonlinear strips. Dual-channel image fusions were created with Delta 2D software. (A) Cells from the exponential growth phase after 8 h of cultivation with little PHB (blue spots) versus cells in the early stationary growth phase after 18 h of cultivation with maximum PHB (orange spots). (B) Cells from the early stationary growth phase after 18 h of cultivation with maximum PHB (blue spots) versus cells in the stationary growth phase after 21 h of cultivation (orange spots). Shortly before sample withdrawal, after 18 h, NH4Cl was added to a concentration of 0.8% (wt/vol), thereby initiating intracellular PHB mobilization. (C) Cells from the stationary growth phase after 21 h of cultivation (blue spots) versus cells in the later stationary growth phase after 33 h of cultivation and still mobilizing PHB (orange spots). The black spots in panels A, B, and C represent equally expressed proteins; the arrows indicate proteins that showed a temporary change in quantity. The proteins in spots 1 to 19 were isolated and analyzed by MALDI-TOF after tryptic digestion. The results of spot identification are listed in Table 2.
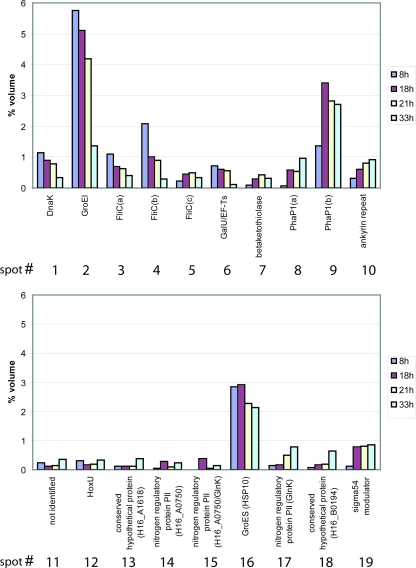
FIG. 4.
Quantification of protein spots in image fusions of 2D PAGE gels shown in Fig. 3. Spot quantities are given as percent volume (representing the relative portion in an individual spot of the total protein present on the respective average fusion image). Quantification was done with Delta 2D software.
TABLE 2.
Analysis of proteins (from 2D gels) whose expression levels were significantly different under two different conditionsa
| Protein no. | Locus tag(s) | pI (theoretical) | Mass (kDa) (theoretical) | Identification |
|---|---|---|---|---|
| 1 | A3089 | 4.9 | 69.9 | DnaK (HSP70; chaperone) |
| 2 | A0706 | 5.0 | 57.3 | GroEL (HSP60; chaperone) |
| 3 + 4 + 5 | B2360 | 4.98 | 44.8 | Flagellin (FliC) |
| 6 | A2752/A2054 | 5.21/5.26 | 32.5/30.9 | UTP-glucose-1-phosphate uridyltransferase (GalU)/protein translation elongation factor TS |
| 7 | B1369 | 6.38 | 43.0 | β-Ketothiolase |
| 8 + 9 | A1381 | 5.96 | 20.0 | PhaP1; isoforms |
| 10 | A1574 | 7.29 | 27.5 | Ankyrin repeat protein |
| 11 | Not identified | |||
| 12 | PHG089 | 6.82 | 26.2 | NAD-reducing hydrogenase diaphorase moiety small subunit (HoxU) |
| 13 | A1618 | 6.05 | 16.6 | Conserved hypothetical protein |
| 14 | A0750 | 5.7 | 12.3 | Nitrogen-regulatory protein PII |
| 15 | A0750/A0320 | 5.87/5.7 | 12.2/12.3 | Nitrogen-regulatory protein PII/nitrogen-regulatory protein PII (GlnK) |
| 16 | A0705 | 5.8 | 10.5 | Cochaperonin GroES (HSP10) |
| 17 | A0320 | 5.87 | 12.2 | Nitrogen-regulatory protein PII (GlnK) |
| 18 | B0194 | 7.93 | 12.6 | Conserved hypothetical protein |
| 19 | A0386 | 7.30 | 13.6 | Sigma 54 modulation protein S30EA |
Identification of isolated proteins was done by tryptic digestion, MALDI-TOF, and, finally, comparison of the resulting mass spectrum data with hypothetical spectra out of the R. eutropha H16 data bank by in silico digestion.
Heat shock proteins (Hsp), including DnaK (Fig. 3, spot 1; H16_A3089; HSP70 homologue) and GroEL (Fig. 3, spot 2; H16_A0706; HSP60 homologue), were among the assigned proteins. After 8 h of cultivation, large amounts of these chaperons were detected in the 2D gels (DnaK, 1.1%; GroEL, 5.8%). The amounts of Hsp slightly increased from 8 h to 18 h and 21 h. During the course of the stationary growth phase (21 h to 33 h), the relative quantities of Hsp decreased (DnaK. 0.3%; GroEL, 1.4%). GroES (Fig. 3, spot 16), the cochaperone of GroEL, formed a prominent spot in all gels.
Spots 8 and 9 represented isoforms of PhaP1 (H16_A1381), which is the major phasin protein in R. eutropha. Increased expression is strictly associated with the accumulation of PHB, when phasins constitute the major component of the boundary layer at the surfaces of PHB granules. After 18 h of cultivation, when the PHB content of the cells had reached its maximum, the predominant isoform, PhaP1(b) (Fig. 3, spot 9), reached up to 3.4% of the relative volume of proteins in the cells. After induced PHB mobilization, the amount slightly decreased. Unlike PhaP1(b), PhaP1(a) (Fig. 3, spot 8) increased from 8 h (0.06%) to 18 h (0.6%), stagnated at 21 h, and increased at 33 h to 1%.
