Abstract
Human histo-blood group antigens (HBGA) have been identified previously as candidate receptors for human norovirus (NOR). Type A, type H1, and Lewis HBGA in humans have been identified as major HBGA for NOR binding. We have found that pig stomach (gastric) mucin (PGM) contains blood group A, H1, and Lewis b HBGA and binds to multiple strains of NOR more broadly than do specific antibodies to NOR. Both genogroup I (GGI) and GGII NOR strains were recovered by PGM-conjugated magnetic beads. A fecal sample containing GGII NOR was detected at a dilution of 1:1,000,000 by the standard RNA extraction procedure, whereas NOR in a 1:100,000,000 dilution could be concentrated by PGM-conjugated magnetic beads and NOR in spiked food samples (e.g., oyster extract, strawberry, raspberry, and lettuce) was captured by PGM, thus minimizing the reverse transcription-PCR inhibitors in food and increasing sensitivity.
Noroviruses (NOR) cause millions of cases of sporadic and epidemic gastrointestinal disease in the United States annually, accounting for 50 to 67% of food-borne illnesses (4, 6). NOR are the most frequent cause of outbreaks of acute gastroenteritis following the ingestion of raw shellfish (3, 14). In addition, various common-source food-borne vehicles that have been implicated in NOR outbreaks include bakery frosting, salad, celery, melon, vermicelli, consommé, fruit salad, coleslaw, frozen raspberries, sandwiches, lettuce, cold cooked ham, commercial ice, and water (15). In most outbreaks of food-borne viral gastroenteritis involving, for example, fresh produce, it has been challenging to detect the viruses directly from the food samples by current methods, presumably due to the low concentration of virus and the inhibition of PCR methods of detection by inhibitors in food samples.
The virus cannot be grown in cell culture; therefore, reverse transcription (RT)-PCR is the primary tool for the detection of these viruses (9). Therefore, the low concentration of virus in food samples requires steps to remove inhibitors from food samples for RT-PCR (2). Depending on the type of food sample, various complicated nucleic acid extraction methods are required to concentrate viral RNA and remove RT-PCR inhibitors (2, 3, 11, 12, 14). Most methods involve multiple extraction and elution procedures that create the potential for viral RNA loss at each step.
We have described previously an immuno-PCR (I-PCR) method for the detection of capsid protein of NOR in food samples by using strain-specific polyclonal antibodies. The method has proven to be excellent for removing NOR from inhibitors in food samples such as fruits, vegetables, and oysters. However, the virus capsid proteins are captured by strain-specific antibodies, thus requiring that multiple antibodies be produced and characterized for the appropriate specificity and then pooled in order to capture multiple strains of NOR. Human histo-blood group antigens (HBGA) have been reported previously as candidate receptors for human NOR. Eight distinct patterns of binding between different NOR and HBGA have been identified (7). NOR binding to type A HBGA has been identified in four of five binding patterns that involve the AB blood group antigens (group A = GalNAcα1-3Gal [Fucα1-2] β1-4 GlcNAc-R; group B = Galα1-3Gal [Fucα1-2] β1-4GlcNAc-R). NOR binding to Lewis y HBGA (Fucα1-2Gal β1-4GlcNAc [α1-3Fuc] β1-3Gal-R) has been identified in two of three binding patterns that involve the Lewis antigens. Ideally, mixed HBGA could be a good candidate for binding multiple NOR strains. However, there will be technical and ethical issues in using human HBGA for NOR detection. Previously, we have shown that pigs have at least some of the same HBGA that occur in humans. Individual pigs have either type A or type H1 HBGA (16). Pig stomach (gastric) mucin (PGM) contains mixed type A, type H1, and Lewis b HBGA and binds to recombinant NOR (rNOR) of genotypes I and II (16). We report in this study the conjugation of HBGA from PGM to magnetic beads and the immunomagnetic separation of multiple NOR strains from clinical and food samples.
MATERIALS AND METHODS
NOR and viral RNA extraction.
