Abstract
Allelic replacement in the Burkholderia genus has been problematic due to the lack of appropriate counter-selectable and selectable markers. The counter-selectable marker sacB, commonly used in gram-negative bacteria, is nonselective on sucrose in many Burkholderia species. In addition, the use of antibiotic resistance markers of clinical importance for the selection of desirable genetic traits is prohibited in the United States for two potential bioterrorism agents, Burkholderia mallei and Burkholderia pseudomallei. Here, we engineered a mutated counter-selectable marker based on the B. pseudomallei PheS (the α-subunit of phenylalanyl tRNA synthase) protein and tested its effectiveness in three different Burkholderia species. The mutant PheS protein effectively killed 100% of the bacteria in the presence of 0.1% p-chlorophenylalanine. We assembled the mutant pheS on several allelic replacement vectors, in addition to constructing selectable markers based on tellurite (Telr) and trimethoprim (Tpr) resistance that are excisable by flanking unique FLP recombination target (FRT) sequences. As a proof of concept, we utilized one of these gene replacement vectors (pBAKA) and the Telr-FRT cassette to produce a chromosomal mutation in the Burkholderia thailandensis betBA operon, which codes for betaine aldehyde dehydrogenase and choline dehydrogenase. Chromosomal resistance markers could be excised by the introduction of pFLP-AB5 (Tpr), which is one of two constructed flp-containing plasmids, pFLP-AB4 (Telr) and pFLP-AB5 (Tpr). These flp-containing plasmids harbor the mutant pheS gene and allow self curing on media that contain p-chlorophenylalanine after Flp-FRT excision. The characterization of the ΔbetBA::Telr-FRT and ΔbetBA::FRT mutants indicated a defect in growth with choline as a sole carbon source, while these mutants grew as well as the wild type with succinate and glucose as alternative carbon sources.
Members of the Burkholderia genus are ubiquitous in the soil (i.e., rhizosphere) and water environments (31). Besides acting as saprophytes, several species associate beneficially with plants, while others cause plant diseases. Some (e.g., Burkholderia cenocepacia) are opportunists in human and animal infections. Others, such as Burkholderia mallei and Burkholderia pseudomallei, cause deadly infections in humans and animals (6, 33).
B. pseudomallei is the causative agent of melioidosis, an emerging global infectious disease, and this microbe is a potential bioterrorism agent of national biodefense concern. This disease has been predominantly studied, identified, and diagnosed in Thailand and northern Australia. However, melioidosis is considered endemic in many countries, including Thailand, Australia, Malaysia, Singapore, Vietnam, Burma, and possibly also Brunei, China, Hong Kong, Cambodia, Laos, India, and Taiwan (34). Cases have occurred in other areas of Asia, Africa, the Americas (e.g., Brazil and Puerto Rico), the Caribbean, the Middle East, and the Pacific (2, 9, 14, 15, 17, 21, 22, 24, 35). In the United States, B. pseudomallei and B. mallei (a clonal derivative of B. pseudomallei) are classified by the CDC as category B select agents because of the seriousness of the disease, the high fatality rates, the ease of isolation in soil throughout the tropics, and the historical use of the clonal derivative B. mallei during World Wars I and II by Germany (11).
The numerous genome sequences now available for several Burkholderia species, including the two potential bioterrorism agents, B. mallei and B. pseudomallei, should aid in the study to yield genetic, physiological, and pathogenic insights into members of this genus. For B. mallei and B. pseudomallei, the CDC restricts the use of resistance markers to clinically important antibiotics, which results in the need for non-antibiotic-selectable markers. Non-antibiotic-selectable markers based on Telr resistance could be useful for various Burkholderia spp., especially B. mallei and B. pseudomallei. The Telr marker, which is based on the genes kilA, telA, and telB, has been utilized successfully as a non-antibiotic-selectable marker in Pseudomonas putida and Pseudomonas fluorescens (16, 20, 26), and it could serve as an alternative selectable marker for Burkholderia species. For allelic replacement, Telr cassettes can be utilized only once without being coupled to the Flp recombination target (Flp-FRT) excision of the chromosomally located Telr-FRT cassette, which allows for the recycling of its use (13). Coupling the Telr selectable markers to the Flp-FRT system will allow numerous cycles of allelic replacement in Burkholderia spp. In addition, several counterselectable markers have been used for different bacteria, including rpsL, tetAR, pheS, thyA, gata-1, ccdB, and the most common, sacB (23). Although published works suggest that the sacB counter-selectable marker is suitable for some Burkholderia spp. (4, 5, 10, 30), it is an inappropriate and leaky counter-selectable marker for many laboratories, resulting in the need for an alternative counter-selectable marker. In addition, rpsL-based plasmids have been described for counter selection during allelic replacement (28), but such plasmids are limited to strains with chromosomal mutations in rpsL and the aminoglycoside efflux pump (7). The further development of non-antibiotic-selectable markers and counter-selectable markers should serve as useful genetic tools to perform molecular genetics, pathogenesis, and bacterium-host interaction studies for the discovery of novel vaccines, therapeutics, and diagnostic targets, as well as the environmental significance of various Burkholderia species.
This study describes novel genetic tools for making repetitive rounds of allelic replacements. A broad-host-range counter-selectable marker, together with several gene replacement vectors, has been engineered for use in Burkholderia species, and it will aid future allelic replacement experiments. Antibiotic and nonantibiotic resistance markers based on unique FRT sequences were developed for repetitive rounds of gene replacement, reducing the risks of undesirable deletions or genome rearrangements (1). These novel antibiotic and nonantibiotic FRT cassettes will be useful for gene replacement in both nonselect agents and category B select agents and should be useful in many laboratories that work on Burkholderia species of clinical and environmental importance.
MATERIALS AND METHODS
Bacterial strains, media, and culturing conditions.
