Abstract
A novel, soluble cytochrome with an unusual visible spectral signature at 579 nm (Cyt579) has been characterized after isolation from several different microbial biofilms collected in an extremely acidic ecosystem. Previous proteogenomic studies of an Fe(II)-oxidizing community indicated that this abundant red cytochrome could be extracted from the biofilms with dilute sulfuric acid. Here, we found that the Fe(II)-dependent reduction of Cyt579 was thermodynamically favorable at a pH of >3, raising the possibility that Cyt579 acts as an accessory protein for electron transfer. The results of transmission electron microscopy of immunogold-labeled biofilm indicated that Cyt579 is localized near the bacterial cell surface, consistent with periplasmic localization. The results of further protein analysis of Cyt579, using preparative chromatofocusing and sodium dodecyl sulfate-polyacrylamide gel electrophoresis, revealed three forms of the protein that correspond to different N-terminal truncations of the amino acid sequence. The results of intact-protein analysis corroborated the posttranslational modifications of these forms and identified a genomically uncharacterized Cyt579 variant. Homology modeling was used to predict the overall cytochrome structure and heme binding site; the positions of nine amino acid substitutions found in three Cyt579 variants all map to the surface of the protein and away from the heme group. Based on this detailed characterization of Cyt579, we propose that Cyt579 acts as an electron transfer protein, shuttling electrons derived from Fe(II) oxidation to support critical metabolic functions in the acidophilic microbial community.
Biological oxidation of Fe(II) by acidophilic microbial communities found in mines with exposed pyrite ore accelerates the dissolution of FeS2 and acidification of the mine water, resulting in acid mine drainage (AMD), a global environmental problem (8). One of the most-intensively studied AMD sites is the Richmond Mine at Iron Mountain, CA, where copious biofilm communities are found in extremely low-pH (0.5 to 1.0) solutions (2). Most of these communities are pink biofilms dominated by Leptospirillum group II bacteria, with lower abundances of Leptospirillum group III bacteria and several archaeal species (4). A Leptospirillum group II bacterium-dominated biofilm was collected at the “5-way” site at the Richmond Mine (11) and analyzed by metagenomic sequencing (5-way community genomics data set [24]). Proteomic characterization by mass spectrometry (MS) of a similar biofilm isolated from the “AB end” site of the Richmond Mine identified an abundant extracellular protein from Leptospirillum group II bacteria, encoded by gene 20 on sequencing scaffold 20 (gene 14-20), that has a CXXCH heme binding motif common to c-type cytochromes but otherwise insignificant sequence similarity to known proteins (17). The results of gel electrophoresis and N-terminal sequencing confirmed that this protein contained heme and was abundant in the extracellular fraction. The first 40 amino acids deduced from the environmental genomic sequence were nearly identical to the N-terminal sequence deduced for the Fe(II)-oxidizing cytochrome 579 (Cyt579) purified from an isolate of Leptospirillum ferriphilum. The reduction potential of L. ferriphilum Cyt579 was estimated to be ≥660 mV, and the cytochrome was fully reduced in the presence of excess Fe(II) at pH 2.0 (17). A cytochrome with very similar spectral and pH-dependent-redox properties had also been isolated from Leptospirillum ferrooxidans (10). The ability of L. ferriphilum and L. ferrooxidans Cyt579 to oxidize Fe(II) at low pH led to the hypothesis that this novel cytochrome identified in the biofilm acted as the primary Fe(II) oxidant for Leptospirillum group II bacteria.
Here we report the purification and characterization of Cyt579 from a Leptospirillum group II bacterium-dominated biofilm collected at Richmond Mine. The results of detailed biochemical and MS studies of Cyt579 from the biofilm suggest that it functions as a periplasmic electron transfer protein.
MATERIALS AND METHODS
Isolation of extracellular proteins.
Richmond Mine biofilm samples were collected in 50-ml conical Falcon tubes (BD Biosciences, San Jose, CA), frozen at the site on dry ice, and later stored at −80°C. Biofilm samples were collected from the AB end site (near the junction of the “A drift” and B drift) in January 2004; from the C drift site (15 m beyond the AMD dam) in November 2005; and from the “UBA” site (in the A drift) in November 2005. A map describing the field site can be found in online supplementary information of reference 11. To obtain the extracellular fraction, the biofilm was thawed, suspended in 110 ml 0.2 M H2SO4 (pH 1.1), and homogenized in a glass tube by using several vigorous strokes of a tight-fitting, round, glass pestle. The resulting homogeneous cell suspension was stirred for 2 h at 4°C and then centrifuged at 24,000 × g for 12 min. The supernatant is the extracellular fraction used for cytochrome purification. For proteomic analysis, proteins from a 10-ml sample of the extracellular fraction of the biofilm from the C drift were precipitated with 10% trichloroacetic acid and the precipitate was collected by centrifugation, rinsed twice with cold methanol, and air dried.
Purification of Cyt579.
Proteins in the extracellular fraction (150 ml) were precipitated with (NH4)2SO4 and redissolved in ∼5 ml sample buffer (SB) containing 20 mM H2SO4 and 100 mM NH4(SO4)2 at pH 2.2. A light red precipitate at 45% NH4(SO4)2 saturation was gelatinous, indicating the presence of exopolysaccharides. A deeper red precipitate at 95% NH4(SO4)2 contained 75 to 80% of the protein found in the extracellular fraction. This precipitate was dialyzed for 16 h at 4°C against 1 liter SB. The dialysate was loaded onto an SP-Sepharose FF column (5 ml) preequilibrated in SB. The column was washed with 2 column volumes of SB, and the red fraction (9 ml; 4 mg total protein) eluted with 100 mM sodium acetate (NaOAc), pH 5.0. Visible spectroscopy indicated that the pH 5.0 fraction was highly enriched in Cyt579. The remaining protein was removed with a 0 to 2 M NaCl gradient (30 ml) in pH 5.0 buffer. Between 1.2 M and 2.0 M NaCl, light yellow fractions (3 ml each; 2 mg total protein) eluted that had visible spectra consistent with the presence of c-type cytochromes (α-band at 552 nm for reduced samples).