NOR-positive clinical samples were kindly provided by David Schnurr (State of California Department of Health Services). Genogroups of the virus were determined by real-time PCR (10). A genogroup II (GGII) and a GGI NOR from outbreaks in California were used for spiking of and detection of NOR in food samples. Viral RNA was extracted with a QIAamp viral RNA mini kit (catalog no. 52906; Qiagen, Valencia, CA) by following the protocol of the manufacturer.
Making PGM-conjugated magnetic beads (PGM-MB) for a capture assay.
Type III PGM (catalog no. M-1778; Sigma, St. Louis, MI) was cross-linked to MagnaBind carboxyl derivatized beads (Pierce Biotechnology, Rockford, IL) by following the manufacturer's protocol. Bead assays involved adding 40 ml of phosphate-buffered saline (PBS) to sterile 50-ml v-bottom centrifuge tubes. One hundred forty microliters of PBS-diluted fecal sample was then added. One hundred forty microliters of sample was saved from each tube as the input. One hundred microliters of PGM-conjugated beads was washed three times with PBS and added to the tube, which was then rotated for 1 h with a Labquake rotisserie (Barnstead International, Chicago, IL). Beads were collected by inserting the tube into a magnetic separation rack (New England BioLabs, Ipswich, MA) for 1 h. The beads were transferred to an Eppendorf tube and washed three times with 1 ml of PBS. The beads were resuspended in 140 μl of PBS. Captured viral RNA was released by heating the tube at 95°C for 5 min (13).
NOR (0.01 to 100 RT-PCR units [RTU]) was added either to a fruit and vegetable wash mixture or to the surface of fresh produce or vegetable. In the latter case, after complete drying, fruit or vegetable samples were washed by vigorous shaking in 40 ml of PBS in a 50-ml Eppendorf tube. The debris was removed by filtration through filter paper in a sterile Buchner funnel. One hundred microliters of PGM-conjugated beads was added to the tube, which was rotated for 1 h with a Labquake rotisserie. Beads were collected and washed, and viral RNA was heat released as described in the previous paragraph.
Live pacific oysters (Crassostrea gigas) were obtained by overnight shipment from Willapa Oysters (Oysterville, WA). Each oyster was shucked and dissected within 2 days of harvest to obtain oyster stomach and digestive tissue. One gram of oyster stomach and digestive tissue was homogenized in 10 ml of PBS (pH 7.4) in an Omni TissueMaster homogenizer (Omni International, Marietta, GA). Homogenized samples were centrifuged at 3,000 × g for 15 min at 4°C. One milliliter of supernatant from a mixed sample of oysters was added to 40 ml of PBS. NOR (0.01 to 100 RTU) was added to oyster samples, and detection of NOR bound to beads was performed as described above.
Real-time RT-PCR for detection of NOR.
A OneStep RT-PCR kit was purchased from Qiagen (Valencia, CA). Real-time RT-PCR was performed on a Stratagene MX3000P QPCR system (Stratagene, La Jolla, CA) with primers and probes described previously (10) and a OneStep RT-PCR kit (Qiagen, Valencia, CA). The primers (10) used for GGI NOR included COGIF (5′ CGY TGG ATG CGN TTY CAT GA 3′), COGIR (5′ CTT AGA CGC CAT CAT CAT TYA C 3′), GI-P1 (5′ FAM-AGA TYG CGA TCY CCT GTC CA-IB FQ 3′), and GI-P1b (5′ FAM-AGA TCG CGG TCT CCT GTC CA-IB FQ 3′, where FAM is 6-carboxyfluorescein and IB FQ is Iowa Black fluorescence quencher). The primers (10) used for GGII NOR include COGIIF (5′ CAR GAR BCN ATG TTY AGR TGG ATG AG 3′), COGIIR (5′ TCG ACG CCA TCT TCA TTC ACA 3′), and GII-P (5′ Cy3-TGG GAG GGC GAT CGC AAT CT-IB RQ 3′). Each 25-μl reaction mixture consisted of 5 μl of 5× buffer, 1.5 μl of 25 mM MgCl2, 1 μl of 10 mM deoxynucleoside triphosphates, 0.75 μl of each primer (COGIF, COGIR, COGIIF, and COGIIR) at 10 μM, 0.25 μl of each probe (GI-P1-FAM, GI-P1B-FAM, and GII-P-CY3) at 10 μM, 1 μl of an enzyme mixture, 0.25 μl of 5 U/μl RNase inhibitor, and 3 μl of extracted RNA. Cycling times and temperatures were 50°C for 50 min and 95°C for 15 min, followed by 45 cycles of 95°C for 10 s, 53°C for 25 s, and 62°C for 70 s.