All strains and plasmids involved in this study are listed in Table 1. Escherichia coli EPMax10B (Bio-Rad) was routinely used as a cloning strain. E. coli strain EPMax10B-pir116-Δasd::Gm (E1345) was used for the cloning of asd-complementing vectors (e.g., pBAKA; the Pseudomonas aeruginosa asd [asdPa] gene codes for aspartate semialdehyde dehydrogenase). The E. coli conjugal and suicidal strain EPMax10B-pir116-Δasd-mob-Km-Δtrp::Gm (E1354) was used for plasmid mobilization into B. thailandensis through conjugation. Luria-Bertani (LB) medium (Difco) was used to culture all of the E. coli strains. Burkholderia strains (B. thailandensis, B. cenocepacia K56-2, and B. dolosa) and their derivatives were cultured in LB or 1× M9 minimal medium supplemented with 20 mM glucose. Alternative carbon sources (e.g., 20 mM succinate or 1% Casamino Acids) may be used; however, citrate should not be used, as we have observed a reduction in the number of CFU for some species when grown on citrate. B. cenocepacia J2315 and derivatives were cultured in LB or 1× M9 minimal medium containing 20 mM glucose and 0.125% (wt/vol) yeast extract. Antibiotics and nonantibiotic bacteriocides were added to the media for selection and plasmid maintenance as follows: for E. coli, Gm at 15 μg/ml, Ap at 110 μg/ml, Tel at 30 μg/ml, Tet at 10 μg/ml, Km at 35 μg/ml, and Tp at 50 μg/ml; for B. thailandensis, Km at 500 μg/ml, Tel at 125 μg/ml, and Tp at 300 μg/ml. Also, Tp at 200, 700, and 500 μg/ml were used for B. cenocepacia K56-2, B. cenocepacia J2315, and B. dolosa, respectively. For the growth of E. coli Δasd strains EPMax10B-pir116-Δasd::Gm and EPMax10B-pir116-Δasd-mob-Km-Δtrp::Gm (E1345 and E1354 of Table 1), 100 μg/ml of diaminopimelic acid (DAP; Sigma) was supplied, unless the strain was complemented by the asdPa gene on the plasmid (e.g., pBAKA). DAP was dissolved in 1 M NaOH to make a 100-mg/ml stock solution. E. coli strain E1345 was used for all cloning steps, and strain E1354 was used for the mobilization of oriT-containing vectors by conjugation (Table 1). For strains E1345 and E1354, DAP (100 μg/ml) was added only during growth for competent cell preparation and the 1 h of recovery after transformation, but DAP was not added to medium plates for the selection of plasmid pBAKA and asdPa-containing plasmid derivatives. For the counter selection of pheS, 0.1% (wt/vol) p-chlorophenylalanine (cPhe; dl-4-chlorophenylalanine; Acros Organics) was autoclaved with the media.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Lab IDa | Relevant property(ies) | Source or reference |
|---|---|---|---|
| Strains | |||
| E. coli | |||
| EPMax10B | E1231 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK rpsL nupG λ− | Bio-Rad |
| EPMax10B-pir116-Δasd::Gm | E1345 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK rpsL nupG λ−Tn-pir116-FRT2 Δasd::Gm-FRT | -b |
| EPMax10B-pir116-Δasd-mob-Km- Δtrp::Gm | E1354 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK rpsL nupG λ−Tn-pir116-FRT2 Δasd::FRT Δtrp::Gm-FRT5 mob[recA::RP4-2 Tc::Mu-Km] | -b |
| B. thailandensis | |||
| E264 | E1298 | Prototroph; environmental isolate | 3 |
| E264-ΔbetBA::Telr-FRT | E1669 | Telr; E264 with Telr cassette inserted in betBA operon | This study |
| E264-ΔbetBA::FRT | E1671 | B. thailandensis ΔbetBA::FRT mutant | This study |
| B. cenocepacia K56-2 | E1554 | Prototroph; cystic fibrosis isolate | P. Sokol |
| B. cenocepacia J2315 | E1553 | Prototroph | J. Goldberg |
| B. dolosa AU0158 | E1551 | Prototroph | J. Goldberg |
| Plasmids | |||
| pUC57-pheS | E1510 | Apr; cloning vector harboring engineered pheS | This study |
| pBBR1MCS-Km | E1277 | Kmr; broad-host-range cloning vector | 18 |
| pBBR1MCS-Km-pheS | E1511 | Kmr; engineered pheS cloned into pBBR1MCS-Km | This study |
| pBBR1MCS-Km-Tp-pheS | E1558 | Kmr, Tpr; Tpr cassette cloned into pBBR1MCS-Km-pheS | This study |
| pBBR1MCS-Km-Tp | E1579 | Kmr, Tpr; pheS removed from pBBR1MCS-Km-Tp-pheS | This study |
| pMLBAD | E1642 | Tpr; cloning vector containing Tpr cassette | 19 |
| pAM2B-PS12 | E1297 | asdPa and PS12-merTPA-containing vector | -b |
| pUC57-pheS-asdPa | E1608 | Apr; asdPa cloned into pUC57-pheS | This study |
| pEX18Gm-pheS-asdPa | E1614 | Apr; pheS-asdPa cloned into pEX18Gm backbone | This study |
| pBAKA | E1624 | Gene replacement vector based on asdPa | This study |
| pBAKA-betBA | E1628 | B. thailandensis betBA operon cloned into pBAKA | This study |
| pBAKA-ΔbetBA::Telr-FRT | E1640 | Telr-FRT cassette inserted into betBA operon | This study |
| pEX18Ap | E0055 | Apr; sacB-based gene replacement vector | 13 |
| pEX18Ap-pheS | E1618 | Apr; gene replacement vector based on pheS and Apr | This study |
| pEX18Gm | E0062 | Gmr; sacB-based gene replacement vector | 13 |
| pEX18Gm-pheS | E1616 | Gmr; gene replacement vector based on pheS and Gmr | This study |
| pEX18Km-pheS | E1753 | Kmr; gene replacement vector based on pheS and Kmr | This study |
| pEX18Tet | E0064 | Tetr; sacB-based gene replacement vector | 13 |
| pEX18Tet-pheS | E1620 | Tetr; gene replacement vector based on pheS and Tetr | This study |
| pEX18Tp-pheS | E1653 | Tpr; gene replacement vector based on pheS and Tpr | This study |
| pPS856-ΔXbas | E1044 | Gmr, Apr; Gmr cassette flanked by wild-type FRTs | -b |
| pBTB-6 | E1508 | Telr; Telr cassette containing plasmid | 20 |
| pBTB-SDM | E1555 | Telr; pBTB-6 with SmaI, XhoI, and EcoRI sites mutated | This study |
| pwFRT-Merr | E1557 | Merr, Apr; Merr cassette flanked by wild-type FRT | -b |
| pwFRT-Telrc | E1584 | Telr, Apr; Telr cassette flanked by wild-type FRT | This study |
| pwFRT-Tprc | E1659 | Tpr, Apr; Tpr cassette flanked by wild-type FRT | This study |
| pFLP2 | E0067 | Apr; flp-containing plasmid | 13 |
| pFLP-AB2a | E1565 | Telr, sacB+; pFLP2 with Apr replaced with Telr cassette | This study |
| pFLP-AB4a | E1569 | Telr, pheS+; sacB on pFLP-AB2a replaced with pheS | This study |
| pFLP-AB4 | E1661 | Telr; Flp-containing plasmid with mutated Telr cassette | This study |
| pFLP-AB5 | E1662 | Tpr; pFLP-AB4 with Telr replaced with Tpr cassette | This study |
Please use the laboratory identification number (Lab ID) when requesting strains and plasmids.
Details on the engineering of these strains and plasmids are to be published elsewhere.
Four other FRT mutants exist for each of these plasmids, in which each plasmid has the identical FRT spacer sequence flanking each selectable marker and the only sequence differences in the mutant plasmids (pmFRT-Telr, pmFRT-Tpr, pFRT1-Telr, pFRT1-Tpr, pFRT2-Telr, pFRT2-Tpr, pFRT3-Telr, and pFRT3-Tpr) relative to the sequences of pwFRT-Telr and pwFRT-TelTpr are within the spacer sequence of each FRT.
Molecular methods and reagents.
Unless otherwise indicated, restriction enzymes, deoxynucleoside triphosphates, T4 DNA polymerase, T4 polynucleotide kinase, and T4 DNA ligase were purchased from New England Biolabs and used as recommended by the supplier. Plasmids and DNA gel bands were isolated using Zyppy Plasmid Miniprep kit I and Zymoclean gel DNA recovery kit, respectively, both from Zymo Research Corporation. Chemically competent cell (e.g., E1354) preparations and other molecular techniques were followed according to Sambrook and Russell (25). Oligonucleotide primers (Table 2) were synthesized by Integrated DNA Technologies. Pfu was purchased from Stratagene. Generally, we performed the various PCRs by initial denaturation for 1 min at 94°C and 34 cycles of 45 s at 94°C, 30 s at 58°C, and 1 min/kb at 72°C, and a final step of 10 min at 72°C was included.
TABLE 2.