Spot 7 was assigned as one of the β-ketothiolases (H16_B1369; β-ketothiolase homologue). The amount of this enzyme increased significantly from the exponential to the stationary growth phase (maximum at 21 h, 0.42%) but finally decreased to 0.3% (33 h). It should be emphasized that this β-ketothiolase is not the enzyme encoded by the phaCAB operon (34, 49) or by bktB (53). The protein yielding spot 10 was identified as an ankyrin repeat protein (H16_A1574). Over the whole time course of the experiment, a significantly and steadily increasing expression of this protein was observed (8 h, 0.32%; 33 h, 0.9%). Spot 14, which was identified as nitrogen regulatory protein PII (H16_0750), showed alternating expression. While the amount of PII increased from 8 h (0.06%) to 18 h (0.28%), it decreased at 21 h (0.09%) but increased again at 33 h (0.23%). Spot 15 consisted of two proteins, isoforms of the nitrogen-regulatory protein PII (H16_0750) and of the nitrogen-regulatory protein PII GlnK (H16_A0320). Whereas no proteins were observed after 8 h, the volume of this spot varied between 0.37% (18 h), 0.04% (21 h), and 0.15% (33 h). An isoform of GlnK was also detected (Fig. 3, spot 17); its quantity increased from 0.14% (8 h) to 0.18% (18 h) and from 0.5% (21 h) to 0.8% (33 h). Spot 19 was assigned as the σ54 modulation protein (H16_A0386; S30EA ribosomal-protein homologue). After 8 h of cultivation, it occurred only at a very low level (0.13%), but it was upregulated in the stationary growth phase (18 h, 0.77%; 21 h, 0.81%; 33 h, 0.86%).
Spot 6 contained two proteins, the UTP-glucose-1-phosphate uridyltransferase (GalU; H16_A2752) and the protein elongation factor TS (H16_2054). Spot 11 could not be identified by MALDI-TOF. The protein that formed spot 12 was identified as HoxU. Spot 13 (H16_A1618) and spot 18 were identified as a conserved hypothetical protein (H16_B0194).
Three proteins were assigned as flagellin (Fig. 3, spots 3, 4, and 5) and are isoforms of flagellin (H16_B2360; FliC homologue). Spots 3 [FliC(a)] and 4 [FliC(b)] exhibited identical MWs but different pIs. Both proteins showed maximal expression in the exponential phase after 8 h (Fig. 3, spot 3, 1.1%; spot 4, 2.1%). While values decreased considerably (Fig. 3, spot 3, 0.7%; spot 4, 1.0%) to the early stationary phase (18 h), only a small decrease was observed after 21 h and 3 hours after induction of PHB mobilization. After 21 h of cultivation, the relative volumes of the proteins FliC(a) and Flic(b) in the cells constituted 0.63% (a decrease of 8.5% compared to 18 h) and 0.98% (a decrease of 11% compared to 18 h), respectively. In the later stationary phase after 33 h of cultivation and 15 h of PHB-mobilizing conditions, significant decreases in the quantities of both proteins occurred. The amount of FliC(a) was reduced by about 37% in comparison to 21 h, to a value of 0.4%, and the amount of FliC(b) decreased about 67.5% in comparison to 21 h, to a value of 0.3%. While in the exponential growth phase (8 h) very large amounts, and in the early stationary growth phase (18 h and 21 h) large amounts, of these two isoforms of flagellin were present, only very small amounts of flagellin could be detected after 33 h of cultivation. In addition, a third flagellin isoform [FliC(c)] showing a lower pI and minor Mw, indicating degradative products of regular flagellin, was identified (Fig. 3, spot 5). The amounts of FliC(c) increased from exponential phase to early stationary phase but slightly decreased to late stationary phase (8 h, 0.22%; 18 h, 0.44%; 21 h, 0.49%; 33 h, 0.34%).
Comparative proteome analysis of the wild type and mutants, depending on the culture medium and cultivation phase.
Successful characterization of the R. eutropha H16 proteome and analysis results encouraged further investigations, including a comparison of the proteomes of the wild type and different mutants. Because a significant decrease in FliC(a) and FliC(b) occurred in the stationary phase after induction of PHB degradation at the later sampling point (33 h), we focused our investigations on cells of the later stationary phase. Initially, all available phasin deletion mutants—the four R. eutropha single mutants (ΔphaP1, ΔphaP2, ΔphaP3, and ΔphaP4); one double mutant (ΔphaP12); one triplet mutant (ΔphaP123); and the quadruple mutant (ΔphaP1234)—were chosen to explore the effects of these phasins and to reveal possible further functions of phasins in the wild type. 2D gels were prepared with proteins extracted from cells after 64 or 72 h of cultivation in MM containing 1.0% (wt/vol) sodium gluconate as a carbon source and 0.1% (wt/vol) NH4Cl as a nitrogen source; at 64 h, 0.1% NH4Cl was fed to induce PHB degradation. By this procedure, cells were obtained that were cultivated under conditions permissive for PHB accumulation (64 h) or permissive for intracellular mobilization of the storage compound (72 h), respectively. The resulting 2D gels, shown in Fig. 5, were prepared from the 72-h samples. These studies confirmed the surprising finding that the amount of flagellin protein detectable in cells cultivated for several hours under conditions permissive for PHB mobilization was significantly reduced (Fig. 3C). Whereas cells of the wild type possessed a lot of flagellin protein after 64 h (Fig. 6A), the protein was absent after 72 h (8 h postinduction). In contrast, all phasin mutants possessed flagellin in the 64-h, as well as in the 72-h, sample. Thus, a relationship between PHB metabolism, phasins, and flagellation in R. eutropha was suggested.
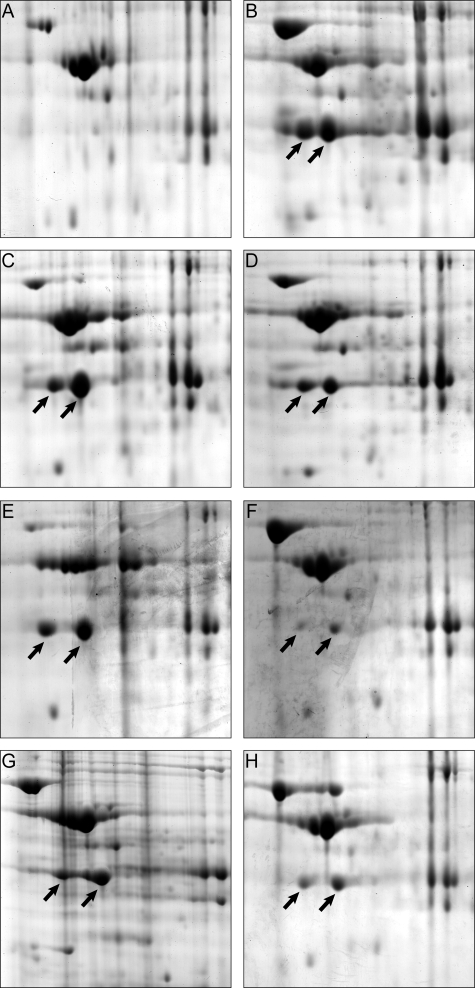
FIG. 5.
