Abstract
To breed resistance to an assortment of infectious phages, continuous cultures of Escherichia coli JM109 grown in a chemostat were exposed to phage mixtures prepared from sewage influent. Four sequential chemostat-grown cultures were each infected with a different phage mixture. At the end of a chemostat run, one phage-resistant colony was isolated and used to inoculate the subsequent culture. This process was repeated, and increased phage resistance of the input bacterial strain resulted from the successive challenges with different phage cocktails. Multiple mutations apparently accumulated progressively. A mutant isolated at the end of the four runs, designated D198, showed resistance to 38 of 40 phages that infect the parent strain, JM109. D198 produced less outer membrane protein C (OmpC) than JM109. However, restoration of the OmpC protein by plasmid-mediated complementation did not completely restore the susceptibility of D198 to the 38 phages. Therefore, alterations beyond the level of OmpC protein production contribute to the phage resistance of D198. PCR-based genetic analysis revealed that D198 has a genome that is 209 kbp (about 200 genes) smaller than JM109. The deletion includes the chromosomal section from ompC to wbbL that encodes the rhamnosyl transferase involved in lipopolysaccharide biosynthesis. Strains D198 and JM109 were comparable in their growth characteristics and their abilities to express a recombinant protein.
Among the many systems available for heterologous protein expression, the gram-negative bacterium Escherichia coli remains one of the most attractive because of its ability to grow rapidly and at high density on inexpensive substrates, its well-characterized genetics, and the availability of a large number of cloning vectors (13, 19). However, phage infection of E. coli cultures can lead to serious problems, including a complete loss of the desired bioproduct and the spread of the destructive bacteriophages. Such problems with culture lysis may reappear suddenly and frequently within the same laboratory. Decontamination is especially difficult in a large factory. If a phage propagated in a bioreactor can spread throughout the plant, it may survive for a long period of time and cause recurrent problems. Although the deleterious effects of bacteriophages are well recognized, there are relatively few published reports addressing this problem.
Extensive work has been conducted to select or breed phage-resistant strains in the dairy industry (7). Dairy fermentation remains susceptible to phage infection, since pasteurized milk is not completely sterilized. In recent years, genetic strategies to improve the phage resistance of bacterial strains developed from knowledge about natural phage defense systems (4, 11, 23). Major categories of natural phage defenses include adsorption barriers, abortive infection mechanisms, and DNA restriction and modification systems (12). One long-term protection strategy is to select phage-resistant mutants with altered adsorption characteristics. These mutants can result from mechanisms that change carbohydrate composition or alter specific phage protein receptors. Nevertheless, in most strains isolated so far, the underlying mutations have not been well characterized. Few alterations in phage receptors have been correlated with a resistance phenotype at the molecular level (16, 17, 18, 24). Therefore, for this type of phage defense strategy to provide protection against different phage species that can use alternative receptors, it is important to understand common and essential features of the phage adsorption process.
Previously, we investigated the interaction between E. coli O157:H7 and its specific bacteriophage PP01 in chemostat continuous culture (15). Following PP01 phage addition, the observed E. coli O157:H7 cell lysis was greater than 4 orders of magnitude. However, the appearance of a series of phage-resistant E. coli organisms, which showed reduced efficiency of plating when PP01 phage was used, led to an increase in the cell concentration in the culture. This observation led us to the idea of strengthening an E. coli strain against phage attack by enrichment methods during continuous culture in a chemostat. As detailed in this report, the sequential exposures to four different phage mixtures prepared from sewage influent succeeded in producing a multiple-phage-resistant E. coli strain. This study describes the methodology for breeding phage resistance and the genetic nature of the resultant E. coli mutant.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and preparation of phage mixtures.
E. coli JM109 was used as a model strain for breeding phage resistance. E. coli (ME8305) RK4784 {ompC deletion mutant of E. coli strain K-12 [F− Δ(argF-lac) recA1 deoC1 ΔbtuB ΔompC]} and E. coli (ME8307) RK4784 {ompA deletion mutant of E. coli strain K-12 [F− Δ(argF-lac) recA1 deoC1 ΔbtuB ΔompA]} were kindly provided by the National Institute of Genetics, Japan. In batch culture, E. coli was cultured overnight in 2 ml of Luria-Bertani (LB) broth at 37°C with shaking (120 rpm). The optical density of the medium at 600 nm (OD600) was measured using a Klett spectrophotometer (Hitachi High-Technologies Corp.) to estimate the cell concentration. Thirty-eight bacteriophages used in this study, IS01 to IS33, IP008, IP052, SP13, EP16, and ECV43, were screened from sewage influent and activated sludge sampled from a wastewater treatment plant in Japan. Samples for phage mixture preparations were taken from an influent to the wastewater treatment plant. The number of inhabitants served by this plant was about 200,000. Most of the contamination was of human origin. The plant treated no effluent from animal farms or industries. The daily volume of the influent was 28,000 m3. The representative sewage influent qualities were as follows: biological oxygen demand, 260 mg/liter; suspended solids, 190 mg/liter; and coliform bacterial count, 600,000 cells/ml. The sewage sample was centrifuged (11,100 × g, 5 min). Then, the obtained supernatant (10 ml) was carefully transferred to a new sterilized tube, mixed with chloroform (100 μl), incubated for 20 min by shaking (120 rpm), centrifuged (11,100 × g, 5 min), and then filtered through a 0.22-μm filter. Thirteen samples of phage mixtures (A to M) were prepared in this way on different days.