RESULTS
Comparison of sensitivities of detection of NOR by standard virus extraction procedure and PGM-MB.
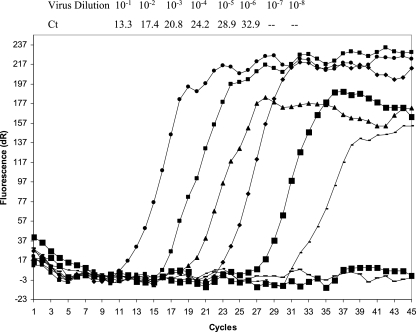
To determine the sensitivity of the PGM-MB method, RNA was extracted from serially diluted GGII NOR and detected by real-time RT-PCR. The cycle threshold (CT) values were 13.28, 17.38, 20.82, 24.20, 28.91, and 32.97 for virus dilutions of 10−1 to 10−6, respectively (Fig. 1). Therefore, the virus load at a dilution of 10−6 was defined as 1 RTU.
FIG. 1.
Detection limit for detection of NOR by real time RT-PCR. NOR virus was diluted serially to different concentrations. Viral RNA was extracted from each dilution and measured by the real time RT-PCR method. Virus dilutions and Ct values are shown at the top.
NOR is captured and concentrated by the PGM-MB method.
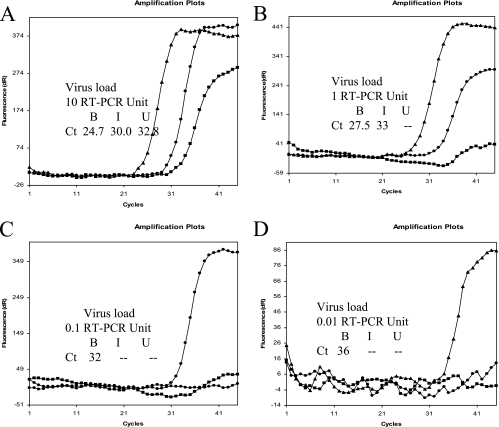
Various concentrations of the different viruses were concentrated further by the PGM-MB method. The GGII virus was diluted to final concentrations of 10 (Fig. 2A), 1 (Fig. 2B), 0.1 (Fig. 2C), and 0.01 (Fig. 2D) RTU. One hundred forty microliters of each sample was added separately to 40 ml of PBS in different tubes. One hundred forty microliters of each sample was saved as the input. One hundred forty microliters of each sample was obtained after the virus was captured on beads to measure the unbound virus. The tube was removed from the magnet, and beads were resuspended in PBS. Virus RNAs were extracted and quantified by real-time RT-PCR. At a virus load of 10 RTU (Fig. 2A), the CT values for the input (I), unbound (U), and bead-concentrated (B) samples were 30.0, 0 (below sensitivity of the assay), and 24.7, respectively. Similar results were obtained at a virus load of 1 RTU (Fig. 2B). The CT values for I, U, and B were 33, 0 and 27.7, respectively. The difference between the CT values (5.3 to 5.5) of the I and B samples indicates that the viruses were concentrated by approximately 2 logs. At virus loads of 0.1 and 0.01 RTU, viruses in the input (I) sample could not be detected (Fig. 2C and D), as expected. However, virus could be detected in the bead (B) sample. Similar results were observed when GGI viruses were tested (data not shown).
FIG. 2.
Concentration of NOR by PGM-MB. Samples containing various amounts of NOR were concentrated by PGM-MB. I (triangle), B (filled circle), and U (square) represent input, beads recovered, and unbound, respectively. Panels A, B, C, and D represent virus loads of 10, 1, 0.1, and 0.01 RT-PCR unit, respectively. The difference in Ct values between beads recovered and input B indicates that viruses are concentrated by using PGM-MB. A 3.3-fold difference in Ct values represents a 10-fold difference in concentrations. NOR can be concentrated at least 100-fold.