Immunogold labeling of Cyt579 and transmission electron microscopy (TEM) of biofilms.
Polyclonal antibodies were produced in rabbits (Covance, Denver, PA) by using the cation-exchange fraction of Cyt579 as the antigen. Prior to immunization, the antigen was concentrated by using MicroCon spin filters (10-kDa-molecular-mass cutoff; Millipore, Billerica, MA) and resuspended in phosphate-buffered saline. Immunoblotting of a biofilm lysate indicated a high specificity of the antibody preparation for Cyt579 (data not shown). Antibodies were purified from serum by using a Melon gel antibody purification kit (Pierce, Rockville, IL).
A biofilm sample was frozen under high pressure (Bal-tec HPM 010) and freeze substituted in 0.2% glutaraldehyde and 0.1% uranyl acetate in acetone. The sample was then rinsed in acetone and embedded in LR White resin. Microtomed sections (∼70 nm thick) were mounted on carbon-coated, Formvar film-covered nickel grids and blocked with bovine serum albumin and cold-water-fish gelatin (Sigma Aldrich, St. Louis, MO). The anti-Cyt579 antibody was used as the primary antibody, and goat anti-rabbit antibody conjugated with 10-nm gold particles was used as the secondary antibody. After being labeled, samples were fixed in 0.5% glutaraldehyde. Prior to analysis, all samples were stained with uranyl acetate and lead citrate. Samples were observed with an FEI Tecnai 12 TEM operated at 120 kV. Images were recorded on film, and the negatives were scanned and digitally processed to optimize contrast by using Adobe Photoshop.
Separation of forms of Cyt579.
The fraction enriched in Cyt579 (3 mg) from the C drift biofilm in 100 mM NaOAc, pH 5.0, was concentrated (as described above) to ∼1 ml and dialyzed for 16 h against 1 liter of 25 mM l-histidine-HCl, pH 6.2. The dialyzed Cyt579 fraction was loaded onto a 1- by 30-cm chromatofocusing column (PBE 94 Polybuffer exchange; Amersham Biosciences, Piscataway, NJ) preequilibrated with 2 column volumes of pH 6.2 buffer and eluted with PBE 74 Polybuffer, pH 5.0. Two red fractions eluted from the column at pH 5.5 (0.3 mg) and pH 5.1 (1.0 mg), and a third fraction was eluted with 1 M NaCl in 100 mM NaOAc, pH 5.0 (1.5 mg).
pH-dependent Fe(II) oxidation of Cyt579.
The C drift site Cyt579 fraction (1.5 mg/ml) was diluted 1:10 in 100 mM glycine-200 mM SO42−, pH 2.0, and oxidized with a small amount of Fe2(SO4)3 in pH 2.0 buffer. The oxidized Cyt579 was then diluted 1:10 further in buffer that contained 30 mM FeSO4-200 mM SO42− in a 1.5-ml quartz cuvette, and the visible spectrum was obtained after 1 min. Low-pH buffers (pH 1.2 to 4.0) were prepared according to the method described by Schnaitman et al. using glycine and β-alanine (19). The spectrum was retaken after 10 min to ensure that the reaction had reached equilibrium. In all cases, the reaction was >95% equilibrated after 1 min.
Protein MS.
Intact-protein characterization was performed by high-resolution Fourier transform ion cyclotron resonance (FTICR)-MS analysis. All FTICR mass spectra were acquired with a Varian 9.4-Tesla HiRes electrospray FTICR-MS. Micromolar solutions of the purified Cyt579 proteins were prepared in 50:50 water-acetonitrile (with ∼0.1% acetic acid added). Using a syringe pump (flow rate of 1.75 μl/min), the analyte was directly infused into a Z-type electrospray ion source. After generation, ions were accumulated in an external hexapole for 1 s and then transferred into the high-vacuum region with a quadrupole lens system. Detection then followed in the cylindrical analyzer cell of the MS. Calibration of the MS was accomplished externally with the various charge states of the protein ubiquitin, resulting in a mass accuracy of plus or minus 3 to 5 ppm and mass resolutions of 50,000 to 160,000 Da (full width at half maximum), as previously described (7). Ion dissociation was accomplished by infrared multiphoton dissociation (IRMPD) with a Synrad carbon dioxide laser (75-W maximum power and 10.2-μm wavelength). For this experiment, the desired parent ion was isolated by ejecting all other ons from the analyzer cell and dissociated with infrared laser irradiation (30% maximum laser power for 1.5 s), and the resulting fragment ions were measured at high resolution in the FTICR analyzer cell.
To verify amino acid differences in Cyt579 variants, purified samples were denatured, reduced, and digested with trypsin (sequencing grade; Promega, Madison, WI). Peptides were analyzed by using one-dimensional liquid chromatography-tandem MS (LC-MS-MS) on a Thermo Fisher linear-trapping quadrupole instrument. All MS-MS spectra were searched with DBDigger (22) against a database of all proteins predicted by genomic sequencing of biofilm samples (15, 24), as well as all potential amino acid variants of Cyt579. The output data files were then filtered and sorted with the DTA Select algorithm (21) using the following parameters: fully tryptic peptides only; delta correlation value of at least 0.08; cross-correlation scores of at least 25 (+1 ions), 30 (+2 ions), and 45 (+3 ions); and at least two unique peptides per protein.