Phage infection in continuous culture.
E. coli JM109 was precultured in 2 ml of LB broth at 37°C with shaking (120 rpm). In continuous culture, 150 μl of precultured broth was inoculated into 15 ml of fresh LB broth in a 30-ml culture flask. The culture was mixed with a stir bar. A peristaltic pump was used to supply fresh medium to and remove spent medium from the culture flask at the same flow rate (15 ml/h). The dilution rate (D, 1.0 h−1) was adjusted by changing the pump running speed. Filter-sterilized air was introduced into the headspace of the culture flask at a rate of 2 liters/min. The culture was kept at 37°C with stirring and maintained overnight to establish a steady-state condition. A phage mixture (300 μl) prepared from sewage influent was then added to the culture. The continuous culture was periodically sampled to determine the concentrations of bacteria and phages. The samples were centrifuged at 11,100 × g for 5 min at 4°C to separate the supernatant and cell pellets. The phage titer of the supernatant was determined by serial dilution with sterile SM buffer (10 mM MgSO4, 100 mM NaCl, 0.01% gelatin, and 50 mM Tris-HCl [pH, 7.5]), followed by a plaque assay on lawns of E. coli JM109. The cell pellets were washed and resuspended in and diluted with phosphate-buffered saline, and the viability of the cells was determined by plating them onto LB agar. All assays were done in triplicate. After 150 to 200 h of chemostat continuous culturing, samples of the cultures were plated onto LB agar, and a colony was used as a seed for the next run. This manipulation was repeated four times (chemostat runs A to D).
Construction of the ompC expression plasmid.
The ompC DNA fragment was PCR amplified from chromosomal DNA of E. coli K-12 with primers +OmpC and −OmpC*. The oligonucleotide primers used for PCR are listed in Table 1. The primers contain recognition sites of the restriction enzymes NcoI and SalI, respectively. The PCR products were digested with NcoI/SalI and inserted into the NcoI/SalI-digested pTV118N (Takara, Kyoto, Japan) to produce plasmid pOmpC.
TABLE 1.
Oligonucleotide primers used for PCR
| Primera | Location (start point)b | Sequence (5′-3′)c |
|---|---|---|
| Inside ompC | ||
| +OmpC | 5′ end | CATGCCATGGGCATGAAAGTTAAAGTACTGTCC |
| −OmpC | 3′ end | CCGCTCGAAGAACTGGTAAACCAGACCCA |
| −OmpC* | 3′ end | CCGGTCGACTGATTATCCTCATGCGAAC |
| Upstream of ompC | ||
| +U1 | U (350 bp) | CGGGATCCAAGTTAATGATGATAGCGGGAGT |
| +U2 | U (770 bp) | CGGGATCCGGCGTGGCCGTTGTCTC |
| +U3 | U (1,150 bp) | CGGGATCCGCGCCCACAATGTGTC |
| +U4 | U (7 kbp) | CGGGATCCACGTATTTTCATTCCGGCGG |
| +U5 | U (11 kbp) | CGGGATCCGACGTTGAGCGCACAA |
| +U6 | U (14 kbp) | GCGCCCCGGCGAAGCTTTCGGTGCTGGACGCTA |
| −U1 | U (start codon) | CCCAAGCTTGGGACAGTACTTTAACTTTCAT |
| −U2 | U (1 kbp) | CCCAAGCTTACAACTCAACGCTTGAGATGA |
| −U3 | U (7 kbp) | CCCAAGCTTACGCCATCAGCGGTACGC |
| −U4 | U (11 kbp) | CCCAAGCTTCGGCACCATATAGGCCAGT |
| Downstream of ompC | ||
| −D1 | D (100 bp) | CCCGTCGACTGATTATCCTCATGCGAACG |
| −D2 | D (1 kbp) | CCCAAGCTTATCCCAGGCATCGGCTTC |
| −D3 | D (4 kbp) | CCCAAGCTTACGCAGCTCCAGCGTTTGC |
| −D4 | D (7 kbp) | CCCAAGCTTACACCGCTGTGAAGTGCTC |
| −D5 | D (10 kbp) | CCCAAGCTTCGTATACTCGGCAACGAAGG |
+, sense; −, antisense.
U, upstream; D, downstream.
Restriction sites are underlined.
Spot test.
Seventy strains of E. coli, screened from sewage influent from February to April in 2004 using Chromocult coliform agar medium (Merck), were used for the selection of 38 phages (IS01 to IS33, IP008, IP052, SP13, EP16, ECV43) (Table 2). E. coli was mixed with 0.5% agar and overlaid on an LB plate. Then, 2 μl of phage lysate (>108 PFU/ml) was dropped on that plate and incubated overnight. Each spot test was conducted once. The 38 phages were discriminated by the sampling day, place, and host range.