Comparison of the proteomes of R. eutropha H16 (wild type) and of phasin mutants. Sections of 2D PAGE gels (pH ca. 5 to 6; mass, ca. 70 to 35 kDa) loaded with protein extracts obtained from cells after 72 h of cultivation in MM (1% [wt/vol] sodium gluconate, 0.1% [wt/vol] NH4Cl) are shown. After 64 h of cultivation, NH4Cl was added to the media to a concentration of 0.1% (wt/vol), thereby inducing PHB mobilization. The sections represent zones in which the flagellin protein occurred when expressed. The arrows indicate the two isoforms of flagellin. Strains of R. eutropha were as follows: H16 (A), ΔphaP1 (B), ΔphaP2 (C), ΔphaP3 (D), ΔphaP4 (E), ΔphaP12 (F), ΔphaP123 (G), and ΔphaP1234 (H).
FIG. 6.
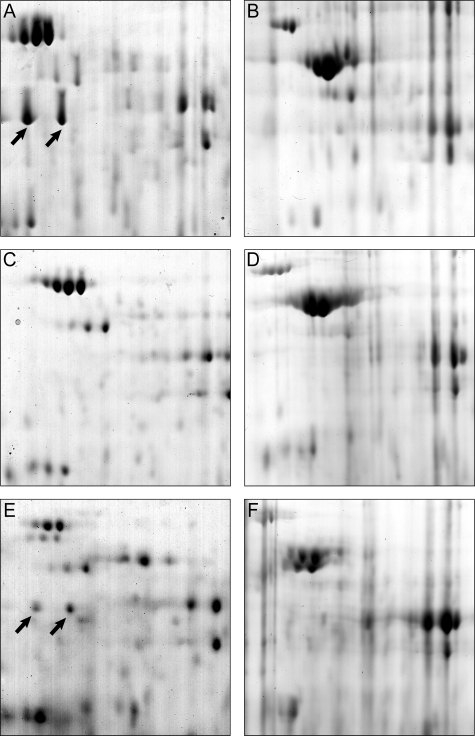
Comparison of the proteomes of R. eutropha H16 (wild type), R. eutropha PHB−4, and R. eutropha HF09. Sections of 2D PAGE gels (pH ca. 5 to 6; mass, ca. 70 to 35 kDa) loaded with protein extracts resulting from cells are shown. The sections represent zones in which the flagellin protein occurred when expressed. The arrows indicate the two isoforms of flagellin. The strains of R. eutropha were as follows: H16 (A and B), PHB−4 (C and D), and HF09 (E and F). The cells for the gels shown on the left were withdrawn after 64 h of cultivation in MM (1% [wt/vol] sodium gluconate, 0.1% [wt/vol] NH4Cl), and conditions were permissive for PHB accumulation. The cells for the gels shown on the right were withdrawn after 72 h of cultivation in this medium; after 64 h, NH4Cl was added to the medium to a concentration of 0.1% (wt/vol).
To obtain more information about the parameters and factors affecting flagellation in R. eutropha, additional experiments were done. In a third experiment, the wild-type strain H16, as well as the ΔphaP1 and ΔphaP1234 strains as representatives of the phasin mutants, were analyzed, together with the PHB-negative mutant R. eutropha PHB−4. The last was used as a negative control to see whether changes in protein expression were caused by general, life cycle-dependent regulation or whether they were specifically due to PHB metabolism. In addition, the rpoN mutant HF09 (41), defective in the alternative sigma factor σ54, was also chosen to investigate a putative involvement of σ54 in the regulation of flagellar genes in R. eutropha H16. Cells were also cultivated in nutrient-rich TSB medium, which effectively suppresses PHB accumulation in the stationary phase (63). 2D gels of the proteomes obtained from cells after 64 and 72 h of cultivation in MM or in TSB medium are shown in Fig. 6 and 7, respectively. To verify the findings regarding flagellation, cells were also investigated by electron microscopy.
FIG. 7.
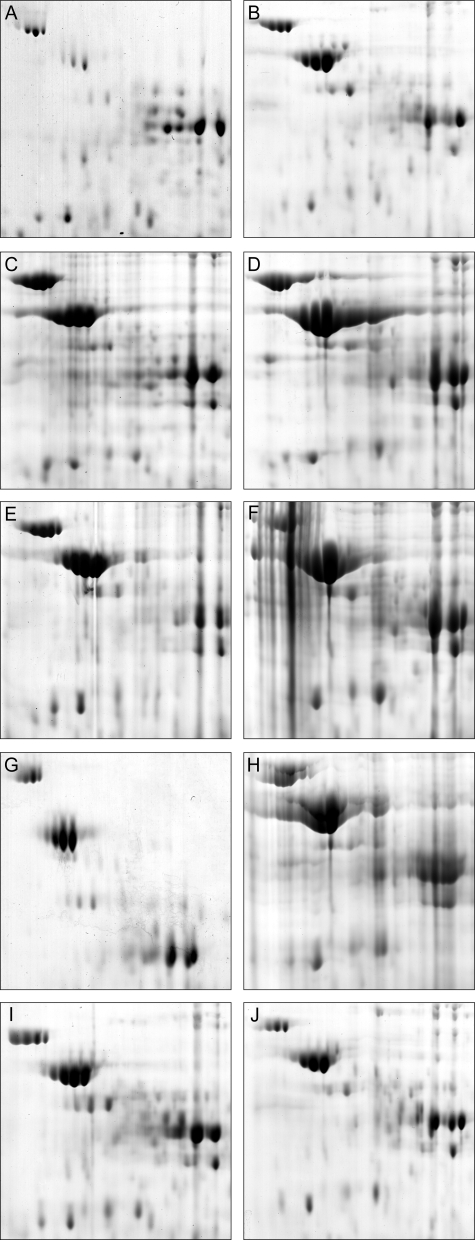
Comparison of the proteomes of R. eutropha PHB−4 and various mutants. Sections of 2D PAGE gels (pH ca. 5 to 6; mass, ca. 70 to 35 kDa) loaded with protein extracts from the cells after 64 h and 72 h of cultivation in TSB medium supplemented with 0.5% (wt/vol) sodium gluconate are shown. After withdrawal of the first sample after 64 h of cultivation, NH4Cl was added to a concentration of 0.1% (wt/vol). A second sample was withdrawn after 72 h of cultivation. The sections represent zones in which the flagellin protein occurred when expressed. The strains of R. eutropha were as follows: H16, 64 h (A); H16, 72 h (B); ΔphaP1, 64 h (C); ΔphaP1, 72 h (D); ΔphaP1234, 64 h (E); ΔphaP1234, 72 h (F); PHB−4, 64 h (G); PHB−4, 72 h (H); HF09, 64 h (I); and HF09, 72 h (J).