Amino acid variants were also verified from crude extracellular fractions of biofilms from the A bend, C drift, and UBA sites. Extracellular proteins were denatured, reduced, trypsin digested, and analyzed by using two-dimensional LC-MS-MS on a linear-trapping quadrupole instrument as previously described (11, 15, 17). The MS-MS spectra were searched and filtered by using the same method as described above for the purified protein.
Structural modeling of Cyt579.
For the best possible results of homology modeling, several different techniques were combined (9) with our high-throughput computational system, AS2TS (29). Pairwise sequence alignments using both Smith-Waterman (20) and FASTA (16) and multiple sequence alignments using PSI-BLAST (1) and CLUSTALW (23) were carried out. PSI-BLAST analyses were performed on the nonredundant set of protein sequences in the NCBI database, with an E-value threshold of 0.001. After five iterations on NR sequences, the final PSI-BLAST run was restricted to sequences corresponding to PDB structures.
Secondary structure predictions were tested by using PSIPRED (12) and PHD (18). Structural alignments between all identified templates and preliminary models were calculated by LGA (28), and these results were used to further guide the process of three-dimensional (3D) model construction. Regions of insertion-deletion and uncertain sequence-structure alignments were built as loops. These regions were modeled using LGA (28) by “grafting” in suitable fragments from related structures in PDB. Finally, SCWRL (5) was used to add coordinates for missing side chain atoms.
General methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method of Laemmli (14). The protein concentration was estimated according to the method of Bradford (6). Trypsin digestion and N-terminal sequencing of proteins were performed as described previously (17). Gel filtration was performed on a 1- by 30-cm Superdex 75 column (Amersham Biosciences, Piscataway, NJ) equilibrated with 100 mM NaOAc, pH 5.0, containing 150 mM NaCl. Bovine serum albumin (67 kDa), ovalbumin (43 kDa), chymotrypsin (25 kDa), and RNase A (13 kDa) were used as molecular-mass standards.
RESULTS
Environmental genomic data indicate three distinct Cyt579 genes.
In addition to the previous metagenomics data for the 5-way site, we examined a second genomic data set obtained from a biofilm collected at the UBA site, which was dominated by a Leptospirillum group II species closely related to the characterized species from the 5-way site (15). Two homologs of Cyt579 were identified. One is encoded by gene 8062-147, with an amino acid sequence 99% identical to the amino acid sequence encoded by gene 14-20 from the AB end site; the amino acid sequence encoded by a paralog of this gene, 8062-372, is 83% identical to that encoded by 14-20 (Fig. 1).
FIG. 1.
Alignment of amino acid sequences of Cyt579 from the 5-way and UBA genomic datasets. One gene from the 5-way site (5wayCG14-20) and two from the UBA site (UBA8062-147 and UBA8062-372) encode variants of Cyt579. Three N-terminal start sites observed by sequencing isolated proteins are indicated in gray, as are the predicted heme binding residues Cys68, Cys71, His72, and Met121 (see model in Fig. 8B). The line above the alignment indicates that portion of Cyt579 used for structural modeling. Arrows indicate the N-terminal signal cleavage site and the observed C terminus.
Cyt579 purification from biofilms.
Cyt579 was purified from the acidic wash of the C drift biofilm by using ammonium sulfate precipitation and cation-exchange chromatography at low pH. Visible spectroscopy of the deep red band that eluted at pH 5.0 confirmed the characteristic absorption peak at 579 nm, consistent with the assignment of this cytochrome as Cyt579. Examination of the purified protein by circular dichroism (CD) spectroscopy indicated a structure that is 70% α-helical, 3% β-strand, 8% turn, and 20% disordered when compared with the structures indicated by reference CD spectra (data not shown). These results were distinctly different from those of similar analyses of a purified membrane cytochrome, Cyt572, which consists largely of β-strands (11).
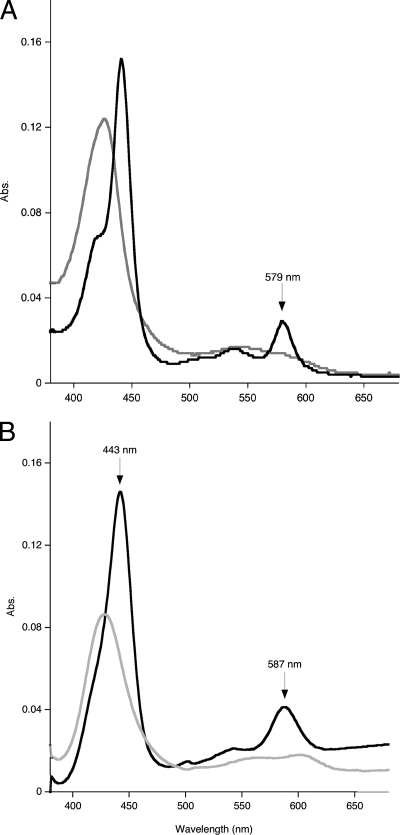
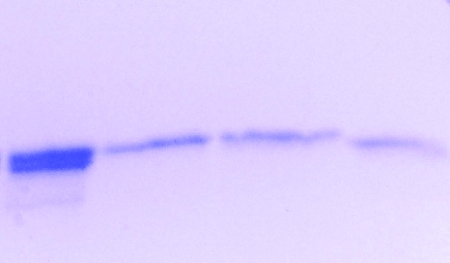
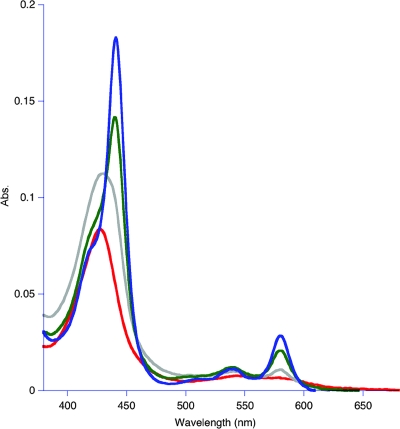