RT-PCR inhibitors can be removed from food samples by the PGM-MB method.
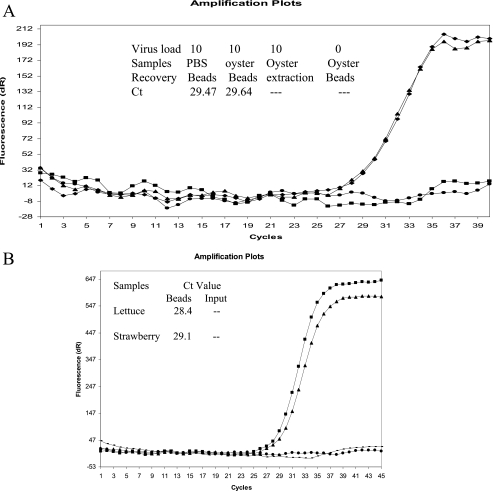
When NOR was used to spike various food samples and then the PGM-MB concentration method was applied, the virus and RT-PCR inhibitors were both removed efficiently. A representative amplification plot of these results is shown in Fig. 3. When 10 RTU of virus was used to spike a 1% oyster homogenate, virus recovery from the oyster homogenate was as effective as recovery from a PBS control, resulting in CT values of 29.64 and 29.47, respectively. No CT value was obtained when virus was extracted directly from a spiked oyster sample, and the RNA was only partially detected in the RT-PCR, indicating inhibition of the reaction by compounds in the sample (Fig. 3A). Similar results were observed when viruses were used to spike fresh food samples such as strawberries and romaine lettuce (Fig. 3B).
FIG. 3.
Detection of NOR in spiked oyster, lettuce, and strawberry samples. A. NOR virus was spiked in PBS (diamond) or in oyster (triangle) samples at a concentration of 10 RT-PCR units and was concentrated by using PGM-MB. An equivalent amount of NOR virus was spiked into oyster samples, and virus RNA was extracted by the standard RNA extraction method (square). Un-spiked oyster samples were used as negative controls (circle). Similar Ct values for recovery of NOR from PBS and oyster samples suggest that inhibitors are removed by PGM-MB. The presence of inhibitors in oyster samples was confirmed in oyster samples where viral RNA was extracted directly (square). B. NOR virus was spiked into lettuce (diamond) or strawberry (triangle) samples at a concentration of 10 RT-PCR units and was concentrated by using PGM-MB. An equivalent amount of NOR virus was spiked into lettuce (circle) and strawberry (square), and virus RNA was extracted by the standard RNA extraction method as an input control.
DISCUSSION
The amount of the virus present in NOR-contaminated ready-to-eat food is usually lower than the concentration of virus that can be detected efficiently by current RT-PCR methods. Additionally, the inhibitors present in many food samples must be removed prior to RT-PCR, thus necessitating complicated concentration and purification procedures. Schwab et al. developed a method to detect NOR in food samples (14). The method involves washing food samples with a guanidium-phenol-based reagent, extraction with chloroform, and precipitation in isopropanol. It was reported that 10 to 100 RTU could be detected with virus in spiked foods. Kingsley and Richards (11) reported an extraction method for the detection of hepatitis A virus (HAV) and NOR in shellfish. The method involves extraction with glycine buffer, polyethylene glycol precipitation, TRI reagent (Sigma, St. Louis, MO), and purification of viral RNA with magnetic poly(dT) beads. The sensitivity reported for NOR-seeded shellfish extracts was 22.4 RTU. An immunomagnetic separation (IMS) method was reported previously to concentrate HAV and minimize inhibitors.
Abd el-Galil et al. developed an IMS method for the detection of HAV in environmental samples with a detection limit of 20 PFU (1). Gilpatrick et al. developed an IMS capture RT-PCR assay for the detection of NOR in stool samples (5). Previously, we developed a sensitive real-time I-PCR (rtI-PCR) method to detect NOR in food samples. The viral antigens were captured by two polyclonal antisera against recombinant Norwalk virus (17). The sensitivity of the rtI-PCR was >1,000-fold higher than that of the standard enzyme-linked immunosorbent assay due to the minimal impact of the PCR inhibitors present in the food samples on antigen capture; the inhibitors were removed by multiple wash steps during the rtI-PCR procedure. The major drawback of this method, however, is that multiple specific antibodies are required to bind different strains, thus requiring a major investment in the production and characterization of reagents for NOR.