TABLE 2.
Susceptibility of phage-resistant E. coli strains against 40 phagesa
| Phage | Plaque formation on indicated host strainb
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JM109 | A54 | A102 | A150 | B54 | B102 | B150 | B174 | C54 | C102 | C150 | C292 | D54 | D102 | D150 | D198 | D198/ OmpCc | K-12 ΔompC | |
| IS01 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| IS02 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| IS03 | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||
| IS04 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||
| IS05 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||||
| IS06 | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||
| IS07 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||
| IS08 | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||
| IS09 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS10 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||
| IS11 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||||
| IS12 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||
| IS13 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS14 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS15 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||
| IS16 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||||
| IS17 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS18 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS19 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||
| IS20 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||
| IS21 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||
| IS22 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||
| IS23 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||
| IS24 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||||
| IS25 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||
| IS26 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS27 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS28 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS29 | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||
| IS30 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||
| IS31 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||
| IS32 | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||
| IS33 | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||
| IP008 | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||
| IP052 | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||
| SP13 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| EP16 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||
| ECV43 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||
| T2 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| T4 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||||
The phages are classified based on their receptors. Group A phages (IS03 and IS24) produced plaques on the lawn of D198/OmpC, and group B phages (IS01 to IS02, IS04, IS06, IS08, IS15, IS25 to IS27, IS29 to IS33) produced plaques on the lawn of K-12 ΔompC. Group C phages (IS05, IS07, IS09 to IS14, IS16 to IS20, T2, T4) produced plaques on the lawns of both D198/OmpC and K-12 ΔompC, while group D phages (IS21 to IS23, EP16, ECV043) produced plaques on the lawns of neither mutant.
○, plaque formed.
Transformant of D198 by pOmpC.
Analysis of the outer membrane components of E. coli.
Outer membrane proteins were purified as previously described (16). Following denaturation with sodium dodecyl sulfate (SDS), the cell outer membrane proteins were separated on a 12% polyacrylamide gel containing SDS (SDS-12% PAGE) and stained with Coomassie brilliant blue R-250. Lipopolysaccharide (LPS) was prepared by hot phenol-water extraction (20) and by proteinase K digestion (10). LPS was separated on an SDS-15% PAGE gel and detected by using Sil-Best Stain-Neo (Nacalai Tesque, Inc., Japan). Silver staining of LPS gels enabled detection of O-antigen-free LPS, i.e., LPS containing only the R-core region and the lipid A complex.
Cell growth and β-galactosidase assay.
E. coli cells were grown in 4 ml LB or M9G medium (6 g of Na2HPO4, 3 g of KH2PO4, 0.2 g of MgSO4·H2O, 0.5 g of NaCl, 1 g of NH4Cl, 10 g of Casamino Acids, and 2 g of glucose [per liter of water]) at 37°C with shaking. Cell concentrations were estimated by measuring the OD660 of the medium. The OD660 was measured using a biophotorecorder (model TVS062CA; Advantec Corp., Japan) to estimate the cell concentration.
To analyze the production of a recombinant protein, the phage-resistant strain (D198) and the parent strain (JM109) were each transformed by the plasmid pUC118 and incubated in LB medium supplemented with ampicillin (50 mg/liter) and IPTG (isopropyl-β-d-1-thiogalactopyranoside; 10 mM). When the OD600 reached 0.6, the cells were separated by centrifugation, and β-galactosidase activity was assayed (14).
RESULTS
Growth of E. coli JM109 and sewage phage samples in continuous culture.
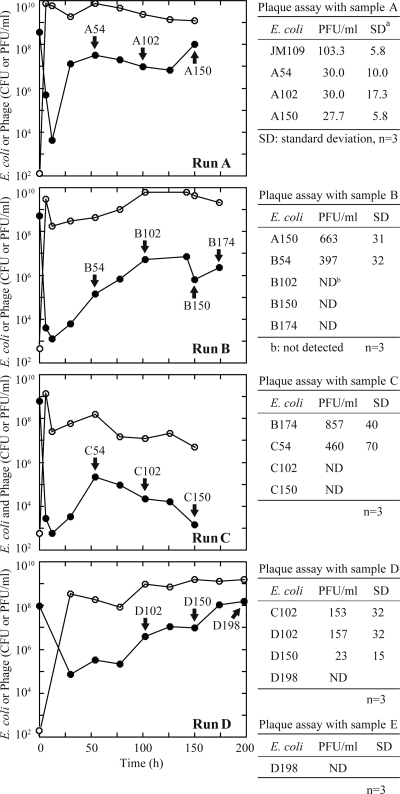
Chemostat continuous culture (D, 1.0 h−1) was employed for analyzing the effect of phage mixtures on E. coli growth (Fig. 1). Before the addition of phage samples, E. coli cells were incubated overnight and achieved a cell concentration of approximately 5 × 108 CFU/ml. Next, a phage mixture prepared from sewage influent, designated sample A, was added to the flask at time zero. The phage concentration in sample A, when assayed on E. coli JM109, was 103.3 PFU/ml. Initially after infection with phage sample A, there was an approximately 105-fold decrease in bacterial concentration that was accompanied by an approximately 108-fold increase in phage concentration. However, E. coli was not washed out of the system. The cell concentration ascended again to a level of 107 CFU/ml 10 hours after phage infection.