The visible spectrum of purified Cyt579 oxidized with Fe(III) at pH 2.0 exhibited a Soret band at 427 nm. In addition, a weak absorption band at 695 nm characteristic of an axial methionine ligand was observed in concentrated solutions (>0.2 mM) of oxidized Cyt579 (data not shown). Upon reduction of isolated Cyt579 with 500 μM sodium ascorbate, the Soret band shifted to 441 nm and β (539 nm) and α (579 nm) bands were observed (Fig. 2A). The Soret band of the reduced spectrum also had a distinct shoulder at 419 nm, a feature absent in the spectrum of reduced Cyt579 isolated from L. ferriphilum (17). The alkaline pyridine hemochrome spectrum had a Soret band at 443 nm and an α band at 587 nm (Fig. 2B). The results of SDS-PAGE of this fraction revealed two closely spaced protein bands at ∼16 kDa (Fig. 3). Since MS proteomics of this fraction digested with trypsin indicated that >98% of the peptides were from Cyt579, we concluded that two protein species represented different forms of Cyt579 (data not shown). The results of Edman degradation identified two N-terminal sequences of Cyt579 from the C drift biofilm (AELDILKPRV and ILKPRVPAD) that corresponded to the predicted amino acid sequence for all the Cyt579 variants. Identical N-terminal sequences were obtained for a Cyt579 preparation from the AB end site, the original proteomic sample (data not shown). The predicted N-terminal cleavage site to give the N-terminal sequence AELDILKPRV of signal peptidase I is between residues 23 and 24 for the variant sequences of Cyt579. The Cyt579 fraction eluted as a single band at an apparent molecular mass of 20 kDa from a Superdex 75 gel filtration column, consistent with the assignment of Cyt579 as a monomer.
FIG. 2.
Visible spectroscopy of Cyt579. (A) Cyt579 (0.015 mg/ml) isolated from the C drift biofilm in 100 mM glycine-200 mM SO42−, pH 2.0, was treated separately with 5 μl of 10% Fe2(SO4)3 [23% Fe(III)] (gray line) and 5 μl of 1 mM sodium ascorbate (black line) in quartz cuvettes. The spectra were compared to those of the same solutions lacking Cyt579. (B) Cyt579 (1.5 mg/ml) was diluted by adding 50 μl into 450 μl of 0.2 M NaOH, 500 μM sodium ferricyanide (gray line) or 2 mM sodium dithionite (black line), and 500 μl of pyridine was added. Abs., absorbance.
FIG. 3.
Separation of different forms of Cyt579. Chromatofocusing was used to fractionate a Cyt579 sample, and proteins were analyzed on a 10 to 20% acrylamide gel using SDS-PAGE. First lane, C drift biofilm Cyt579 fraction; second lane, C1 fraction; third lane, C2 fraction; and fourth lane, C3 fraction.
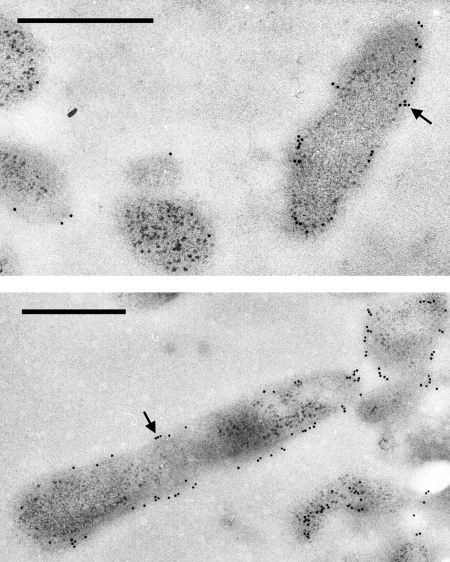
Cyt579 was localized in Leptospirillum group II cells by TEM imaging of a thin section of the C drift biofilm that had been treated with polyclonal antibodies raised against Cyt579 and a secondary gold-labeled antibody. Visualization of the antibody-treated thin section by TEM indicated that Cyt579 was localized on the exterior of the Leptospirillum group II cells and was not distributed throughout the biofilm (Fig. 4). Since Cyt579 contains a signal peptide and has no other hydrophobic regions in its amino acid sequence, we hypothesize that it is located in the periplasm of Leptospirillum group II cells.
FIG. 4.
TEM images of immunogold-labeled biofilm. Ultrathin section of biofilm showing Cyt579 distribution on the edges of cells, possibly in the periplasm, and along the exterior of cells. Two representative fields are shown. Black arrows show gold particles; scale bars show 500 nm.
Multiple forms of Cyt579 separated by chromatofocusing.
As mentioned above, the results of SDS-PAGE indicated that multiple forms of Cyt579 were present in the purified fraction. The forms were too close in molecular weight to separate successfully by gel filtration. However, the forms of Cyt579 were separated by using a preparative chromatofocusing column. Two red bands were eluted at pH 5.5 (C1) and pH 5.1 (C2) in a pH gradient of 6.2 to 5.0. The red fraction remaining on the column was eluted with pH 5.0 1 M NaCl buffer (C3). All three red fractions had nearly identical visible spectra; however, C1 had a Soret band for the oxidized Cyt579 that was shifted to 425 nm, compared to 428 nm for C2 and C3. The results of SDS-PAGE of the separated Cyt579 fractions confirmed that the pH 5.5 and pH 5.1 fractions represented the higher band in the crude Cyt579 fraction, while the pH 5.0 1 M NaCl fraction represented the lower band (Fig. 3). N-terminal sequencing of the individual bands revealed different start sites for each (Table 1). Cyt579-specific polyclonal antibodies detected all three forms of the protein.