The proteomes from R. eutropha H16 and the ΔphaP1 and ΔphaP1234 strains after cultivation in MM were identical with those from previous experiments described above (Fig. 5) in regard to flagellin protein. While the wild type expressed large amounts of flagellin protein after 64 h (Fig. 6A), it was absent after 72 h (Fig. 6B). In contrast, the proteomes of either phasin mutant contained flagellin in both samples (not shown). The σ54 mutant HF09 expressed only small quantities of flagellin after 64 h of cultivation in MM (Fig. 6E), and after 72 h of cultivation, flagellin was absent, as in the wild type (Fig. 6F). The proteome of R. eutropha PHB−4 never contained any detectable flagellin protein after 64 or 72 h of cultivation in MM (Fig. 6C and D). None of the investigated strains possessed detectable flagellin protein at any time when cultivated in TSB (Fig. 7). In summary, the following can be said. Apparently, the wild type was strongly flagellated under conditions permissive for PHB accumulation but lost flagella when PHB degradation was induced in the stationary phase. In contrast, PHB-storing cells of all phasin mutants retained their flagella when PHB degradation was induced in the stationary phase. When the wild type, as well as the phasin mutants, did not accumulate PHB (cultivation in TSB in late stationary phase), none of the cells were flagellated. The PHB-negative mutant PHB−4 never did express flagellin when cultivated both in MM and in TSB. The σ54 mutant HF09 behaved quite similarly to the wild type, but expression of flagellin in PHB-storing cells (64 h; MM) was reduced.
Electron microscopy.
Electron microscopy pictures of cells of R. eutropha H16, the ΔphaP1 strain, and HF09 cultivated in MM or in TSB were prepared to analyze the extents of flagellation (Fig. 8). Three different categories in regard to the extent of flagellation were distinguished: (i) strongly flagellated, (ii) sporadically flagellated, and (iii) nonflagellated. Electron microscopy revealed the extent of flagellation in much more detail than 2D gel electrophoresis of the proteins. In principle, however, the electron-microscopic examinations of cells confirmed the observations made in the 2D gels.
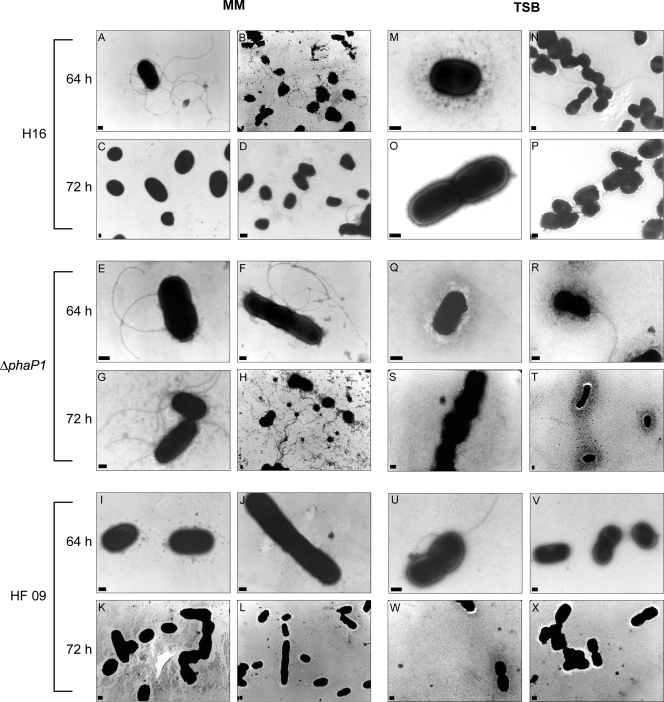
FIG. 8.
Changes in flagellation of R. eutropha H16 and of mutants during the time course of the experiment as revealed by electron microscopy. (Left) Cells were cultivated in MM (1% [wt/vol] sodium gluconate, 0.1% [wt/vol] NH4Cl) under conditions permissive for PHB accumulation. The first samples were withdrawn after 64 h. Then, NH4Cl was added to the medium to a concentration of 0.1% (wt/vol), thereby inducing PHB mobilization. The second samples were withdrawn after 72 h of cultivation. (A and B) Cells of H16 after 64 h of cultivation. (C and D) Cells of H16 after 72 h of cultivation. (E and F) Cells of the ΔphaP1 mutant after 64 h of cultivation. (G and H) Cells of the ΔphaP1 mutant after 72 h of cultivation. (I and J) Cells of HF09 after 64 h of cultivation. (K and L) Cells of mutant HF09 after 72 h of cultivation. (Right) Cells were cultivated in TSB medium (with 0.5% [wt/vol] sodium gluconate addition). Sample withdrawal time points were as described above. (M and N) Cells of H16 after 64 h of cultivation. (O and P) Cells of H16 after 72 h of cultivation. (Q and R) Cells of the ΔphaP1 mutant after 64 h of cultivation. (S and T) Cells of the ΔphaP1 mutant after 72 h of cultivation. (U and V) Cells of mutant HF09 after 64 h of cultivation. (W and X) Cells of mutant HF09 after 72 h of cultivation. The electron micrographs were obtained as described in Materials and Methods. The bars represent 0.1 μm.