Oligonucleotide primers used in this study
| Primer no. and name | Sequencea |
|---|---|
| 459; sacB-end | 5′-CAACGTTTGCGCCTAGCTTC-3′ |
| 463; GmR-RT | 5′-GAGCAGCCGCGTAGTGAG-3′ |
| 715; pPS854-XhoI | 5′-AAGCTCGAGCTAATTCC-3′ |
| 716; pPS854-Cla-EcoRV | 5′-CAATATCGATATCCATTGCTGTTGACAAAG-3′ |
| 732; asdPa-BglII | 5′-CAATAGATCTCCGATCAGCGCTCCAGCAG-3′ |
| 758; asdPa-BamHI | 5′-ATGAGGATCCAGTTGCGATGAAGCGTGTAGG-3′ |
| 792; merT-reverse | 5′-CAAGCCCTCCAGTGAAGA-3′ |
| 815; pBAD-ScaI | 5′-CTGTAGTACTCCAAAAAAACGGGTATGGAGA-3′ |
| 821; dhfr-SpeI | 5′-ACGCACTAGTGGAACGAAATCGATGAG-3′ |
| 826; oriT-ScaI | 5′-GCTTGCCCTCAGTACTGTTACGCCGGCG-3′ |
| 827; tel-ΔSmaI | 5′-GGGAACGACCCTGGCCGCGTGCA-3′ |
| 828; tel-ΔXhoI | 5′-GTAGGCGGCCTGGAGGCCCGAG-3′ |
| 829; tel-ΔEcoRI | 5′-TCGGGGGTGAGTTCTGCGGGTT-3′ |
| 834; tel-XhoI | 5′-CCTCCTCGAGCAGAAAGTCAAAAGCCTC-3′ |
| 836; tel-start | 5′-CTTTAAGAAGGAGATATACCATGGAAGAACAAAGCGTGAA-3′ |
| 837; PCS12 | 5′-ATCAGCCGTTGACTTAGTTGGTATTTCCGGAATATCATGCTGGGTTCCGAATAATTTTGTTTAACTTTAAGAAGGAGATATACC-3′ |
| 861; Bt-betB-HindIII | 5′-CCCGCAAGCTTGCCGGCAA-3′ |
| 862; Bt-betA-KpnI | 5′-GACCGGTACCCGGCGGGCGGGGATAT-3′ |
| 867; Bt-betB-up | 5′-GCACATCAAGCCGGACCAG-3′ |
| 868; Bt-betA-down | 5′-CCGGGCCGAATATCGACGG-3′ |
Restriction enzyme sites utilized in this study are underlined.
Preparation of high-efficiency E. coli Δasd-competent cells.
For the cloning and maintaining of the asdPa-containing vectors (e.g., pBAKA), we prepared highly efficient E. coli Δasd competent cells of strain E1345. E. coli Δasd strain E1345 was grown in LB medium containing 15 μg/ml Gm (LB-Gm-15) and supplemented with DAP (100 μg/ml) overnight at 37°C. Next, the overnight culture was diluted 100-fold into 250 ml of fresh LB-DAP and shaken at 37°C and 250 rpm. When the optical density at 600 nm (OD600) reached ∼0.3 (at approximately 3 h), an equal volume (250 ml) of fresh prewarmed (37°C) LB-DAP-40 mM glucose was added to the growing culture. The culture was grown to an OD600 of ∼0.7 (∼1 h) and then transferred to ice. Cells were harvested immediately by centrifugation at 4°C and 8,000 rpm for 10 min. Supernatant was discarded, and cells were resuspended in a 1-ml total volume of 1 mM HEPES buffer (pH 7.0) and transferred to a chilled microcentrifuge tube. Cells were pelleted and washed four times with ice-cold 1 mM HEPES buffer (pH 7.0) in a refrigerated microcentrifuge at 9,000 rpm. The volume of cell pellets from the final spin were estimated, and the sample was gently resuspended in fresh 1 mM HEPES buffer with a 1/3 volume of the cell pellet size. A 40-μl volume of the competent cells was used for each electroporation. For long-term storage, the final concentration of 10% glycerol was added, and 45-μl aliquots of the competent cells were frozen at −80°C. When needed, frozen cells were thawed on ice prior to electroporation. All other E. coli strains could be prepared similarly to create highly competent cells.
Engineering of pUC57-pheS and construction of pBBR1MCS-Km-Tp and pBBR1MCS-Km-Tp-pheS.
We submitted the mutated pheS gene sequence to GenScript Corporation, which synthesized and cloned the sequence into pUC57 to yield pUC57-pheS. This vector contains the mutated B. pseudomallei pheS gene with altered DNA sequences (see Fig. S3 in the supplemental material), which is driven by an upstream PS12 promoter of the B. pseudomallei rpsL gene. Using pUC57-pheS, we constructed pBBR1MCS-Km-Tp-pheS and pBBR1MCS-Km-Tp in several steps. First, pUC57-pheS was digested with SmaI and SacI, and the 1.1-kb PS12-pheS fragment was cloned into pBBR1MCS-Km digested with the same enzymes, yielding pBBR1MCS-Km-pheS. Next, the 0.6-kb Tpr cassette was obtained from pMLBAD by SalI and EcoRV digestion and was cloned into pBBR1MCS-Km-pheS digested with the same enzymes to construct pBBR1MCS-Km-Tp-pheS. Finally, pBBR1MCS-Km-Tp-pheS was digested with NdeI and XbaI and blunt ended, and the 5.8-kb backbone was self ligated to delete pheS, resulting in pBBR1MCS-Km-Tp.
Construction of gene replacement vectors.
We constructed the gene replacement pBAKA vector in several steps, using the P. aeruginosa aspartate semialdehyde dehydrogenase (asdPa) gene as a non-antibiotic-selectable marker. asdPa was PCR amplified from pAM2B-PS12 using oligonucleotides 732 and 758, and the 1.1-kb asdPa fragment was digested with BamHI and BglII and ligated with pUC57-pheS that had been digested with BglII to construct pUC57-pheS-asdPa. This generated a construct with pheS and asdPa in the same orientation. Next, the pEX18Gm plasmid backbone was PCR amplified with oligonucleotides 826 and 463 to delete the sacB-Gmr fragment, and the resulting 2.8-kb PCR product was digested with ScaI and BglII. The 2.2-kb pheS-asdPa fragment (removed from pUC57-pheS-asdPa with ScaI and BglII) was cloned into the pEX18Gm backbone, resulting in pEX18Gm-pheS-asdPa. Finally, pEX18Gm-pheS-asdPa was digested with BspHI and BglII, blunt ended, and self ligated to remove the remaining 0.4-kb Gmr fragment, yielding pBAKA.
Five different gene replacement vectors, pEX18Ap-pheS, pEX18Gm-pheS, pEX18Km-pheS, pEX18Tc-pheS, and pEX18Tp-pheS, which harbored different antibiotic resistance cassettes, were constructed as described below. To replace the sacB gene with pheS, the plasmid backbones of pEX18Ap, pEX18Gm, and pEX18Tc were amplified with oligonucleotides 459 and 826. The pheS gene from pUC57-pheS (digested with ScaI and BglII and blunt ended) was ligated into these PCR products to create pEX18Ap-pheS, pEX18Gm-pheS, and pEX18Tc-pheS, respectively. For pEX18Tp-pheS, a larger 2-kb fragment, containing the dihydrofolate reductase gene that codes for Tp resistance, was amplified from pMLBAD with oligonucleotides 815 and 821. This PCR product was digested with NsiI and blunt ended. The 0.7-kb Tpr cassette recovered from the gel was ligated into the pEX18Ap-pheS backbone, which was cut with BspHI and blunt ended to yield pEX18Tp-pheS. To create pEX18Km-pheS, pBBR1MCS-Km-Tp-pheS was digested with SspI and BglII and blunt ended, and the 1-kb Kmr fragment was cloned into the pEX18Gm-pheS backbone (digested with BsrGI and SacII and blunt ended), resulting in pEX18Km-pheS.