TABLE 1.
Forms of Cyt579 identified by N-terminal sequence and intact mass
| Cyt579 fraction | N-terminal sequencea | Avg molecular mass (Da) of species
|
Sequence start and end | |
|---|---|---|---|---|
| Major | Minorb | |||
| C1 | AELDILKPRV | 16,058.97 | 16,045.60 | AELD… LKPE |
| C2 | ILKPRVPAD | 15,691.10 | 15,705.87 | ILKP… . LKPE |
| C3 | AKAMPPFV | 14,316.57 | 14,332.22 | AKAM… LKPE |
| 14,571.63 | − | LAAK… LKPE | ||
N-terminal sequences determined in each fraction by Edman degradation. Overlap in sequences is indicated in bold type.
−, species not detected by MS.
Mass spectrometry of separated Cyt579 forms.
To determine the accurate molecular masses and fragmentation products for the individual forms of C drift biofilm Cyt579, the separated proteins were examined by FTICR-MS. The measured average molecular masses of the peaks in each Cyt579 fraction are given in Table 1.
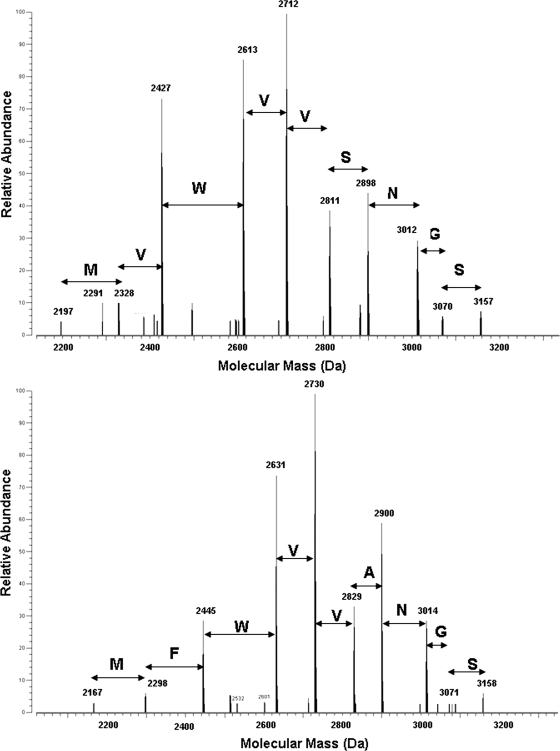
The amino acid sequences of each of these proteins were examined by MS-based fragmentation techniques. Isolation and IRMPD fragmentation of the (M + 13H)13+ ion for the 16,060-Da species revealed a variety of fragment ions, including a sequence tag, MVWVVSNGS, which is representative of the 8062-147-encoded sequence (Fig. 5, upper panel). The larger b-type fragment ions verified the presence of a truncated N terminus, supporting the experimentally determined N terminus, AELDILKPRV, and provided sequence information for the first 110 amino acids of the mature protein. Interestingly, some of the smaller y-type fragment ions revealed truncation of the C terminus, indicating that this form of Cyt579 corresponds to the sequence AELD… . LKPE of the product of gene 8062-147 lacking the C-terminal eight amino acids. The observed mass is also consistent with removal of the heme group from the protein. However, the predicted average molecular mass of this species at 16,075.26 Da is 16 Da heavier than the measured value stated above. Further studies will determine if the discrepancy between the observed and calculated molecular masses of C1 is due to posttranslational modification or is an artifact of purification and mass spectrometry analysis.
FIG. 5.
Sequence tags of Cyt579 obtained by IRMPD dissociation of the molecular species. The upper panel shows C1, 16,060 Da, and the lower panel shows C2, 15,690 Da. Amino acids are presented in the single-letter code above the spectra, and these indicate the difference in sequence between the two variants.
The 15,691 Da (C2) and 14,317 Da (C3) species most closely corresponded to the 8062-372-encoded sequences ILKPR… . LKPE and AKAMP… . LKPE, respectively, based on the observed N-terminal sequences (see Table 1). The additional satellite peak at 14,572 Da in C3 was assigned to the sequence LAAAK… . LKPE, although this N-terminal sequence was not observed by Edman degradation. Based on the measured molecular masses, the C-terminal truncations are identical in the C1 to C3 samples. In each of these cases, the predicted average molecular mass based on the predicted sequence of 8062-372 was 32 Da heavier than the observed mass. To determine if an amino acid variation could account for this difference, the relevant ions from these species were isolated and fragmented by IRMPD as described above. In both C2 and C3, the fragmentation revealed a sequence tag corresponding to the amino acid sequence MFWVVANGS (Fig. 5, lower panel). This sequence was identical to the sequence encoded by gene 8062-372, MFWVVSNGS, except for the Ser to Ala (S112A) variation (in bold), which accounts for a difference of 16 Da. The S112A variation was confirmed by PCR amplification and sequencing of the 8062-372 gene from the C drift biofilm (data not shown). The amino acid variant was also confirmed by the results of two-dimensional LC-MS-MS analyses of the crude extracellular fraction of the C drift biofilm (Table 1). The S112A variation accounts for the observation of the minor species at 15,706 Da (C2) and 14,332 Da (C3). The major species in C2 (15,691 Da) and C3 (14,317 Da and 14,572 Da) may arise from the same posttranslational process as the C1 species.