FIG. 1.
Continuous cultures of E. coli JM109 and its derivatives with phage mixtures prepared from sewage. Phage sample A was added to a chemostat inoculated with JM109 (run A). Different phage mixtures were added to the chemostats inoculated with isolates from each previous run; strain A150 was grown with sample B (run B), strain B174 with sample C (run C), and strain C102 with sample D (run D). The number following the run designation (A to D) indicates the time of incubation after the phage addition. Bacteria (filled circles) were first allowed to reach equilibrium density in LB medium at 37°C and a dilution rate of 1.0 h−1. At time zero, a phage mixture was added to the culture (open circles) (concentration determined by assay with JM109). At the times indicated by arrows, culture medium was used to isolate phage-resistant strains. The tables show plaque assay data with each phage sample on a lawn of the strain indicated. Standard deviations (SD) and the number of titrations (n) are shown.
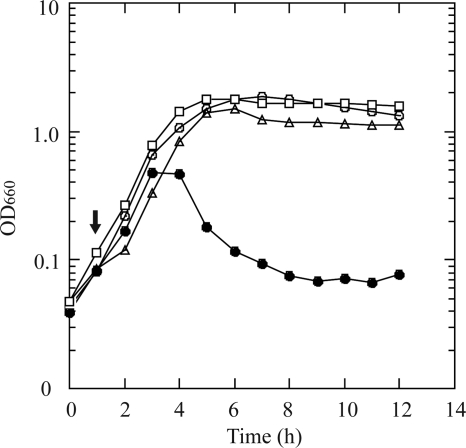
Three E. coli colonies were isolated from the spent culture medium, and the corresponding strains were designated A54, A102, and A150. The number following “A” in the designations indicates the time after phage addition (in hours) at which the bacteria were removed from the chemostat. Plaque assays were conducted by using phage sample A on lawns of E. coli JM109 (wild type), A54, A102, and A150 (Fig. 1, tables). The PFU values for the last three strains were all less than 30% that of the parent strain, JM109. However, plaque assays indicated that A54, A102, and A150 were not completely resistant to all phages present in sample A. Some of the phages in the mixture could have been washed out during the early period of the chemostat run, and they might remain infectious to A54, A102, and A150. Phage resistance of these E. coli strains was further assessed by determining growth curves during batch culturing (Fig. 2). After the addition of phage sample A to the exponentially growing E. coli wild type, JM109, the marked decrease in OD660 was presumably due to cell lysis. However, no such decrease in the culture turbidity was observed with the phage-resistant strains, A54, A102, and A150.
FIG. 2.
Growth of E. coli JM109 and its derivative strains after infection with phage sample A. Overnight LB medium-grown cultures of bacteria were diluted to about 107 CFU/ml in fresh LB medium (5 ml) and incubated at 37°C with shaking (120 rpm) for 1 hour. The phage mixture (100 μl) was then added to the culture (arrow). Bacterial growth or lysis was monitored by measuring the culture turbidity (660 nm). E. coli strains JM109 (filled circles), A54 (open circles), A102 (open triangles), and A150 (open squares) were used.
Strain A150 was used to inoculate the second chemostat culture (run B), to which a different sewage-derived phage mixture, sample B, was added. The phage concentration in sample B, when assayed with E. coli strain A150, was 663 PFU/ml. Chemostat continuous culture was applied for further breeding of phage-resistant cells. As occurred during the first chemostat run, when the phage sample was added to the continuous culture, there was a sudden drop in the E. coli cell concentration and a concomitant increase in the phage concentration (assayed by infection of E. coli JM109 cells). Then, recovery of cell concentration was observed, as in the case of run A. Four phage-resistant derivatives were purified from the continuous culture (strains B54, B102, B150, and B174) and used as the host strains in the plaque assay with phage sample B. No plaques were detected with strain B102, B150, or B174. The same procedure was applied twice more using different phage mixtures, samples C and D, in chemostat runs inoculated with strains B174 and C102, respectively (Fig. 1). A phage-resistant strain, designated D198, was finally isolated from the final run, run D. D198 produced no plaques when assayed with another phage mixture, sample E.
Susceptibility of E. coli isolates to different phages.
The transition of E. coli isolates from being phage susceptible to being phage resistant in this experiment was analyzed by a phage spot test (Table 2). The phages used were isolated from sewage influent or activated sludge of a wastewater treatment plant. These phages showed a different host range among E. coli cells isolated from sewage influent (data not shown). Three strains that were isolated from run A, A54, A102, and A150 (Fig. 1), were susceptible to all of the individual phages tested. As the chemostat continuous culturing progressed, the E. coli isolates acquired resistance to an increasing number of individual phages. The mutations responsible for the phage resistance most likely accumulated. However, in some cases, a strain isolated at a later time was susceptible to a phage to which an earlier isolate was resistant. For example, D54 was resistant to IS01, yet D102 was susceptible to the same phage. D198 was resistant to all phages except SP16 and T2.