Construction of FRT vectors.
pFRT plasmids, containing either a Telr or Tpr marker flanked by two wild-type FRT genes, were constructed in this study. pPS856-ΔXbas was amplified with oligonucleotides 715 and 716, and then the 3.1-kb fragment was digested with EcoRV and XhoI and used as the plasmid backbone for the construction of pwFRT-Telr (wFRT indicates the wild-type FRT). Site-directed mutagenesis was performed on the Telr cassette (kilA telAB) of pBTB-6 with oligonucleotides 827, 828, and 829, resulting in pBTB-SDM, which had the SmaI, XhoI, and EcoRI sites removed from the telA and telB genes. Next, two-step PCR was performed to introduce the sequence of the promoter of the B. cenocepacia rpsL gene (PCS12) upstream of the Telr cassette. Oligonucleotides 836 and 834 were used in the first PCR to amplify the Telr cassette from pBTB-SDM. This yielded a 3.1-kb PCR product, which was used as a template for the second PCR with oligonucleotides 837 and 834. The final product, with PCS12 upstream of the Telr cassette, was digested with XhoI and ligated into the pPS856-ΔXbas backbone as described above to yield pwFRT-Telr.
The construction of pwFRT-Tpr was based on a laboratory plasmid, pwFRT-Merr. PCR was performed on pMLBAD using oligonucleotides 815 and 821 to amplify a larger Tpr cassette, and the 2-kb fragment was digested with EcoRV. The smaller 0.6-kb Tpr cassette was 5′ phosphorylated with T4 polynucleotide kinase and ligated into the pwFRT-PCS12 backbone (which was amplified from pwFRT-Merr by using oligonucleotides 792 and 715). The orientation of the Telr and Tpr cassettes relative to the PCS12 promoter on plasmids pwFRT-Telr and pwFRT-Tpr was confirmed by restriction mapping and DNA sequencing.
Besides these two pwFRT plasmids (pwFRT-Telr and pwFRT-Tpr) with TCTAGAAA as the core spacer of the FRT sequences, we also constructed eight other plasmids of these two resistance markers based on four other unique FRT sequences: pmFRT-Telr and pmFRT-Tpr, with the flanking FRT spacer TGTAGATA; pFRT1-Telr and pFRT1-Tpr, with the TCTTGAAA spacer; pFRT2-Telr and pFRT2-Tpr, with the TCTAGGAA spacer; and pFRT3-Telr and pFRT3-Tpr, with the TCTCGAAA spacer. The underlined bases represent differences in the core spacer sequence yielding unique FRT sequences. These 10 resistance plasmids (pwFRT-Telr, pwFRT-Tpr, pmFRT-Telr, pmFRT-Tpr, pFRT1-Telr, pFRT1-Tpr, pFRT2-Telr, pFRT2-Tpr, pFRT3-Telr, and pFRT3-Tpr) are useful for performing multiple allelic replacement and Flp-FRT excision by using and recycling the same resistance marker without the risk of undesirable genomic deletions or inversions. To construct these eight plasmids, laboratory vectors pmFRT-Gmr, pFRT1-Gmr, pFRT2-Gmr, and pFRT3-Gmr were PCR amplified with oligonucleotides 715 and 716 to obtain plasmid backbones without the Gmr marker. Each plasmid backbone was digested with EcoRV and XhoI, and the Telr and Tpr fragments that were digested with the upstream PCS12 promoter were removed from pwFRT-Telr and pwFRT-Tpr, respectively, with EcoRV and XhoI. These resistance fragments were individually cloned into the EcoRV and XhoI sites of each FRT plasmid backbone. Essentially, the sequences of all four new Telr-FRT plasmids are the same as that for pwFRT-Telr, with the exception of the FRT spacer sequence flanking the resistant cassette; this is also the case for all Tpr-FRT plasmids.
Construction of flp-containing plasmids.
For the excision of chromosome-selectable markers in mutant strains, we engineered two flp-containing vectors, pFLP-AB4 (Telr) and pFLP-AB5 (Tpr). First, we constructed pFLP-AB4a by basing it on the Tel-resistant cassette. The Telr cassette, including kilA, telA, and telB genes, was amplified from pBTB-6 by using oligonucleotides 834 and 836. The blunt-ended 3.1-kb fragment was 5′ phosphorylated and ligated into pFLP2, which was digested with SspI and BsaI and blunt ended, to yield pFLP-AB2a. Next, pFLP-AB2a was digested with XbaI and SphI, and the pheS gene from pUC57-pheS was removed with the same enzymes and cloned into this plasmid, resulting in pFLP-AB4a. The PCS12-Telr (3.1 kb) or PCS12-Tpr (0.6 kb) fragment was obtained from pwFRT-Telr or pwFRT-Tpr by EcoRV and XhoI digestion and then was cloned into the pFLP-AB4a backbone (cut with NcoI, blunt ended, and digested with XhoI). These resulted in pFLP-AB4 and pFLP-AB5, which contain the PCS12 promoter driving the Telr or Tpr cassette, respectively.
Construction of pBAKA-ΔbetBA::Telr-FRT.
The B. thailandensis betBA operon was amplified from chromosomal DNA of strain E264 by using oligonucleotides 861 and 862 (see Fig. 4A). The 4.5-kb fragment was digested with HindIII and KpnI and cloned in pBAKA that had been digested with the same enzymes, yielding pBAKA-betBA. Next, pBAKA-betBA was digested with SmaI, and the 6.6-kb backbone was ligated with the SmaI fragment (Telr-FRT) from pwFRT-Telr, resulting in pBAKA-ΔbetBA::Telr-FRT. For these gene replacement vectors, a 2.2-kb internal region of betBA on pBAKA-betBA was deleted, and the Telr-FRT cassette is in the same orientation as the betBA operon (see Fig. 4A).
FIG. 4.
(A) Gene replacement scheme using a Telr-FRT or Tpr-FRT cassette to inactivate the B. thailandensis betBA operon. We utilized the Telr-FRT cassette. The betBA operon was amplified and cloned into pBAKA with oligonucleotides 861 and 862, and the inactivation and selection procedure performed was described in Materials and Methods. After the Flp excision, the resulting ΔbetBA::FRT mutant has one remaining FRT sequence (∼100 bp) that inactivates the betBA operon. (B) PCR confirmation of the ΔbetBA mutant with outside oligonucleotides 867 and 868. Numbers in circles from 1 to 3 corresponds to lanes 1 to 3. Lane 1, wild-type betBA operon; lane 2, ΔbetBA::Telr-FRT mutant before Flp excision; lane 3, ΔbetBA::FRT mutant after Flp excision; M, 1-kb ladder.
Engineering of unmarked B. thailandensis ΔbetBA::FRT mutants.