The S112A variation of the 8062-372 sequence was not found in the genomic data set for the 5-way or UBA genome. However, reexamination of the LC-MS-MS peptide data obtained for the AB end and UBA biofilm extracellular proteomes identified tryptic peptides corresponding to this sequence (Table 2).
TABLE 2.
Spectral counts obtained for MXWVVXN sequences of Cyt579 from extracellular proteomes
| Cyt579 gene | Sequencea | Spectral countb of sample from:
|
||
|---|---|---|---|---|
| AB end | UBA | C drift | ||
| 8062-147 (14-20) | MVWVVSN | 28 | 48 | 57 |
| 8062-372 | MFWVVSN | 85 | 144 | 148 |
| 8062-372 C drift (S112A) | MFWVVAN | 73 | 7 | 128 |
Spectral counts are derived from peptide R.TAGEMXWVVXNGSPLQPMVGFVSAGQITDK.Q. Amino acid substitutions are indicated in bold type.
Spectral counts refer to the total number of MS-MS spectra taken for the peptide as an indicator of overall abundance. Each count is the average of the results for three technical replicates.
Fe(II) oxidation by Cyt579 forms.
Previous work on Cyt579 purified from L. ferriphilum and L. ferrooxidans demonstrated that the oxidized form was fully reduced with excess Fe(II) at pH 2.0 (10, 17). When subjected to the same conditions as L. ferriphilum Cyt579 (30 mM FeSO4, 0.2 M total SO42−), the C drift Cyt579 fraction before separation by chromatofocusing was ∼30% reduced at pH 2.0, as determined by measuring the amplitude of the 579-nm band, in comparison to reduction with sodium ascorbate (data not shown). Studies of the pH dependence of Fe(II) oxidation by Cyt579 indicated that minimal oxidation occurred at pH 1 to 2, but the equilibrium shifted to reduced Cyt579 at a pH of >3, and Cyt579 was almost fully reduced in the presence of 30 mM Fe(II) at pH 4 (Fig. 6). A nearly identical pH dependence of Fe(II) oxidation was observed for the crude Cyt579 fraction obtained from the AB end biofilm, as well as the separated Cyt579 forms (C1 to C3) obtained by chromatofocusing (data not shown).
FIG. 6.
pH-dependent Fe(II) oxidation by Cyt579. The results of redox experiments are shown as follows: pH 1.2 (red), pH 2.0 (gray), pH 3.0 (green), and pH 4.0 (blue). Abs, absorbance.
Structural model of Cyt579.
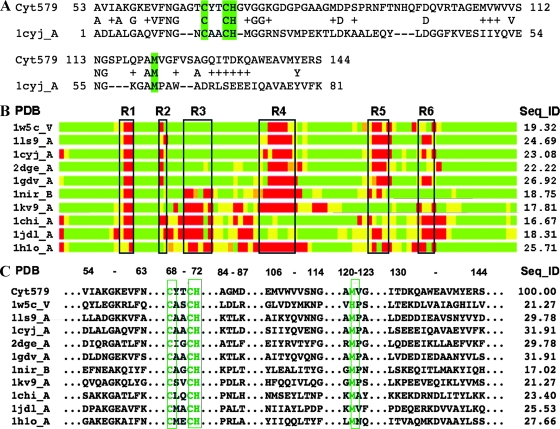
Although no significant homology to Cyt579 was found in protein database searches, over 100 candidate structural templates for modeling Cyt579 were detected, ranging from 7% to 25% sequence identity. Secondary-structure predictions, along with high levels of structural similarities observed between the analyzed templates, narrowed the candidates to 25. An initial 3D model was constructed based on an alignment of the Cyt579 sequence with that of cytochrome c6, 1cyjA (Fig. 7A).
FIG. 7.
Modeling of Cyt579. (A) The initial structural model of Cyt579 was constructed based on sequence alignment with the structure of cytochrome c6, 1cyjA (13). In the alignment, amino acids repeated on the first and second lines are identical, and residues that are chemically similar to those of Cyt579 are indicated by plus symbols. Dashes indicate gaps in the alignment. Highlighted residues Cys68, Cys71, His72, and Met121 form the direct interactions with the heme. (B) Regions in the model having structures similar to those of corresponding regions in the structural templates analyzed are aligned in a schematic bar plot. Structural similarity with these templates is indicated as good (green), intermediate (yellow), and nonhomologous (red). Black boxes (R1 to R6) mark regions of structural deviation, or insertions/deletions, observed in structural templates. The region between R1 and R2 corresponds to the conserved CXXCH heme-binding motif. In Cyt579, the regions R1 to R6 correspond to the following fragments: R1, 65-AGT-67; R2, 73-GV-74; R3, 78-GDGPGA-83; R4, 93-FTNHQFDQ-100; R5, 115-SPLQPA-120; and R6, 126-SAGQI-130. (C) Residue-to-residue correspondences extracted from structurally conserved regions that were identified within a set of the closest structural templates. The results of the analysis of these regions increased confidence in the calculated sequence alignments used in modeling. The results from calculation of sequence identities between the templates and the model in structurally conserved regions are given in the column labeled “Seq_ID”; in most cases these values are higher than the corresponding Seq_IDs calculated for entire structural alignments shown in panel B.