The advantages of using PGM-MB include effective recovery of the majority of known human NOR strains and genogroups from clinical and food samples and an improved method to concentrate NOR from and minimize RT-PCR inhibitors in clinical and food samples. Our method resulted in a 2-log increase in the sensitivity of detection of multiple genogroups of NOR present in complex samples by using PGM-MB capture and separation. NOR can be concentrated at least 100-fold by the PGM-MB method under optimal conditions of virus diluted in PBS (Fig. 2). However, similar results were obtained also with viruses used to spike various food samples including oysters, lettuce, and strawberries. These results indicated that the viruses could be captured and RT-PCR inhibitors removed by PGM-MB. Virus recovery was excellent, considering that no obvious change in CT values occurred compared to the nonfood control (Fig. 3A and B). The CT values for virus recovered from the PBS control and from oyster, lettuce, and strawberry samples were 29.47, 29.64, 28.4, and 29.1, respectively. The general applicability of the method reported here for the detection of NOR in oyster and produce samples was confirmed by our method. However, there is minimal information on the nature of the interactions of viruses, including NOR, with the compounds present in food samples, nor any information that would relate to viruses exposed to food samples, including produce, in the field (preharvest contamination) versus during the many points in the food-processing cycle (postharvest contamination). It is presumed by many food safety professionals that most NOR contamination of food occurs during food preparation by ill humans in the home, restaurants, or other facilities where food is prepared or purchased. The sources of NOR contamination and data regarding the potential fate and transport of NOR associated with food will require more sensitive methods for detection, like that described here. Therefore, the usefulness of our method will require that viruses bound to oysters or to various types of produce be released effectively by vigorous mixing and homogenizing of the samples. Our current studies involve testing the effectiveness of methods to release virus exposed to complex and relevant food samples for various lengths of time and under different conditions for detection and identification.
Only one strain each of GGI and GGII NOR was used in this study. Ideally, if a virus binds to HBGA, it should be recovered by the beads containing the corresponding HBGA. Eight distinct binding patterns have been reported for different NOR strains and HBGA (7, 8). Most NOR strains can bind to one or more of the type A, O, or Lewis antigens. We have found that PGM contains multiple HBGA (type A, H1, and Lewis b antigens) that bind multiple NOR strains (16). To further study if the PGM-MB method can be used for the detection of multiple strains of NOR, five GGI (NV, West Chester, SoV, DSV, and Chiba) and nine GGII (HV, SMV, TV, LV1987, LV1997, FH 2002, FH2002a, Hunter, and Sakai) rNOR strains were selected and their abilities to bind to PGM were tested. Of the five GGI strains tested (NV, West Chester, SoV, DSV, and Chiba), the HBGA binding pattern is known for four (NV, SoV, DSV, and Chiba). All of these four strains prefer binding to type A rather than type H1 HBGA. Our preliminary results suggest that all five GGI rNOR strains were captured by PGM. Of the nine GGII strains tested, the HBGA binding pattern is known for six (TV, LV1987, LV1997, FH 2002, FH2002a, and Hunter). The preliminary results suggest that rNOR representing these six strains was captured by PGM. Of the three rNOR strains that do not bind to known ABO and Lewis antigens, two (HV and Sakai) had positive/negative ratios close to 2.0, as measured by the PGM binding assay. We speculate that these viruses were captured weakly by some unknown oligosaccharides in the PGM. We are in the process of obtaining these viruses to determine if they can be captured by PGM for viral RNA extraction and detection by the method we have described.
Acknowledgments
We thank D. Schnurr (State of California Department of Health Service) for providing clinical samples and William Burkhardt III and Gary Hartman (FDA) for helping with oyster dissection work. The five GGI recombinant baculovirus strains expressing capsid proteins (NV, DSV, Sov, WC, and Chiba) and the nine GGII recombinant baculovirus strains expressing capsid proteins (HV, SMV, LV1997, LV1987, TV, FH2002, FH2002a, Hunter, and Sakai) were kindly provided by Lisa Lindesmith and Ralph Baric (Department of Microbiology and Immunology, University of North Carolina at Chapel Hill).