E. coli strain EPMax10B-pir116-Δasd-mob-Km-Δtrp::Gm (E1354) was used as the conjugal donor to introduce the gene replacement vector pBAKA-ΔbetBA::Telr-FRT into B. thailandensis strain E264. Both recipient and donor strains were grown to log phase prior to conjugation. Conjugation was performed by mixing 0.5 ml of each of the donor and recipient strains at approximately equal cell densities. The tube was centrifuged at 9,000 × g for 1 min, and all of the supernatant was discarded, except for 30 μl, which was used to gently resuspend the cell pellet. The 30-μl mixture of cell suspension was spotted onto cellulose acetate filters (Sartorius), which were prewarmed at 37°C on LB agar plates, and the conjugation plates were incubated at 37°C for 8 h. Conjugation filters were aseptically transferred to a 1.5-ml microcentrifuge tube and vortexed in 1 ml of 1× M9 minimal medium to resuspend cells. One hundred microliters and equal volumes of 10× dilutions were plated on plates containing 1× M9 medium supplemented with 20 mM glucose and 125 μg/ml Tel (Tel-125). Resulting colonies were streaked out on 1× M9 minimal medium-20 mM glucose-0.1% cPhe-Tel-75. Telr mutants were screened by being patched with toothpicks onto plates of MPG (1× M9 minimal medium-0.1% cPhe-20 mM glucose) and MPC (M9 minimal medium-0.1% cPhe-20 mM choline chloride [Sigma]). Mutants growing on glucose but defective in choline degradation were purified once on MPG medium and further screened through PCR using oligonucleotides 867 and 868 (see Fig. 4A), which anneal upstream and downstream of the betBA operon and outside of oligonucleotides 861 and 862, which were used for cloning.
As a proof of concept, we performed Flp excision on the chromosomal Telr marker from the B. thailandensis ΔbetBA::Telr-FRT mutant. To perform Flp excision on the Telr cassette in the ΔbetBA::Telr-FRT mutant, pFLP-AB5 was introduced into B. thailandensis mutant strain E264/ΔbetBA::Telr-FRT through conjugation. The conjugation of E1354, harboring pFLP-AB5, with E264/ΔbetBA::Telr-FRT was performed as described above for allelic replacement. Note that DAP is required when this E. coli strain was used to mobilize pFLP-AB4 and pFLP-AB5, but no DAP is required after conjugation, leading to DAP-less death. After conjugation, cells were plated on 1× M9 minimal medium-20 mM glucose-300 μg/ml Tp (1× M9 minimal medium-20 mM glucose-Tp-300) to select for single colonies of E264/ΔbetBA::Telr-FRT containing pFLP-AB5. pFLP-AB5 then was cured by streaking these single Telr colonies on 1× M9 minimal medium-20 mM glucose-0.1% cPhe. Next, colonies from the cPhe plates were patched onto 1× M9 minimal medium-20 mM glucose with or without Tel-75 to confirm the excision of the Telr cassette, in addition to being patched on a 1× M9 minimal medium-20 mM glucose-Tp-300 plate to ensure that pFLP-AB5 was cured. Similarly, pFLP-AB4 could be used to excise FRT cassettes other than Telr-FRT. Finally, the phenotype of the B. thailandensis ΔbetBA::FRT strain was confirmed by patching it onto 1× M9 minimal medium supplemented with 20 mM glucose or choline chloride along with wild-type strain E264 as a positive control. The inactivation of the betBA operon was characterized by the inability to grow on choline as a sole carbon source.
Characterization of ΔbetBA mutants.
The B. thailandensis ΔbetBA::Telr-FRT and ΔbetBA::FRT mutants were chosen to be further characterized by growth curve analyses, and the results were compared to those for wild-type B. thailandensis with choline, succinate, or glucose as the sole carbon source. These strains were grown overnight at 37°C in LB medium. Overnight cultures were washed once with one volume of 1× M9 minimal medium and resuspended in equal volumes of the same buffer. Resuspended cultures then were diluted 100-fold into fresh 1× M9 minimal medium containing 30 mM succinate, 20 mM glucose, or 30 mM choline chloride and grown at 37°C. At each time point, a measurement was taken at an OD600 for each culture.
Nucleotide sequence accession numbers.
All of the vectors presented in Fig. 2 and 3 were submitted to GenBank. The accession numbers are the following: pUC57-pheS, EU277853; pBAKA, EU277854; pEX18Ap-pheS, EU332346; pEX18Gm-pheS, EU332347; pEX18Km-pheS, EU491017; pEX18Tc-pheS, EU332348; pEX18Tp-pheS, EU334848; pwFRT-Telr, EU329006; pwFRT-Tpr, EU334849; pFLP-AB4, EU329004; and pFLP-AB5, EU334847.
FIG. 2.
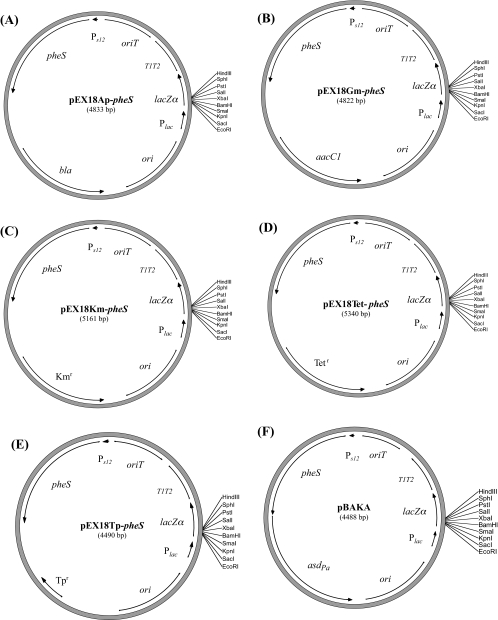
Allelic replacement vectors based on the mutant pheS gene. Each vector contains a different selectable marker for resistance to Ap (A), Gm (B), Km (C), Tet (D), and Tp (E). (F) The asdPa gene as a non-antibiotic-selectable marker. All plasmids can be maintained in regular laboratory E. coli strains, with the exception of pBAKA, which was maintained in E. coli strain E1345 or E1354 (Table 1). aacC1, Gm acetyltransferase-encoding gene; bla, β-lactamase-encoding gene; lacZα, β-galactosidase α-peptide; ori, ColE1 origin of replication; oriT, conjugal origin of transfer; Plac, lac promoter; PS12, the B. pseudomallei rpsL gene promoter; asdPa, P. aeruginosa aspartate semialdehyde dehydrogenase gene; pheS, mutant gene for the α-subunit of phenylalanyl tRNA synthase; and T1T2, transcriptional terminators.
FIG. 3.
Maps of FRT and Flp plasmids. Telr and Tpr FRT cassettes can be removed by restriction digestion from pwFRT-Telr (A) and pwFRT-Tpr (B), respectively. Not shown are four other unique FRT cassettes for each resistance determinant, which yield eight other plasmids (pmFRT-Telr, pmFRT-Tpr, pFRT1-Telr, pFRT1-Tpr, pFRT2-Telr, pFRT2-Tpr, pFRT3-Telr, and pFRT3-Tpr), where each selectable marker is flanked by identical FRTs with unique spacer sequences. The DNA sequences and restriction sites for all five Telr-FRT plasmids are identical, with the exception of the FRT spacer sequences on both sides of the resistant marker; similarly, all Tpr-FRT plasmids have identical DNA sequences, with the exception of the spacers. Two Flp-containing replicative plasmids, pFLP-AB4 (C) and pFLP-AB5 (D), were engineered to excise chromosomal markers based on Telr and Tpr, respectively. cI857, temperature-sensitive λ cI repressor; flp, gene encoding flippase (Flp); Ω, tonB transcriptional terminator; ori1600-rep, broad-host-range replicon; PS12, promoter of the B. pseudomallei rpsL gene; PCS12, promoter of the B. cenocepacia rpsL gene.