After 64 h of cultivation, cells of strain H16 were strongly flagellated when cultivated in MM medium (Fig. 8A and B) whereas cells cultivated in TSB (Fig. 8M and N) were only sporadically flagellated. After 72 h of cultivation in MM, the wild type was also only sporadically flagellated (Fig. 8C and D), whereas this strain appeared to be nonflagellated when cultivated in TSB (Fig. 8O and P).
Cells of the ΔphaP1 phasin mutant were strongly flagellated after 64 h in MM (Fig. 8E and F), whereas flagella occurred only sporadically when it was grown in TSB medium (Fig. 8Q and R). In contrast to the wild type, the ΔphaP1 strain was still strongly flagellated (Fig. 8G and H) after 72 h of cultivation in MM. On the other hand, after 72 h of cultivation in TSB medium, no flagella were observed for cells of the ΔphaP1 strain (Fig. 8S and T), as for cells of the wild type.
Cells of the σ54-negative mutant HF09 demonstrated differences in flagellation. When cultivation was done in MM or in TSB, the cells were sporadically flagellated after 64 h of cultivation (Fig. 8I and J or 8U and V). After 72 h of cultivation in either medium, flagella occurred even more sporadically (Fig. 8K and L or 8W and X).
All results of this study regarding the visibility of flagella in electron microscopy and the occurrence of the relevant proteins in 2D gels, depending on the cultivation conditions and cultivation phase in R. eutropha H16 and in selected mutants, are summarized in Table 3. In addition to the PHB contents of the cells, the table contains information about the occurrence of flagellin in 2D gels, the detection of flagella by electron microscopy, and the state of supply with carbon and nitrogen.
TABLE 3.
Summarized results of the investigations of R. eutropha H16, ΔphaP1 ΔphaP1234, PHB−4, and HF09 strains
| Treatment and R. eutropha strain/genotype | FliC detectiona | Flagellation levelb | PHB (% cell dry weight) | Concn
|
|
|---|---|---|---|---|---|
| C sourcec | N sourced | ||||
| MM | |||||
| 64 h | |||||
| H16 | + | ++ | 34 | − | − |
| ΔphaP1 | + | ++ | 27 | − | − |
| ΔphaP1234 | + | 10 | − | − | |
| PHB−4 | − | 0 | + | − | |
| HF09 | ± | ± | 36 | − | − |
| 72 h | |||||
| H16 | − | ± | 30 | + | + (puls) |
| ΔphaP1 | + | ++ | 22 | + | + (puls) |
| ΔphaP1234 | + | 6 | + | + (puls) | |
| PHB−4 | − | 0 | ± | + (puls) | |
| HF09 | − | ± | 34 | + | + (puls) |
| TSB | |||||
| 64 h | |||||
| H16 | − | ± | 0 | ± | + |
| ΔphaP1 | − | ± | 2 | ± | + |
| ΔphaP1234 | − | 0 | ± | + | |
| PHB−4 | − | 0 | ± | + | |
| HF09 | − | ± | 0 | ± | + |
| 72 h | |||||
| H16 | − | − | 1.5 | ± | ++ (puls) |
| ΔphaP1 | − | − | 2 | ± | ++ (puls) |
| ΔphaP1234 | − | 0 | ± | ++ (puls) | |
| PHB−4 | − | 0 | ± | ++ (puls) | |
| HF09 | − | ± | 0 | ± | ++ (puls) |
+, flagellin detected; ±, eventually small amounts of flagellin detected; −, no flagellin detectable.
++, strongly flagellated; ±, sporadically flagellated; −, nonflagellated.
+, sufficient; ±, concentration hardly predictable; −, depleted.
++; very high; +, high; −, depleted. Puls means provision of NH4Cl to a concentration of 0.1% (wt/vol).
DISCUSSION
A modified protein extraction protocol suitable for 2D PAGE of samples derived from PHB-containing cells was successfully established, and the proteome of R. eutropha H16 was portrayed and analyzed at characteristic cultivation stages with regard to PHB metabolism. Quantitative changes in protein expression were observed for several different proteins. (i) Heat shock proteins (Hsp) play an important role in the protection of proteins under stress (19); furthermore, they support the correct folding of nascent proteins (27). It is therefore understandable that at early stages a higher concentration of these chaperons was found in 2D gels than at later stages (Fig. 3, spots 1, 2, and 16). (ii) Two isoforms (Fig. 3, spots 8 and 9) of the phasin PhaP1 with identical apparent molecular masses but different apparent pIs occurred, as previously found (37). As expected, the concentration of PhaP1 was correlated with the amount of PHB accumulated in the cells. (iii) The β-ketothiolase identified in the gel (Fig. 3, spot 7) was neither PhaA nor BktB, but rather represented another isoenzyme so far not noticed in PHB metabolism. (iv) Expression of the ankyrin repeat protein (spot 10) increased with the time of cultivation. The ankyrin repeat is one of the most common modular protein-protein interaction motifs in nature and is found in proteins with diverse functions, such as transcription initiation, cell cycle regulation, cytoskeletal integrity, ion transport, and cell-cell signaling (31). It occurs in particular in eukaryotes, but also in prokaryotes and viruses (7). In contrast to other protein-protein binding motifs, the ankyrin repeat motif is defined by its folding rather than by its function, as there is no specific sequence or structural motif that is universally recognized by ankyrin repeat proteins (24). The recognition surfaces at the binding sites of the molecule exhibit high variability to accommodate diverse groups of potential binding partners (31). Upregulated synthesis of the ankyrin protein during the time course of the cultivation experiments might be caused by involvement of the protein in cell metabolism during the stationary growth phase or in PHB metabolism. It will be difficult to reveal the particular function, because the structure of the binding site of the R. eutropha ankyrin protein is not known. (v) HoxU (Fig. 3, spot 12) is part of the NAD-reducing hydrogenase of R. eutropha. HoxU is one subunit of this heterotrimeric enzyme and represents the Fe-S-containing flavoprotein, called the diaphorase, which mediates NADH oxidoreductase activity (29). The reason for differential expression of this protein is unclear. (vi) Spots 14, 15, and 17 are isoforms of two different PII nitrogen-regulatory proteins (GlnK and H16_A0750). PII proteins act as sensors of cellular nitrogen status and are regulated by covalent modifications, like uridylation (21). Such modifications could cause a pI shift that might explain the observed isoforms of different pIs. PII proteins sense 2-oxoglutarate and ATP and thus link the state of a central carbon and energy metabolite to the control of nitrogen assimilation. Furthermore, it has been shown that PII proteins regulate the activities of other cytoplasmic proteins, as well as enzymes such as transcription factors, by protein-protein interactions (11, 20). Varying expression and isoforms suggest that the identified PII proteins act as sensors of energy and nitrogen and as part of a signal cascade, perhaps regulating enzymes involved in PHB metabolism.