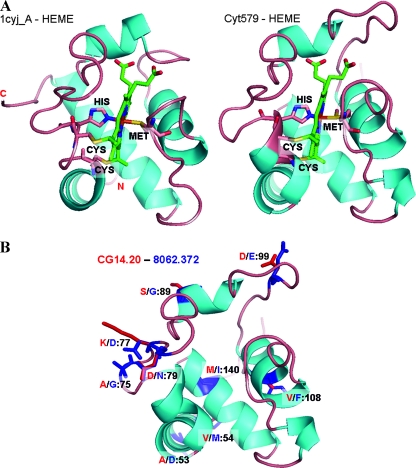
Based on calculated alignments to several structural templates (including RCSB Protein Data Bank accession no. 1cyj, 2dge, 1w5c, 1ls9, 1h1o, 1jdl, 1kv9, and 1nir), the final 3D model was created, including the position of a c-type heme group from cytochrome c6 (13). The heme in Cyt579 is likely to be different, as discussed above, due to the unique spectral character of the cytochrome (see also Discussion). This model was compared by sequence to structure alignments and in 3D plots with selected structural templates (Fig. 7B and C). The heme orientation and structural elements were compared with the cytochrome c6 structure, 1cyj_A (13) (Fig. 8A). Models for the two major genetic variants of Cyt579 were then superimposed to indicate the positions of all nine side chain substitutions, thioether linkages between heme and Cys68 and Cys71, and heme-Fe complex with axial ligands His72 and Met121 (Fig. 8B).
FIG. 8.
Structural comparison and variants of Cyt579. (A) Structure of cytochrome c6, 1cyjA (13), compared with the final model of Cyt579, predicting heme orientation, covalent binding with two Cys residues, and iron coordination complex with axial His and Met residues. (B) Amino acid substitutions are depicted for the two major variants of the Cyt579 gene, CG14-20 (red) and 8062-372 (blue) (see Fig. 1 for sequence alignment).
DISCUSSION
In this study, we have purified the abundant, novel bacterial cytochrome first identified by proteogenomic studies in the acidic-wash fraction of biofilms collected at the Richmond Mine in Iron Mountain, CA. We have confirmed the prediction that this protein is Cyt579, a modified c-type cytochrome that has been implicated as the Fe(II) oxidase in biochemical and physiological studies of Leptospirillum isolates (17). In the initial genomic data set obtained from a biofilm at the Richmond Mine, only one gene was sequenced that coded for Cyt579; however, two paralogs of Cyt579 were sequenced in a genomic data set from a second biofilm (15, 24). The amino acid substitutions observed in these genetic variants can be predicted in a 3D rendering of the protein structure based on homology modeling (Fig. 8B). It is noteworthy that the predicted variant residues are all located on the surface of the protein and in contact with solvent and thus do not appear to impose any perturbance to structural elements or to the putative interactions with heme. Modeling also predicts a His-Met axial ligation for Cyt579 that is consistent with the observation of an absorption band at 695 nm and a mostly helical protein structure that is corroborated by CD spectroscopy.
Detailed biochemical studies of Cyt579 isolated from the biofilms have revealed some unexpected features of Cyt579. The alkaline pyridine hemochrome spectrum closely resembles the spectrum of heme A (Soret band, 430 nm, and α band, 587 nm), suggesting that the heme in Cyt579 may contain a formyl group (3). The spectrum is also consistent with the removal of the heme from the protein, since the α band is red shifted from 579 nm to 587 nm. The presence of a CXXCH amino acid motif and the periplasmic localization of Cyt579 are evidence that this unusual heme is covalently bound to the protein, so its removal under alkaline pyridine conditions is unexpected. One interpretation of this result is that the covalent thioether linkages of the modified heme in Cyt579 are more sensitive to alkaline pH than those of conventional c-type cytochromes.
The second unexpected feature of Cyt579 was the isolation of three forms of the protein, truncated at different sites on the N terminus. One of these forms, with a detected N-terminal sequence of AELDILKPRV, was consistent with removal of the predicted signal peptide; however, the other two forms may result from additional proteolysis. Cyt579 from L. ferriphilum was isolated in one form, corresponding to an N terminus of AELDILKPRV, that is identical to the highest-molecular-weight form of Cyt579 from the biofilm (17). These truncations may be due to proteolytic activity during the preparation of Cyt579; however, identical N-terminal sequences were observed for Cyt579 preparations from the AB end and C drift biofilms, suggesting that the cleavages are not random and are posttranslational modifications that occur in vivo. N-terminal cleavage sites of Cyt579 have been correlated with the different stages of the biofilm life cycle, establishing their ecological relevance (S. W. Singer and M. P. Thelen, unpublished results).
Accurate molecular-mass values for each of the forms of Cyt579 were determined by intact-protein analysis using MS. This confirmed the N-terminal cleavage sites observed by Edman degradation and revealed a C-terminal cleavage site. A particularly significant finding was that a sequence variant of Cyt579 in the C drift sample was not observed in environmental genomic sequences obtained from Richmond Mine biofilms. The sequence was identified by fragmenting the intact protein and isolating a sequence tag that contained an Ala to Ser variation. The presence of the sequence variant was verified by MS-MS analysis of tryptic peptides. High-resolution intact-protein MS will be invaluable in discriminating between variants of the protein isolated from the environment, allowing the correlation of protein variation with changes in environmental conditions.
The third unexpected feature of Cyt579 was that Fe(II) oxidation was not favored thermodynamically at a pH of <3. This result is inconsistent with the results of previous studies with Leptospirillum isolates, where complete reduction of Cyt579 in the presence of 30 mM Fe(II) was observed at pH 2, and casts doubt on the proposed role of Cyt579 as the Fe(II) oxidase for Leptospirillum group II bacteria (10, 17).