RESULTS AND DISCUSSION
Characterization of the B. pseudomallei mutant pheS as a counter-selectable marker.
The lack of an appropriate counter-selectable marker for a broader range of Burkholderia species and strains has hampered various molecular genetic studies of this genus. Existing tools for allelic replacement based on rpsL and aminoglycoside efflux pump mutations (7, 28), although useful, have been limited to specific strains. Previously, the utilization of the mutant (A294G) E. coli pheS gene was successful for allelic replacement in E. coli (12). Recent data from our laboratory (unpublished) indicate that the P. aeruginosa pheS gene carrying a similar mutation (A305G) killed P. aeruginosa in the presence of cPhe. However, we observed that neither the E. coli nor the P. aeruginosa mutant pheS gene efficiently killed Burkholderia species in the presence of cPhe (data not shown). We reasoned that the interactions between PheS (α-subunit) and PheT (β-subunit) of the functional multisubunit complex phenylalanyl tRNA synthase could be genus or species specific. The alignment of various PheS proteins from different Burkholderia species indicated high amino acid identity (93 to 97%) between members of this genus (see Fig. S1 in the supplemental material), suggesting that the interactions between PheS and PheT in different Burkholderia species are highly conserved. Thus, the similar A304G mutation of a Burkholderia (e.g., B. pseudomallei) pheS gene may be widely applicable for this genus. However, the high level of conservation of the pheS gene among the Burkholderia species at the DNA level (see Fig. S2 in the supplemental material) may present problems such as an undesirable homologous recombination at the pheS locus during allelic replacement. Therefore, we engineered a new B. pseudomallei pheS gene by switching alternative codons that existed on the same protein while conserving the amino acid sequence and introducing the critical A304G mutation (see Fig. S3 in the supplemental material). Although not yet tested, we reasoned that this strategy alters the DNA sequence of the B. pseudomallei pheS gene significantly enough to possibly reduce aberrant and undesirable recombination during allelic replacement at this locus.
To initially test the effectiveness of the engineered B. pseudomallei mutant pheS in other Burkholderia species, we constructed two replicative plasmids, pBBR1MCS-Km-Tp and pBBR1MCS-Km-Tp-pheS. Plasmid pBBR1MCS-Km-Tp-pheS was transformed into B. cenocepacia K56-2, B. dolosa, B. cenocepacia J2315, and B. thailandensis to test for killing on cPhe. The results indicate that the engineered B. pseudomallei pheS mutant effectively killed 100% of cells (∼50,000 to 100,000 CFU) that were plated on medium containing 0.1% cPhe (Fig. 1), demonstrating its effectiveness as a counter-selectable marker. In addition, no spontaneous cPhe-resistant colonies were observed for strains harboring the mutant pheS gene, even after 2 weeks of incubation.
FIG. 1.
Killing of four different Burkholderia species by a pheS plasmid (pBBR1MCS-Km-Tp-pheS) in the presence of 0.1% cPhe. The control plasmid used was pBBR1MCS-Km-Tp. Tp was used at 200 μg/ml for B. cenocepacia K56-2 (A), 500 μg/ml for B. dolosa (B), and 700 μg/ml for B. cenocepacia J2315 (C); kanamycin (Km) was used at 500 μg/ml for B. thailandensis (D). The same numbers of CFU were plated for each strain on three different media, 1× M9-glucose medium (MG), 1× M9-glucose medium containing Tp (MG+Tp), and 1× M9-glucose medium containing Tp and cPhe (MG+Tp+cPhe); more bacteria were plated in the bottom picture for each species to show effective killing at a high number of CFU. The four Burkholderia species containing the mutant pheS died in the presence of cPhe. No synergistic effect of Tp/Km and cPhe was observed. There was no effect on the number of CFU when each control strain was grown on cPhe-Tp or cPhe-Km.
Construction of gene replacement vectors and FRT-based selectable markers.
We next constructed several gene replacement vectors based on the B. pseudomallei mutant pheS gene (Fig. 2). Plasmids pEX18Ap-pheS, pEX18Gm-pheS, pEX18Km-pheS, pEX18Tc-pheS, and pEX18Tp-pheS will be useful for allelic replacement in various Burkholderia species. However, for the two restricted category B select agents, B. mallei and B. pseudomallei, we constructed pBAKA to prevent the further introduction of antibiotic resistance into these two organisms. Instead of antibiotic resistance selection, pBAKA contains the asdPa gene (encoding aspartate semialdehyde dehydrogenase from P. aeruginosa) as a selectable marker for the cloning and manipulation of E. coli Δasd strains. The Δasd E. coli cloning and mobilizable strains E1345 and E1354 (Table 1), respectively, were routinely used in our laboratory for pBAKA and other asd-containing plasmids. All allelic replacement vectors contain the B. pseudomallei mutant pheS gene driven by PS12, the promoter of the B. pseudomallei rpsL gene, which was previously described (36). The addition of this conserved rpsL promoter sequence (AGCTGTTGACTCGCTTGGGATTTTCGGAATATCATGCCGGGT; the −35 and −10 regions are underlined) ensures the sufficient expression of the mutant pheS in Burkholderia species to outcompete the native chromosomal copy for PheT.
We engineered two selectable markers to contain flanking FRT sequences, which will be useful for Burkholderia species (Fig. 3A to C). FRT cassettes have been very beneficial for multiple rounds of allelic replacement and the recycling of useful selectable markers (8, 13). Therefore, we constructed pwFRT-Telr and pwFRT-Tpr, which encode resistance to Tel and Tp, respectively (Fig. 3A and B). Unique restriction sites flanking these cassettes allow easy manipulations, and site-directed mutagenesis was performed to eliminate all repetitive restriction sites from within these resistance cassettes (see Materials and Methods). Tp resistance has been shown to be useful for various Burkholderia species (27). However, for restricted category B select agents (e.g., B. mallei and B. pseudomallei), the Telr-FRT cassette will be more appropriate for preventing the engineering of strains resistant to clinically important antibiotics. Each of the two cassettes is driven by the B. cenocepacia rpsL PCS12 promoter (AGCCGTTGACTTAGTTGGTATTTCCGGAATATCATGCTGGGT), with the −35 and −10 underlined regions conserved from the B. pseudomallei PS12 promoter described above. However, the intervening sequences between these two regions of the PCS12 and PS12 promoters are very different (36), which prevents any possible recombination between these two promoter sequences during allelic replacement.
Although the Flp excision of FRT cassettes is useful for recycling selectable markers for subsequent rounds of allelic replacement (13), each round of mutation and Flp excision results in one chromosomal FRT scar at the replaced locus (Fig. 4A). As a result, a previous study showed that multiple rounds of sequential allelic replacement with an identical wild-type FRT could lead to the Flp-catalyzed inversion of large chromosomal pieces (1). In addition, creating multiple closely linked mutations with one identical FRT cassette yielded Flp-catalyzed deletions of chromosomal genes between the two loci (unpublished data). Since wild-type Flp protein does not recombine FRT sequences of different spacer sequences and only recombines identical FRT sequences (29, 32), we have utilized unique FRT sequences with altered core spacer sequences, which prevented undesirable deletions of chromosomal fragments in multiple loci allelic replacements (unpublished data). We have used other resistance markers based on these unique FRT sequences for multiple mutations in several species, including E. coli, Sphingomonas chlorophenolica, and P. aeruginosa, without any undesirable deletions or rearrangements (unpublished data). Accordingly, to prevent undesirable chromosomal deletions and rearrangements, we have engineered each of the Telr and Tpr cassettes flanked by five unique sets of FRTs (Fig. 3A and B; also see Materials and Methods). Therefore, when performing multiple allelic replacements on the chromosomes of the same bacteria, we recommend using Telr or Tpr cassettes flanked by unique FRTs, which allows for multiple rounds of allelic replacement and the recycling of these resistance determinants.