Spot 6 consisted of two different proteins and therefore prevented a clear interpretation of the results. An interpretation of the varying expressions of the conserved hypothetical proteins that formed spots 13 and 18 is not possible.
The most unexpected and remarkable result revealed in this study concerns the changes in flagellin concentrations (Fig. 3, spots 3 and 4) and flagellation of the cells, which suggest a linkage between metabolism and flagellation. Previous studies reported a decrease in flagellation (the percentage of cells with flagella) during the exponential growth phase of R. eutropha H16 cultivated in MM with fructose as a carbon source and 0.1% (wt/vol) NH4Cl as a nitrogen source (4). However, these investigations excluded observations of cells at later growth phases and also did not examine PHB metabolism. The results of our study provide unequivocal evidence for striking changes in flagellation depending on the life cycle, the nutrient supply, and, in particular, the state of PHB metabolism. They suggest that cells are strongly flagellated in the exponential growth phase and lose flagella significantly during transition to the stationary phase. In the stationary phase under conditions permissive for PHB biosynthesis (an excess of the extracellular carbon source), flagellation of cells admittedly stagnates. However, under conditions permissive for intracellular PHB mobilization (addition of a nitrogen source to cells that are carbon deprived but with full PHB accumulation), cells significantly increase the degradation of their flagella, or at least stop flagellin synthesis while normal degradation continues. The possibility that the bacteria simply discarded their flagella could be excluded because the protein extraction was performed directly from the MM culture broth; therefore, proteins of flagella released from the cells would also have been seen.
Interestingly, and in contrast to the wild type, cells of all phasin-negative mutants remained flagellated under conditions permissive of PHB mobilization (Fig. 5B to H, MM). The absence of phasins in the presence of PHB obviously produces generally flagellated cells. At the moment, the explicit reasons for this causal relationship or correlation remain unknown. It is noticeable that this situation is caused by deletion of either phasin. If the sole function of all phasins was only to cover the granule surface, these responses would not be explainable. PhaR regulates expression of the predominant phasin, PhaP1, and also of PhaP3. Synthesis of these phasins in the cells is only stopped if the entire surfaces of granules are covered with proteins (36, 37). Therefore, PhaP1 and PhaP3 should easily substitute for the minor phasins PhaP2 and PhaP4 in the R. eutropha mutant ΔphaP2 and ΔphaP4 strains to cover the entire surfaces of granules. However, the results of this study indicate that at least some of the phasins (most probably PhaP2 and PhaP4) mediate so-far-unknown functions and are not solely, or even not at all, required to shield the PHB granule surface from the cytoplasm.
R. eutropha PHB−4 did not show flagellation after 64 h or after 72 h (MM). While nitrogen became limited with the commencement of stationary phase, carbon should still have been present in the medium because no carbon source was consumed for the accumulation of PHB. In the stationary phase, a sufficient supply of a carbon source (with H16 after 72 h in MM by PHB mobilization and with PHB−4 after both 64 h and 72 h in MM) apparently results in the loss of flagella. On the other hand, cells of the wild type were flagellated after 64 h under nitrogen- and carbon-limited conditions. Therefore, carbon limitation in the stationary phase seems to suppress loss of flagella, whereas nitrogen limitation alone does not result in the loss of flagella.
R. eutropha H16 and all of the investigated mutants were not flagellated when cultivated in complex and nutrient-rich TSB medium, where cells were not exposed to nutrient limitation and obviously did not require motility. Under these cultivation conditions, flagellar biosynthesis does not occur, or else previously existing flagella are discarded and degraded, as during cultivation in MM after the addition of ammonium and when carbon originates from mobilization of the storage polyester after 72 h. When the phasin mutants were cultivated in TSB and accumulation of PHB did not occur, the bacteria showed no flagellation, in contrast to cultivation in MM, where flagella were observed under PHB storage conditions, as well as under PHB-mobilizing conditions. This supports the assumption that a linkage exists between the loss of phasins in the presence of PHB granules and flagellation.
R. eutropha HF09 showed behavior similar to that of the wild type concerning growth and PHB accumulation; however, with regard to flagellation, it behaved quite differently (Fig. 7I to L, MM). In HF09, flagellar biosynthesis was not totally prevented. This shows that flagellar biosynthesis in R. eutropha is dependent only to an extent on the σ54-encoding rpoN gene.