The properties of Cyt579 from Leptospirillum group II bacteria are analogous to those of rusticyanin, a periplasmic Cu-containing protein expressed by Acidithiobacillus ferrooxidans, an acidophilic Fe(II)-oxidizing bacterium found in environments similar to those where members of Leptospirillum group II are found. Biochemical and transcriptomic evidence has implicated rusticyanin as the initial electron acceptor for an outer membrane-bound c-type cytochrome, Cyc2, which is the proposed Fe(II) oxidase for A. ferrooxidans (25-27). In support of this analogy, we have recently purified a novel membrane cytochrome, Cyt572, that is expressed by Leptospirillum group II in the Richmond biofilms (11). In contrast to Cyt579, Cyt572 oxidizes Fe(II) at low pH and may donate electrons to Cyt579. Efforts to reconstruct the Fe(II)-dependent electron transfer pathway in Leptospirillum group II bacteria and clarify the role of Cyt579 in this pathway are currently under way.
Acknowledgments
Funding was provided by the U.S. Department of Energy, Office of Science, from the Genomics: GTL Program, grant DE-FG02-05ER64134, to J.F.B., R.L.H., and M.P.T. Work at LLNL was performed under the auspices of the U.S. Department of Energy under contract DE-AC52-07NA27344.
We are grateful to the Banfield lab members for obtaining biofilm samples and to T. W. Arman, President, Iron Mountain Mines, R. Sugarek, EPA, and R. Carver for site access and on-site assistance. We also thank Chris Jeans and Anna Siebers for CD spectroscopy and assistance in biochemical studies on biofilm proteins at LLNL; Kent MacDonald and Reena Zalpuri for assistance with sample preparation at the Electron Microscope Laboratory, University of California, Berkeley; Mary Ann Gawinowicz at the Columbia University Protein Core Facility for protein sequence analyses; and Brian Erickson for assistance in acquiring IRMPD mass spectra at ORNL.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 3.Berry, E. A., and B. L. Trumpower. 1987. Simultaneous determination of hemes-a, b and c from pyridine hemochrome spectra. Anal. Biochem. 161:1-15. [DOI] [PubMed] [Google Scholar]
- 4.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bower, M. J., F. E. Cohen, and R. L. Dunbrack. 1997. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J. Mol. Biol. 267:1268-1282. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Connelly, H. M., D. A. Pelletier, T. Y. Lu, P. K. Lankford, and R. L. Hettich. 2006. Characterization of pII family (GlnK1, GlnK2, and GlnB) protein uridylylation in response to nitrogen availability for Rhodopseudomonas palustris. Anal. Biochem. 357:93-104. [DOI] [PubMed] [Google Scholar]
- 8.Druschel, G. K., B. J. Baker, T. M. Gihring, and J. F. Banfield. 2004. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 5:13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginalski, K., N. V. Grishin, A. Godzik, and L. Rychlewski. 2005. Practical lessons from protein structure prediction. Nucleic Acids Res. 33:1874-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart, A., J. C. Murrell, R. K. Poole, and P. R. Norris. 1991. An acid-stable cytochrome in iron-oxidizing Leptospirillum ferrooxidans. FEMS Microbiol. Lett. 81:89-94. [Google Scholar]
- 11.Jeans, C., S. W. Singer, C. S. Chan, N. C. VerBerkmoes, M. Shah, R. L. Hettich, J. F. Banfield, and M. P. Thelen. 2008. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J. 2:542-550. [DOI] [PubMed] [Google Scholar]
- 12.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 13.Kerfeld, C. A., H. P. Anwar, R. Interrante, S. Merchant, and T. O. Yeates. 1995. The structure of chloroplast cytochrome-c6 at 1.9 angstrom resolution: evidence for functional oligomerization. J. Mol. Biol. 250:627-647. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lo, I., V. J. Denef, N. C. VerBerkmoes, M. B. Shah, D. Goltsman, G. DiBartolo, G. W. Tyson, E. E. Allen, R. J. Ram, J. C. Detter, P. Richardson, M. P. Thelen, R. L. Hettich, and J. F. Banfield. 2007. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446:537-541. [DOI] [PubMed] [Google Scholar]
- 16.Pearson, W. R. 1991. Searching protein-sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics 11:635-650. [DOI] [PubMed] [Google Scholar]
- 17.Ram, R. J., N. C. VerBerkmoes, M. P. Thelen, G. W. Tyson, B. J. Baker, R. C. Blake, M. Shah, R. L. Hettich, and J. F. Banfield. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915-1920. [PubMed] [Google Scholar]
- 18.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70-percent accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 19.Schnaitman, C. A., M. S. Korczynski, and D. G. Lundgren. 1969. Kinetic studies of iron oxidation by whole cells of Ferrobacillus ferrooxidans. J. Bacteriol. 99:552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 21.Tabb, D. L., W. H. McDonald, and J. R. Yates. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabb, D. L., C. Narasimhan, M. B. Strader, and R. L. Hettich. 2005. DBDigger: reorganized proteomic database identification that improves flexibility and speed. Anal. Chem. 77:2464-2474. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 25.Yarzabal, A., C. Appia-Ayme, J. Ratouchniak, and V. Bonnefoy. 2004. Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiology 150:2113-2123. [DOI] [PubMed] [Google Scholar]
- 26.Yarzabal, A., G. Brasseur, and V. Bonnefoy. 2002. Cytochromes c of Acidithiobacillus ferrooxidans. FEMS Microbiol. Lett. 209:189-195. [DOI] [PubMed] [Google Scholar]
- 27.Yarzabal, A., G. Brasseur, J. Ratouchniak, K. Lund, D. Lemesle-Meunier, J. A. DeMoss, and V. Bonnefoy. 2002. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 184:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zemla, A. 2003. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 31:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemla, A., C. E. Zhou, T. Slezak, T. Kuczmarski, D. Rama, C. Torres, D. Sawicka, and D. Barsky. 2005. AS2TS system for protein structure modeling and analysis. Nucleic Acids Res. 33:W111-W115. [DOI] [PMC free article] [PubMed] [Google Scholar]