Engineering of B. thailandensis ΔbetBA::Telr-FRT chromosomal mutants.
As a proof of concept of the developed system, we constructed a deletion in the B. thailandensis betBA operon by using pBAKA and the Telr-FRT cassette. pBAKA-ΔbetBA::Telr-FRT was constructed in E. coli strain E1345. These plasmids were transformed into another E. coli strain, E1354, and mobilized into B. thailandensis by routine conjugation, as described in Materials and Methods. The selection of exconjugants (merodiploids) (Fig. 4A) was performed on 1× M9 minimal medium-20 mM glucose-Tel-125. The E. coli conjugal donor strain E1354 cannot grow on this minimal medium due to the lack of tryptophan, and we observed no E. coli in the background. We routinely obtained 100% of mutants after the counter-selection step on cPhe. However, it is critical that the counter-selection medium, in the presence of cPhe, contains no competing phenylalanine for clean counter selection. Other saccharides or succinate could be substituted for glucose if required. For fastidious Burkholderia species requiring amino acids in addition to 1× M9 minimal medium-20 mM glucose, the addition of 1 mM amino acid mix lacking phenylalanine and tryptophan often is appropriate for pheS counterselection. Alternatively, 1.25 g/liter of yeast extract, in addition to 1× M9 minimal medium-20 mM glucose, works well for fastidious Burkholderia species (e.g., B. cenocepacia). The initial screening of potential mutants on MPG and MPC yielded a 100% choline-auxotrophic phenotype. Further screening by PCR with oligonucleotides 867 and 868 showed that all were mutants (data not shown), and one of these mutants is shown in Fig. 4B.
As a minor note, the concentration of Tel used is cell density dependent in our experience. Typically, a higher concentration of Tel (125 μg/ml) was used to select for merodiploids or pFLP plasmids from the conjugation mix, in which cell densities are high. However, the purification of the resulting merodiploids or colonies harboring replicative pFLP plasmids must be done at a lower Tel (75 μg/ml) concentration. This lower Tel concentration should also be used in the counter selection on 0.1% cPhe to obtain double-crossover mutants and the curing of pFLP plasmids on plates. We also recommend this lower concentration of Tel (75 μg/ml) for cultures grown in liquid medium from single colonies.
Flp excision of chromosomal resistance markers.
We successfully performed Flp excision on the chromosomal Telr cassette in the B. thailandensis ΔbetBA::Telr-FRT mutant (Fig. 4A). The introduction of pFLP-AB5 (Fig. 3D) into this mutant resulted in the excision of the Telr cassette by selection on Tp-300. The excision of the chromosomal Telr cassette was confirmed by PCR (Fig. 4B). The curing of Flp-containing plasmids (pFLP-AB5) was easily performed on 1× M9 minimal medium-20 mM glucose-0.1% cPhe. This strategy allows the recycling and reuse of the same resistance marker for another round of allelic replacement. If the wFRT cassette is used in the first round of gene replacement, then the resistance marker in the second round of allelic replacement should be flanked identically by other FRTs with different spacer sequences (e.g., mFRT, FRT1, FRT2, or FRT3) (Fig. 3). This prevents undesirable chromosomal fragment deletions and rearrangements (1), as mentioned above. In our experience, the native Flp on pFLP-AB4 and pFLP-AB5 yielded an 80 to 100% efficiency of excision.
B. thailandensis ΔbetBA mutants are defective in choline degradation.
To further characterize the B. thailandensis ΔbetBA::Telr-FRT and the Flp-excised ΔbetBA::FRT mutants, we performed growth curve experiments for these mutants and compared the results to those for wild-type B. thailandensis. The results indicated that the betBA operon is involved in choline degradation (Fig. 5A). There are no other betaine aldehyde dehydrogenase (BetB) and choline dehydrogenase (BetA) homologs in B. thailandensis, because the ΔbetBA::Telr-FRT and ΔbetBA::FRT mutants were unable to grow on choline as a sole carbon source. The engineered ΔbetBA::FRT mutation affected choline degradation and not the degradation of other carbon sources, such as succinate and glucose (Fig. 5). However, when grown in succinate and glucose media, death occurred more quickly with the ΔbetBA::Telr-FRT mutant than with wild-type B. thailandensis and the Flp-excised ΔbetBA::FRT mutant. We reasoned that there is a polar effect in the ΔbetBA::Telr-FRT mutant due to a tonB transcriptional terminator in the Telr-FRT cassette that was constructed and used for allelic replacement (Fig. 3A). This polar effect was eliminated after the Flp excision of the tonB transcriptional terminator and the Telr cassette (Fig. 5). The ability to easily create a polar mutation and a derivative nonpolar mutation by using this Flp-FRT system may be beneficial to study the polar effects of different genetic loci. The regulation mechanism of the betBA operon is currently unknown. The future development of genetic tools is required to study the regulation of this betBA operon in B. thailandensis.
FIG. 5.
Growth defect of the ΔbetBA::Telr-FRT and ΔbetBA::FRT mutants in choline, succinate, and glucose media. (A) The ΔbetBA mutation abolished the growth of B. thailandensis in 1× M9 minimal medium-choline, while the wild type grew well on choline as a sole carbon source. The ΔbetBA::FRT nonpolar mutation does not affect growth on the other carbon sources, succinate (B) and glucose (C), compared to that of wild-type B. thailandensis. However, quicker death was observed for the ΔbetBA::Telr-FRT polar mutant (B and C).
Conclusions.
(i) We have engineered a broad-host-range counter-selectable marker, pheS, for Burkholderia species. (ii) Several allelic replacement vectors were constructed that were based on the mutant pheS gene, which will aid in gene replacement. Specifically, for B. mallei and B. pseudomallei, pBAKA along with E. coli cloning and delivery strains (E1345 and E1354) should be useful nonrestricted tools for allelic replacement. (iii) Ten unique FRT cassettes, based on two different resistance markers, allow repetitive rounds of allelic replacement that use the same selectable marker. The five Telr-FRT nonantibiotic cassettes will be particularly useful for B. mallei and B. pseudomallei studies. Although the effective selectable concentration has to be determined, it has been previously shown that B. pseudomallei is very sensitive to Tel (<1 μg/ml) (M. Frazier, K. Choi, A. Kumar, C. Lopez, R. R. Karkhoff-Schweizer, and H. P. Schweizer, 2007 American Society for Microbiology Biodefense and Emerging Diseases Research Meeting, Washington, DC). (iv) Two Flp-containing plasmids were engineered with the mutant pheS for self curing after the chromosomal excision of useful selectable markers. For B. mallei and B. pseudomallei, we recommend the combination of pBAKA and Telr-FRT for allelic replacement, because the whole process excludes the use of any clinically important antibiotics and antibiotic resistance markers. The Flp excision of the chromosomal Telr-FRT cassette could be performed with the select agent-compliant Flp plasmids pFLPe2 and pFLPe4, which were recently described (7). (v) These genetic systems were used to mutate the B. thailandensis betBA operon, demonstrating the function of BetB and BetA in choline degradation.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant R21-AI074608 to T.T.H. Graduate salary support for A.R.B. and M.S.S. was provided by grant P20RR018727 from the National Center for Research Resources (NCRR), a component of the NIH.