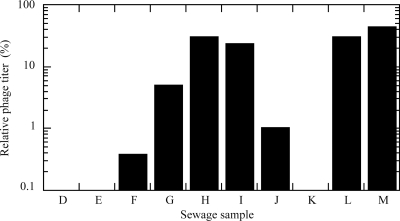
The susceptibility of D198 to different phage mixtures, samples D to M, was analyzed by the relative phage titer, the ratio of the number of plaques produced on a lawn of D198 to that produced on a lawn of JM109 (Fig. 3). Phage mixtures in samples D, E, and K did not produce any plaques on D198. However, phage mixtures in the other samples produced plaques, indicating that D198 was not completely resistant to all phages in the sewage influent. The relative phage titer for sample M was the highest (44%).
FIG. 3.
Analysis of the phage resistance of D198. Plaque assays were conducted on a lawn of strain D198 with phage mixtures prepared from sewage influents (samples D to M). The relative phage titer (%) was defined as the ratio of the number of plaques formed on a lawn of D198 to the number formed on a lawn of its parent strain, JM109.
Outer membrane protein analysis.
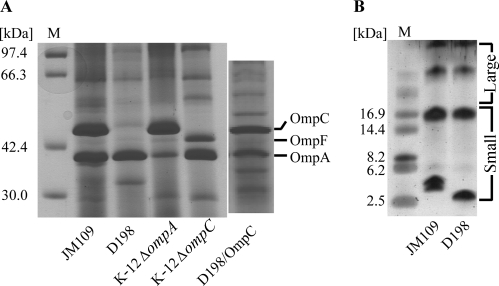
The levels of three E. coli major outer membrane proteins, OmpA, OmpC, and OmpF, were compared in the SDS-12% PAGE profiles of JM109, D198, an ompA deletion mutant, and an ompC deletion mutant. The increase of OmpF production in the ΔompC mutant should be a result of the deletion of micF, which is an inhibitor of OmpF located upstream of ompC in E. coli (2). The production of OmpC was not detected in D198. The loss of OmpC in D198 may account for the reduced affinity of this strain for the sewage phages, since OmpC may serve as a receptor protein for some of these phages. To test this hypothesis, the plasmid pOmpC, which encodes OmpC of E. coli K-12, was constructed and introduced into D198. This transformant (D198/OmpC) was grown in the presence of IPTG to induce OmpC, and SDS-12% PAGE revealed a protein band corresponding in size to OmpC (Fig. 4A). The susceptibility of D198 to the sewage phages was partly restored by this complementation, and 17 phages regained infectivity to strain D198/OmpC (Table 2). However, OmpC complementation did not completely restore susceptibility. The specific deletion of ompC from E. coli K-12 led to the resistance to seven phages, IS03, IS21 to IS24, EP16, and ECV43 (Table 2).
FIG. 4.
Bacterial envelope analysis. (A) Outer membrane protein analysis of JM109, D198, K-12 ΔompA, K-12 ΔompC, and D198/OmpC (a transformant of D198 with pOmpC which encodes OmpC of E. coli K-12). The positions of OmpA, -C, and -F are indicated on the right of the gel. (B) LPS analysis performed by using Sil-Best Stain-Neo. The molecular-size standards are shown on the left (lanes M).
LPS was found in both the large-molecular-size (>30-kDa) and small-molecular-size (<20-kDa) ranges in both JM109 and D198. However, the pattern of small-molecular-size LPS was altered (Fig. 4B).
Identification of the D198 genetic deletion.
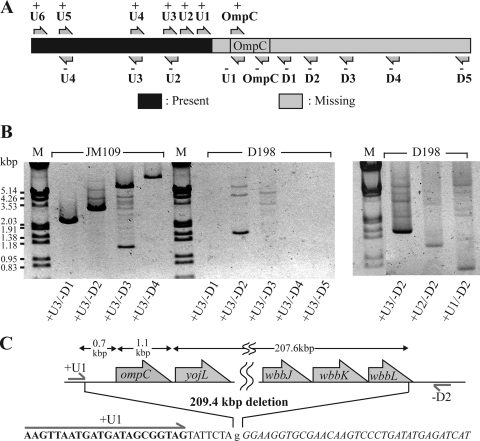
Since D198 did not produce OmpC, genetic analysis of the ompC region was conducted by PCR. The relative locations of the primers used for PCR are summarized in Fig. 5A. The primer set +OmpC/−OmpC did not generate an amplicon with template DNA from D198, suggesting a deletion (data not shown). However, analysis of the region upstream of ompC by PCR with various primer sets (+U6/−U4, +U5/−U3, and +U4/−U2) revealed fragments of the expected sizes with template DNA from either JM109 or D198, indicating that the genome of D198 was unaltered in this region (data not shown).