This project was supported by USDA-ARS CRIS project 5325-42000-044.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Abd El Galil, K. H., M. A. El Sokkary, S. M. Kheira, A. M. Salazar, M. V. Yates, W. Chen, and A. Mulchandani. 2005. Real-time nucleic acid sequence-based amplification assay for detection of hepatitis A virus. Appl. Environ. Microbiol. 71:7113-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmar, R. L., and M. K. Estes. 2001. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin. Microbiol. Rev. 14:15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar, R. L., F. H. Neill, J. L. Romalde, F. Le Guyader, C. M. Woodley, T. G. Metcalf, and M. K. Estes. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 61:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels, N. A., D. A. Bergmire-Sweat, K. J. Schwab, K. A. Hendricks, S. Reddy, S. M. Rowe, R. L. Fankhauser, S. S. Monroe, R. L. Atmar, R. I. Glass, and P. Mead. 2000. A foodborne outbreak of gastroenteritis associated with Norwalk-like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J. Infect. Dis. 181:1467-1470. [DOI] [PubMed] [Google Scholar]
- 5.Gilpatrick, S. G., K. J. Schwab, M. K. Estes, and R. L. Atmar. 2000. Development of an immunomagnetic capture reverse transcription-PCR assay for the detection of Norwalk virus. J. Virol. Methods 90:69-78. [DOI] [PubMed] [Google Scholar]
- 6.Glass, R. I., J. Bresee, B. Jiang, J. Gentsch, T. Ando, R. Fankhauser, J. Noel, U. Parashar, B. Rosen, and S. S. Monroe. 2001. Gastroenteritis viruses: an overview. Novartis Found. Symp. 238:5-25. [DOI] [PubMed] [Google Scholar]
- 7.Huang, P., T. Farkas, W. Zhong, M. Tan, S. Thornton, A. L. Morrow, and X. Jiang. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 79:6714-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutson, A. M., M. K. Estes, and R. L. Atmar. 2004. Re: nosocomial outbreak of norovirus gastroenteritis and investigation of ABO histo-blood group type in infected staff and patients. J. Hosp. Infect. 58:163-164. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, X., J. Wang, D. Y. Graham, and M. K. Estes. 1992. Detection of Norwalk virus in stool by polymerase chain reaction. J. Clin. Microbiol. 30:2529-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kingsley, D. H., and G. P. Richards. 2001. Rapid and efficient extraction method for reverse transcription-PCR detection of hepatitis A and Norwalk-like viruses in shellfish. Appl. Environ. Microbiol. 67:4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sair, A. I., D. H. D'Souza, C. L. Moe, and L. A. Jaykus. 2002. Improved detection of human enteric viruses in foods by RT-PCR. J. Virol. Methods 100:57-69. [DOI] [PubMed] [Google Scholar]
- 13.Schwab, K. J., M. K. Estes, F. H. Neill, and R. L. Atmar. 1997. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwab, K. J., F. H. Neill, R. L. Fankhauser, N. A. Daniels, S. S. Monroe, D. A. Bergmire-Sweat, M. K. Estes, and R. L. Atmar. 2000. Development of methods to detect “Norwalk-like viruses” (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLV outbreak. Appl. Environ. Microbiol. 66:213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seymour, I. J., and H. Appleton. 2001. Foodborne viruses and fresh produce. J. Appl. Microbiol. 91:759-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian, P., X. Jiang, W. Zhong, H. M. Jensen, M. Brandl, A. H. Bates, A. L. Engelbrektson, and R. Mandrell. 2007. Binding of recombinant norovirus like particle to histo-blood group antigen on cells in the lumen of pig duodenum. Res. Vet. Sci. 83:410-418. [DOI] [PubMed] [Google Scholar]
- 17.Tian, P., and R. Mandrell. 2006. Detection of norovirus capsid proteins in faecal and food samples by a real time immuno-PCR method. J. Appl. Microbiol. 100:564-574. [DOI] [PubMed] [Google Scholar]