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR or the NIH.
We gratefully thank Ryan Gill (University of Colorado) for providing pBTB-6, Joanna Goldberg (University of Virginia) for B. cenocepacia J2315 and B. dolosa AU0158, Michael Kovach (Baldwin Wallace College) for pBBR1MCS-Km, Herbert Schweizer (Colorado State University) for the gift of B. thailandensis E264, Pam Sokol (University of Calgary) for providing B. cenocepacia K56-2, and Miguel Valvano (University of Western Ontario) for the gift of pMLBAD.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barekzi, N., K. L. Beinlich, T. T. Hoang, X.-Q. Pham, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2000. The oriC-containing region of the Pseudomonas aeruginosa chromosome undergoes large inversions at high frequency. J. Bacteriol. 182:7070-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borgherini, G., P. Poubeau, F. Paganin, S. Picot, A. Michault, F. Thibault, and C. A. Berod. 2006. Melioidosis: an imported case from Madagascar. J. Travel Med. 13:318-320. [DOI] [PubMed] [Google Scholar]
- 3.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., description of Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317-320. [DOI] [PubMed] [Google Scholar]
- 4.Brown, N. F., C.-A. Logue, J. A. Boddey, R. Scott, R. G. Hirst, and I. R. Beacham. 2004. Identification of a novel two-partner secretion system from Burkholderia pseudomallei. Mol. Gen. Genomics 272:204-215. [DOI] [PubMed] [Google Scholar]
- 5.Chan, Y. Y., and K. L. Chua. 2005. The Burkholderia pseudomallei BpeAB-OprB efflux pump: expression and impact on quorum sensing and virulence. J. Bacteriol. 187:4707-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, K. H., T. Mima, Y. Casart, D. Rholl, A. Kumar, I. R. Beacham, and H. P. Schweizer. 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol. 74:1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman, S. E., V. J. Gill, J. I. Gallin, and S. M. Holland. 1998. Burkholderia pseudomallei infection in a Puerto Rican patient with chronic granulomatous disease: case report and review of occurrences in the Americas. Clin. Infect. Dis. 26:889-894. [DOI] [PubMed] [Google Scholar]
- 10.Essex-Lopresti, A. E., J. A. Boddey, R. Thomas, M. P. Smith, M. G. Hartley, T. Atkins, N. F. Brown, C. H. Tsang, I. R. A. Peak, J. Hill, I. R. Beacham, and R. W. Titball. 2005. A type IV pilin, PilA, contributes to adherence of Burkholderia pseudomallei and virulence in vivo. Infect. Immun. 73:1260-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frischknecht, F. 2003. The history of biological warfare. Eur. Mol. Biol. Org. 4:S47-S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamper, M., and P. Kast. 2005. Strategy for chromosomal gene targeting in RecA-deficient Escherichia coli strains. BioTechniques 38:405-408. [DOI] [PubMed] [Google Scholar]
- 13.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 14.How, H. S., K. H. Ng, H. P. Tee, and A. Shah. 2005. Pediatric melioidosis in Pahang, Malaysia. J. Microbiol. Immunol. Infect. 38:314-319. [PubMed] [Google Scholar]
- 15.Issack, M. I., C. D. Bundhun, and H. Gokhool. 2005. Melioidosis in Mauritius. Emerg. Infect. Dis. 11:139-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jäderlund, L., M. Hellman, I. Sundh, M. J. Bailey, and J. K. Jansson. 2008. Use of a novel nonantibiotic triple marker gene cassette to monitor high survival of Pseudomonas fluorescens SBW25 on winter wheat in the field. FEMS Microbiol. Ecol. 63:156-168. [DOI] [PubMed] [Google Scholar]
- 17.Jesudason, M. V., A. Anbarasu, and T. J. John. 2003. Septicaemic melioidosis in a tertiary care hospital in south India. Indian J. Med. Res. 117:119-121. [PubMed] [Google Scholar]
- 18.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, Jr., and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 19.Lefebre, M. D., and M. A. Valvano. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch, M. D., and R. T. Gill. 2006. Broad host range vectors for stable genomic library construction. Biotech. Bioeng. 94:151-158. [DOI] [PubMed] [Google Scholar]
- 21.Orellana, C. 2004. Melioidosis strikes Singapore. Lancet Infect. Dis. 4:655. [DOI] [PubMed] [Google Scholar]
- 22.Phetsouvanh, R., S. Phongmany, P. Newton, M. Mayxay, A. Ramsay, V. Wuthiekanun, and N. J. White. 2001. Melioidosis and pandora's box in the Lao People's Democratic Republic. Clin. Infect. Dis. 32:653-654. [DOI] [PubMed] [Google Scholar]
- 23.Reyrat, J.-M., V. Pelicic, B. Gicquel, and R. Rappuoli. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 66:4011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rolim, D. B. 2005. Melioidosis, northeastern Brazil. Emerg. Infect. Dis. 11:1458-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Sanchez-Romero, J. M., R. Diaz-Orejas, and V. De Lorenzo. 1998. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl. Environ. Microbiol. 64:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer, H. P., T. R. Klassen, and T. Hoang. 1996. Improved methods for gene analysis and expression in pseudomonas, p. 229-237. In T. Nakazawa, K. Furukawa, D. Haas, and S. Silver (ed.), Molecular biology of pseudomonads. American Society for Microbiology, Washington, DC.
- 28.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 29.Storici, F., M. Coglievina, and C. Bruschi. 1999. A 2-μm DNA-based marker recycling system for multiple gene disruption in the yeast Saccharomyces cerevisiae. Yeast 15:271-283. [DOI] [PubMed] [Google Scholar]
- 30.Ulrich, R. L., K. Amemiya, D. M. Waag, C. J. Roy, and D. DeShazer. 2005. Aerogenic vaccination with a Burkholderia mallei auxotroph protests against aerosol-initiated glanders in mice. Vaccine 23:1986-1992. [DOI] [PubMed] [Google Scholar]
- 31.Vandamme, P., J. Govan, and J. LiPuma. 2007. Diversity and role of Burkholderia spp., p. 1-28. In T. Coenye and P. Vandamme (ed.), Burkholderia: molecular microbiology and genomics. Horizon Scientific, Wymondham, United Kingdom.
- 32.Voziyanov, Y., A. F. Stewart, and M. Jayaram. 2002. A dual reporter screening system identifies the amino acid at position 82 in Flp site-specific recombinase as a determinant for target specificity. Nucleic Acids Res. 30:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitlock, G. C., D. M. Estes, and A. G. Torres. 2007. Glanders: off to the race with Burkholderia mallei. FEMS Microbiol. Lett. 277:115-122. [DOI] [PubMed] [Google Scholar]
- 34.Wiersinga, W. J., T. van der Poll, N. J. White, N. P. Day, and S. J. Peacock. 2006. Melioidosis: insight into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4:272-282. [DOI] [PubMed] [Google Scholar]
- 35.Wuthiekanun, V., N. Pheaktra, H. Putchhat, L. Sin, B. Sen, V. Kumar, S. Langla, S. J. Peacock, and N. P. Day. 2008. Burkholderia pseudomallei antibodies in children, Cambodia. Emerg. Infect. Dis. 14:301-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, M., and J. S. H. Tsang. 2006. Use of ribosomal promoters from Burkholderia cenocepacia and Burkholderia cepacia for improved expression of transporter protein in Escherichia coli. Protein Expr. Purif. 49:219-227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.