FIG. 5.
Genetic analysis of phage-resistant E. coli strain D198. (A) Positions of primers used for PCR-based gene analysis. (B) PCR analysis of JM109 and D198. Primer sets used are indicated at the bottom. (C) DNA sequence of the interrupted region of the D198 genome. The sequence in bold is identical to the sequence of the primer +U1, and the sequence in italics is identical to the region that is 207.6 kbp downstream of ompC. The lowercase “g” represents a nucleotide that is not common to either of them. Arrows indicate putative open reading frames based on the E. coli K-12 genome.
In contrast, the genomic region downstream of and encompassing ompC appeared to be missing in D198, since PCRs with many primer sets (including primer sets +U3/−OmpC, +U3/−U1, and +OmpC/−D1) generated DNA fragments of the expected sizes with template DNA from JM109 but not D198 (data not shown). To define the endpoints of the deletion, the upstream primer +U3 was used with a series of downstream primers (D1 to D5). DNA of the expected size was observed with the JM109 template, whereas only the primer set +U3/−D2 produced a distinct band with the D198 template (Fig. 5B). However, the size of the fragment was consistent with a significant chromosomal deletion in D198. The +U2/−D2 and +U1/−D2 primer sets also generated amplicons with D918 template DNA (Fig. 5B). The PCR-generated fragment produced by primer pair +U2/−D2 was digested with BamHI and HindIII and ligated into a similarly digested pUC118 vector. The DNA sequence of the cloned fragment revealed a segment corresponding to the primer +U1 and its 7-bp downstream region. This segment was fused with DNA corresponding exactly to the sequence of a region of the E. coli K-12 genome (NCBI/GenBank accession no. NC-000913) that is normally 207.6 kbp downstream from ompC. Inadvertently, part of primer −D2 (5′-CCCAAGCTTATCCCAGGCATCGGCTTC-3′) carries a sequence complementary to this region (shown underlined). To summarize the above analysis, phage-resistant E. coli JM109 (i.e., D198) lost a genomic fragment of approximately 209.4 kbp from ompC to wbbL that encodes the rhamnosyl transferase involved in LPS biosynthesis.
Recombinant protein production.
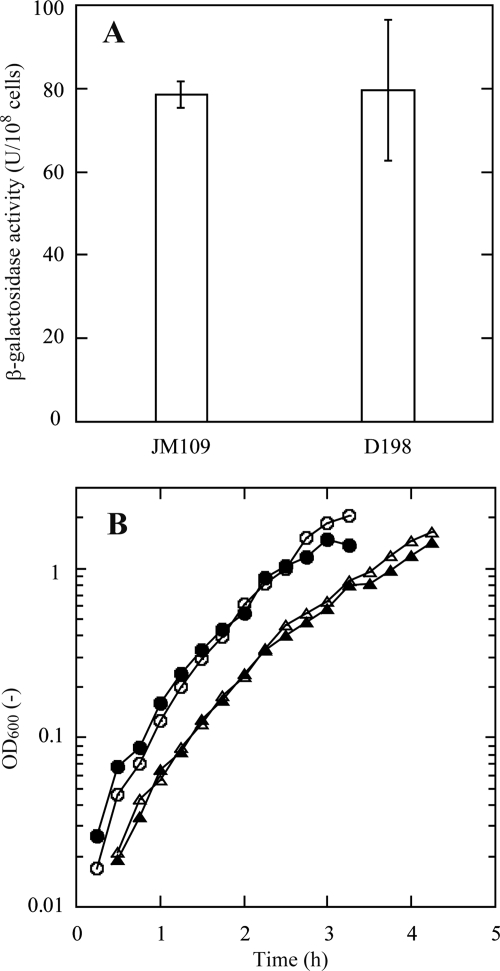
To determine whether the genetic changes affecting phage resistance would impact the potential use of D198 as a host for recombinant plasmids, the growth rates of this strain were compared to those of the parent strain, JM109. As shown in Fig. 6, the growth rates of both strains were indistinguishable on rich medium or minimal medium. To compare the expression levels of a recombinant protein, these two strains were transformed with a pUC118 plasmid carrying lacZ. D198 and JM109 produced comparable levels of β-galactosidase (LacZ) enzyme activity (Fig. 6).
FIG. 6.
Cell growth and β-galactosidase assay. (A) β-Galactosidase activity of E. coli cells (JM109 and D198 transformed by pUC118). I-bars show standard deviations (n = 3). (B) Profiles of E. coli cell growth. JM109 in LB medium (open circles), D198 in LB medium (filled circles), JM109 in M9G medium (open triangles), and D198 in M9G medium (filled triangles) were used.
DISCUSSION
The most likely source of phage contamination in the E. coli culturing process is human, since E. coli is one of the main inhabitants of the gastrointestinal tracts of warm-blooded animals. Previously, we reported that a relatively abundant coliphage, with concentrations of 1,000 to 10,000 PFU/ml determined by using three different E. coli strains, was detected in sewage influent (21). In our current study, the sequential exposure of E. coli to multiple-phage mixtures from sewage was used to develop JM109 derivatives that are resistant to many different phages. This approach builds on previous demonstrations that long phage exposure of E. coli selects for resistant cells (15, 16, 22).