Sigma factor 54-dependent transcription has several distinctive features: the RNA polymerase holoenzyme, consisting of a complex of σ54 and the core RNA polymerase, catalyzes the open promoter complex formation only in the presence of a distinct class of transcriptional activators. The σ54 modulation protein identified in the gels falls into this category. Unlike most eubacterial transcriptional activators, the σ54-dependent activators bind to sites that are effective regardless of distance from and orientation to the start site for transcription (9). These activators interact with the complex of σ54 and the core RNA polymerase from these binding sites (43). The activators themselves are controlled by phosphorylation, interactions with a low-molecular-weight ligand, or interaction with one or several other regulatory proteins (52). Expression of the σ54 modulation protein (Fig. 3, spot 9) increased from the exponential growth phase to the stationary growth phase, suggesting that the modulator is involved in gene regulation at this stage. Its participation in PHB metabolism is likely. The amount of modulator protein was approximately the same under PHB-accumulating and PHB-mobilizing conditions, but usually the status rather than the amount is crucial. However, there was no evidence for a modification of the σ54 modulation protein, such as a shift in pI caused, for example, by phosphorylation of the protein would indicate.
Acknowledgments
This study was supported by a grant provided by the Deutsche Forschungsgemeinschaft (Ste 386/6-4).
The genome sequence data of R. eutropha strain H16 required for proteome analysis were revealed within the framework of the Competence Network Göttingen “Genome research on bacteria” (GenoMik) financed by the German Federal Ministry of Education and Research (BMBF). The genome sequence was obtained and annotated in the Göttingen Genomics Laboratory headed by Gerhard Gottschalk and in the laboratories of B.F., B.B., and A.S., who appreciate very much the contributions of their coworkers who were involved in the project.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Abe, T., T. Kobayashi, and T. Saito. 2005. Properties of a novel intracellular poly(3-hydroxybutyrate) depolymerase with high specific activity (PhaZd) in Wautersia eutropha H16. J. Bacteriol. 187:6982-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aragno, M., A. Walther-Mauruschat, F. Mayer, and H. G. Schlegel. 1977. Micromorphology of Gram-negative hydrogen bacteria. I. Cell morphology and flagellation. Arch. Microbiol. 114:93-100. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, M. (ed.). 1968. Histochemie. Einführung in die Grundlagen und Prinzipien der Methoden. Springer, Berlin, Germany.
- 6.Berth, M., F. M. Moser, M. Kolbe, and J. Bernhardt. 2007. The state of the art in the analysis of two-dimensional gel electrophoresis images. Appl. Microbiol. Biotechnol. 76:1223-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bork, P. 1993. Identification of a ring protein that can interact in vivo with the BRCA1 gene product. Proteins 17:363-374. [DOI] [PubMed] [Google Scholar]
- 8.Brandl, H., R. A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck, M., M. T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagella gene expression to flagella assembly. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Commichau, F. M., K. Forchhammer, and J. Stülke. 2006. Regulatory links between carbon and nitrogen metabolism. Curr. Opin. Microbiol. 9:167-172. [DOI] [PubMed] [Google Scholar]
- 12.Doi, Y., A. Segawa, S. Nakamura, and M. T. Kunioka. 1990. Production of biodegradable copolyesters by Alcaligenes eutrophus, p. 37-48. In E. A. Dawes (ed.), New biosynthetic biodegradable polymers of industrial interest from microorganisms. Kluyver, Dordrecht, The Netherlands.
- 13.Gacesa, P. 1998. Bacterial alginate biosynthesis—recent progress and future prospects. Microbiology 144:1133-1143. [DOI] [PubMed] [Google Scholar]
- 14.Garrett, E. S., D. Perlegas, and D. J. Wozniak. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 181:7401-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Görg, A., W. Weiss, and M. J. Dunn. 2004. Current two-dimensional electrophoresis technology for proteomics. Proteomics 4:3665-3685. [DOI] [PubMed] [Google Scholar]
- 16.Hanna, S. L., N. E. Sherman, M. T. Kinter, and J. B. Goldberg. 2000. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from cystis fibrosis patient: an analysis by 2-D-gel electrophoresis and capillary column liquid chromatography tandem mass spectrometry. Microbiology 146:2495-2508. [DOI] [PubMed] [Google Scholar]
- 17.Haywood, G. W., A. J. Anderson, and E. A. Dawes. 1988. Characterization of two 3-ketothiolases possessing differing substrate specificities in the polyhydroxyalkanoate synthesizing organism Alcaligenes eutrophus. FEMS Microbiol. Lett. 52:91-96. [Google Scholar]
- 18.Haywood, G. W., A. J. Anderson, L. Chu, and E. A. Dawes. 1988. The role of NADH- and NADPH-linked acetoacetyl-CoA reductases in the poly-3-hydroxybutyrate synthesizing organism Alcaligenes eutrophus. FEMS Microbiol. Lett. 52:259-264. [Google Scholar]
- 19.Hendrick, J. P., and F. U. Hartl. 1993. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 62:349-384. [DOI] [PubMed] [Google Scholar]
- 20.Hocking, P. J., and R. H. Marchessault. 1994. Biopolyesters, p. 48-96. In G. J. L. Griffin (ed.), Chemistry and technology of biodegradable polymers. Chapman and Hall, London, United Kingdom.