Susceptibility tests suggest that the mutations responsible for multiple-phage resistance accumulate sequentially (Table 2). The chemostat provides an important device for studying bacterium-phage interactions. In continuous culture, bacteria rapidly evolve resistance to phage infection. Different mutations can produce distinct resistance phenotypes that, for example, determine whether resistance is partial or complete, determine the magnitude of the physiological cost associated with resistance, and determine whether the mutation can be countered by host-range mutation in the phage. These differences determine the ability of a mutant to invade, the effect its invasion has on the population dynamics of susceptible bacteria and of the phage, and the resulting structure of the community (5). As described here, a single colony was picked up randomly and used for the isolation of phage-resistant cells at each sampling time. However, the medium in the chemostat continuous culture contains a variety of phage-resistant cells. Some of the phage-resistant isolates selected in this study could be washed out from the system in long-term studies. On the other hand, resistant cells that were not selected might grow and become dominant in the system later on.
A phage-resistant strain, D198, was obtained after exposure to four different phage mixtures. However, additional sewage samples contained phages infectious to D198 (samples F to J and L to M) (Fig. 3). Although D198 remained susceptible to some phages, the low relative phage titer suggests that the use of D198 might minimize bacteriophage attack. Furthermore, additional exposure of D198 to different phage mixtures might yield increased resistance. The methodology presented here using successive exposures to heterogeneous mixtures of phages might be applicable to the isolation of other types of bacteria, in addition to E. coli, that are resistant to multiple phages.
Phage infection starts with the adsorption of phages on the bacterial cell surface, and the host range is controlled by the interactions of the phage and its receptor. Generally, phages use outer membrane proteins and/or LPS as their receptor. PP01-resistant cells lost ompC expression due to the deletion of a 14-kbp region upstream of ompC (16). Outer membrane analysis of D198 also indicated the loss of OmpC production. Genotypic analysis of D198 revealed a 209.4-kbp deletion that encompasses the whole ompC and genes necessary for LPS production. The deleted genes include wbbJ (O acetyltransferase), wbbK (glucosyltransferase), and wbbL (rhamnosyl transferase) involved in LPS synthesis. LPS consists of lipid A, core oligosaccharide, and O antigen. The biogenesis of LPS is a complex multistep process. The core oligosaccharide is assembled on preformed lipid A by the sequential glycosyl transfer of each monosaccharide. In addition to the ompC deletion, deletions of transferase genes may contribute to the phage-resistant phenotype.
Since OmpC complementation did not completely restore phage susceptibility to D198, additional factors affect the phage resistance properties of this strain. Genes that are included in the 209.4-kbp deletion (besides ompC) and/or other genomic mutations in D198 appear to be involved in phage interactions. Forty phages used in this study could be classified into four groups based on the OmpC complementation test with D198 and the phenotype of an ompC deletion mutant, K-12 ΔompC (Table 2). Phages classified into group A appear to use OmpC as a receptor, since resistance results from the specific deletion of its gene in K-12 ΔompC and since susceptibility was restored in D198/OmpC. Phages classified into group B do not depend on OmpC as a receptor, since its absence alone does not cause resistance. Phages in this group might use a receptor that is not produced (or not functional) in D198 because of the deletion or other unknown mutations. For phages in group C, the complementation of D198 indicates that OmpC can be used as a receptor. However, since the ompC deletion alone does not confer resistance, members of this group may also be able to use another receptor that is not made or not functional in D198. For phages in group D, it appears that OmpC is necessary but not sufficient for infection.
It is known that phage T4 uses the LPS of the outer cell envelope membrane as a receptor (8, 9). The alteration of LPS shown in Fig. 4 and the fact that genes responsible for the production of LPS were borne in the lost DNA fragment suggest that LPS plays an important role in determining the D198 phenotype. However, a complete genetic analysis of D198 has not yet been conducted.
Although the D198 genome is 4.5% smaller than that of JM109, the two strains have comparable growth rates and produce the same levels of recombinant LacZ activity (Fig. 6). As a recombinant host, E. coli is exposed to a limited and controlled set of conditions. Therefore, the genes required for survival in the gut may not be the same ones required for optimum recombinant protein production. Consistent with this theory, improvements in E. coli as a recombinant host have often involved genetic deletions (1, 3, 6). Here, we demonstrate that a large deletion surrounding ompC strengthens phage resistance, and this study provides a suitable foundation for further genomic reduction in E. coli hosts to be used for recombinant protein production.