- 21.Javelle, A., E. Severi, J. Thornton, and M. Merrick. 2004. Ammonium sensing in Escherichia coli. J. Biol. Chem. 279:8530-8538. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, T., K. Uchino, T. Abe, Y. Yamazaky, and T. Saito. 2005. Novel intracellular 3-hydroxybutyrate-oligomer hydrolase in Wautersia eutropha H16. J. Bacteriol. 187:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunioka, M., Y. Nakamura, and Y. Doi. 1988. New bacterial copolyesters produced in Alcaligenes eutrophus from organic acids. Polym. Commun. 29:174-176. [Google Scholar]
- 24.Kuriyan, J., and D. Cowburn. 1997. Structures of Src-family tyrosine kinases. Annu. Rev. Biophys. Biomol. Struct. 26:259-288. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lemoigne, M. 1926. Produits de deshydration et de polymerisation de lácide β-oxybutyrique. Bull. Soc. Chim. Biol. 8:770-782. [Google Scholar]
- 27.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 28.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 55:77-110. [DOI] [PubMed] [Google Scholar]
- 29.Massanz, C., S. Schmidt, and B. Friedrich. 1998. Subforms and in vitro reconstitution of the NAD-reducing hydrogenase of Alcaligenes eutrophus. J. Bacteriol. 180:1023-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAdams, H. H., and L. Shapiro. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301:1874-1877. [DOI] [PubMed] [Google Scholar]
- 31.Mosavi, L. K., D. L. Minor, and Z. Peng. 2002. Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl. Acad. Sci. USA 25:16029-16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller, H. M., and D. Seebach. 1993. Poly(hydroxyfettsäureester), eine fünfte Klasse von physiologisch bedeutsamen organischen Biopolymeren? Angew. Chem. 105:483-509. [Google Scholar]
- 33.Oeding, V., and H. G. Schlegel. 1973. β-Ketothiolase from Hydrogenomonas eutrophus H16 and its significance in the regulation of poly-β-hydroxybutyrate metabolism. Biochemistry 134:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peoples, O. P., and A. J. Sinskey. 1989. Poly-β-hydroxybutyrate biosynthesis in Alcaligenes eutrophus H16. Characterization of the genes encoding β-ketothiolase and acetoacetyl-CoA reductase. J. Biol. Chem. 263:15293-15297. [PubMed] [Google Scholar]
- 35.Pohlmann, A., W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Pötter, E. Schwartz, A. Strittmatter, I. Voß, G. Gottschalk, A. Steinbüchel, B. Friedrich, and B. Bowien. 2006. Hydrogen-based biotechnology: genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257-1262. [DOI] [PubMed] [Google Scholar]
- 36.Pötter, M., M. H. Madkour, F. Mayer, and A. Steinbüchel. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148:2413-2426. [DOI] [PubMed] [Google Scholar]
- 37.Pötter, M., H. Müller, F. Reinecke, R. Wieczorek, F. Fricke, B. Bowien, B. Friedrich, and A. Steinbüchel. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301-2311. [DOI] [PubMed] [Google Scholar]
- 38.Pötter, M., H. Müller, and A. Steinbüchel. 2005. Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology 151:825-833. [DOI] [PubMed] [Google Scholar]
- 39.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 40.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 42.Prüß, B. M., D. J. Kim, S. Forst, R. T. Fleming, K. L. Visick, and A. J. Wolfe. 2005. Global regulatory networks in enteric bacteria. Genomics of flagella. Research Signpost, Kerala, India.
- 43.Reitzer, L., and B. L. Schneider. 2001. Metabolic contest and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Römermann, D., M. Lohmeyer, C. G. Friedrich, and B. Friedrich. 1988. Pleiotropic mutants from Alcaligenes eutrophus defective in the metabolism of hydrogen, nitrate, urea and fumarate. Arch. Microbiol. 149:471-475. [Google Scholar]
- 45.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2001. Cloning of an intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saegusa, H., M. Shiraki, and T. Saito. 2002. Cloning of an intracellular d-(−)-3-hydroxybutyrate oligomer hydrolase gene from Ralstonia eutropha H16 and identification of the active site serine residue by site-directed mutagenesis. J. Biosci. Bioeng. 94:106-112. [DOI] [PubMed] [Google Scholar]
- 47.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209-222.13747777 [Google Scholar]
- 48.Schlegel, H. G., R. Lafferty, and I. Krauss. 1970. The isolation of mutants not accumulating poly-β-hydroxybutyric acid. Arch. Microbiol. 71:283-294. [DOI] [PubMed] [Google Scholar]
- 49.Schubert, P., A. Steinbüchel, and H. G. Schlegel. 1988. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-β-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J. Bacteriol. 170:5837-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz, E., A. Henne, R. Cramm, T., Eitinger, B. Friedrich, and G. Gottschalk. 2003. Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H2-based lithoautotrophy and anaerobiosis. J. Mol. Biol. 332:369-383. [DOI] [PubMed] [Google Scholar]
- 51.Shevchenko, A., O. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 52.Shingler, V. 1996. Signal sensing by sigma 54-dependent regulators: derepression as a control mechanism. Mol. Microbiol. 19:409-416. [DOI] [PubMed] [Google Scholar]
- 53.Slater, S., K. L. Houmiel, M. Tran, T. A. Mitsky, N. B. Taylor, S. R. Padgette, and K. J. Gruys. 1998. Multiple β-kethothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 180:1979-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinbüchel, A. 1991. Polyhydroxyalkanoic acids, p. 123-213. In D. Byrom (ed.), Biomaterials. Macmillan, Basingstoke, United Kingdom.
- 55.Steinbüchel, A., and H. E. Valentin. 1995. Diversity of microbial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219-228. [Google Scholar]
- 56.Timm, A., and A. Steinbüchel. 1990. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl. Environ. Microbiol. 56:3360-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uchino, K., T. Saito, B. Gebauer, and D. Jendrossek. 2007. Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J. Bacteriol. 189:8250-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voigt, B., T. Schweder, M. J. J. B. Sibbald, D. Albrecht, A. Ehrenreich, J. Bernhardt, J. Feesche, K.-H. Maurer, G. Gottschalk, J. M. van Dijl, and M. Hecker. 2006. The extracellular proteome of Bacillus licheniformis grown in different media and under different nutrient starvation conditions. Proteomics 6:268-281. [DOI] [PubMed] [Google Scholar]
- 59.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 60.Wieczorek, R., A. Pries, A. Steinbüchel, and F. Mayer. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkins, M. R., J. C. Sanchez, A. A. Gooley, R. D. Appel, I. Humphery-Smith, D. F. Hochstrasser, and K. L. Williams. 1996. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 13:19-50. [DOI] [PubMed] [Google Scholar]
- 62.Williams, S. F., and D. P. Martin. 2002. Applications of PHAs in medicine and pharmacy, p. 91-127. In Y. Doi and A. Steinbüchel (ed.), Biopolymers, vol. 4. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 63.York, G. M., J. A. Stubbe, and A. J. Sinskey. 2001. New insight of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J. Bacteriol. 183:2394-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.York, G. M., J. Stubbe, and A. J. Sinskey. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.York, G. M., J. Lupberger, J. M. Tian, A. G. Lawrence, J. Stubbe, and A. J. Sinskey. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 185:3788-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]