Acknowledgments
This work was supported by a Grant-In-Aid for Scientific Research from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Andersen, D. C., J. Swartz, T. Ryll, N. Lin, and B. Snedecor. 2001. Metabolic oscillations in an Escherichia coli fermentation. Biotechnol. Bioeng. 75:212-218. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J., N. Delihas, K. Ikenaka, P. J. Green, O. Pines, O. Ilercil, and M. Inouye. 1987. The isolation and characterization of RNA coded by the micF gene in Escherichia coli. Nucleic Acids Res. 15:2089-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessette, P. H., F. Aslund, J. Beckwith, and G. Georgiou. 1999. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. USA 96:13703-13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binetti, A. G., N. B. Bailo, and J. A. Reinheimer. 2007. Spontaneous phage-resistant mutants of Streptococcus thermophilus: isolation and technological characteristics. Int. Dairy J. 17:343-349. [Google Scholar]
- 5.Bohannan, B. J. M., and R. E. Lenski. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3:362-377. [Google Scholar]
- 6.Cebolla, A., J. L. Royo, V. de Lorenzo, and E. Santero. 2002. Improvement of recombinant protein yield by a combination of transcriptional amplification and stabilization of gene expression. Appl. Environ. Microbiol. 68:5034-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christopher, J. P., J. C. Lesley, J. H. W. Lawrence, M. C. Brenda, D. S. Brian, J. T. Marie, A. H. Howard, and M. P. Kayla. 2000. Efficacy of four conjugal lactococcal phage resistance plasmids against phage in commercial Lactococcus lactis subsp. cremoris cheese starter strains. Int. Dairy J. 10:617-625. [Google Scholar]
- 8.Datta, D., A. Bernhard, and U. Henning. 1977. Proteins of the Escherichia coli outer cell envelope membrane as bacteriophage receptors. J. Bacteriol. 131:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawes, J. 1975. Characterization of the bacteriophage T4 receptor site. Nature 256:127-128. [DOI] [PubMed] [Google Scholar]
- 10.Fujimoto, S., Y. Meno, and K. Horikawa. 1998. Heterogeneity in expression of lipopolysaccharide and major outer-membrane proteins by strains of Escherichia coli O157 with different H-serotypes. Microbiol. Immunol. 42:527-531. [DOI] [PubMed] [Google Scholar]
- 11.Guglielmotti, D. M., J. A. Reinheimer, A. G. Binetti, G. Giraffa, D. Carminati, and A. Quiberoni. 2006. Characterization of spontaneous phage-resistant derivatives of Lactobacillus delbrueckii commercial strains. Int. J. Food Microbiol. 111:126-133. [DOI] [PubMed] [Google Scholar]
- 12.Gwen, E. A., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226. [Google Scholar]
- 13.Mergulhão, F. J. M., D. K. Summersb, and G. A. Monteiro. 2005. Recombinant protein secretion in Escherichia coli. Biotechnol. Adv. 23:177-202. [DOI] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 15.Mizoguchi, K., M. Morita, C. R. Fischer, M. Yoichi, Y. Tanji, and H. Unno. 2003. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl. Environ. Microbiol. 69:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita, M., Y. Tanji, K. Mizoguchi, T. Akitsu, N. Kijima, and H. Unno. 2002. Characterization of a virulent bacteriophage specific for Escherichia coli O157:H7 and analysis of its cellular receptor and two tail fiber genes. FEMS Microbiol. Lett. 211:77-83. [DOI] [PubMed] [Google Scholar]
- 17.Riede, I., K. Drexler, M. L. Eschbach, and U. Henning. 1987. DNA sequence of genes 38 encoding a receptor-recognizing protein of bacteriophages T2, K3 and of K3 host range mutants. J. Mol. Biol. 194:31-39. [DOI] [PubMed] [Google Scholar]
- 18.Riede, I., K. Drexler, M. L. Eschbach, and U. Henning. 1987. T-even-type bacteriophages use an adhesin for recognition of cellular receptors. J. Mol. Biol. 194:23-30. [DOI] [PubMed] [Google Scholar]
- 19.Sharma, S. S., F. R. Blattner, and S. W. Harcum. 2007. Recombinant protein production in an Escherichia coli reduced genome strain. Metab. Eng. 9:133-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slauch, J. M., M. J. Mahan, P. Michetti, M. R. Neutra, and J. J. Mekalanos. 1995. Acetylation (O-factor 5) affects the structural and immunological properties of Salmonella typhimurium lipopolysaccharide O antigen. Infect. Immun. 63:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanji, Y., K. Mizoguchi, M. Yoichi, M. Morita, K. Hori, and H. Unno. 2002. Fate of coliphage in a wastewater treatment process. J. Biosci. Bioeng. 94:172-174. [DOI] [PubMed] [Google Scholar]
- 22.Tanji, Y., T. Shimada, M. Yoichi, K. Miyanaga, K. Hori, and H. Unno. 2004. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 64:270-274. [DOI] [PubMed] [Google Scholar]
- 23.Viscardi, M., R. Capparelli, and D. Iannelli. 2003. Rapid selection of phage-resistant mutants in Streptococcus thermophilus by immunoselection and cell sorting. Int. J. Food Microbiol. 89:223-231. [DOI] [PubMed] [Google Scholar]
- 24.Xiong, X., J. N. Deeter, and R. Misra. 1996. Assembly-defective OmpC mutants of Escherichia coli K-12. J. Bacteriol. 178:1213-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]