Abstract
Gene expression in biofilms is dependent on bacterial responses to the local environmental conditions. Most techniques for studying bacterial gene expression in biofilms characterize average values across the entire population. Here, we describe the use of laser capture microdissection microscopy (LCMM) combined with multiplex quantitative real-time reverse transcriptase PCR (qRT-PCR) to isolate and quantify RNA transcripts from small groups of cells at spatially resolved sites within biofilms. The approach was first tested and analytical parameters were determined for Pseudomonas aeruginosa containing an isopropyl-β-d-thiogalactopyranoside-inducible gene for the green fluorescent protein (gfp). The results show that the amounts of gfp mRNA were greatest in the top zones of the biofilms, and that gfp mRNA levels correlated with the zone of active green fluorescent protein fluorescence. The method then was used to quantify transcripts from wild-type P. aeruginosa biofilms for a housekeeping gene, acpP; the 16S rRNA; and two genes regulated by quorum sensing, phzA1 and aprA. The results demonstrated that the amount of acpP mRNA was greatest in the top 30 μm of the biofilm, with little or no mRNA for this gene at the base of the biofilms. In contrast, 16S rRNA amounts were relatively uniform throughout biofilm strata. Using this strategy, the RNA amounts of individual genes were determined, and therefore the results are dependent on both gene expression and the half-life of the transcripts. Therefore, the uniform amount of rRNA throughout the biofilms likely is due to the stability of the rRNA within ribosomes. The levels of aprA mRNA showed stratification, with the largest amounts in the upper 30-μm zone of these biofilms. The results demonstrate that mRNA levels for individual genes are not uniformly distributed throughout biofilms but may vary by orders of magnitude over small distances. The LCMM/qRT-PCR technique can be used to resolve and quantify this RNA variability at high spatial resolution.
Bacteria thrive in spatially defined microenvironments, and therefore they are exposed to local environmental conditions that may vary on the micrometer scale (53). When growing on surfaces in biofilms, bacteria typically are distributed in a heterogeneous manner. The cell distribution reflects heterogeneities in the properties of the surface and the concentration gradients of chemicals dissolved in the interstitial fluid within the biofilm matrix (22, 27, 38, 54). These physical and chemical heterogeneities in the biofilms promote differences in bacterial enzymatic activities at different regions (25, 61, 64) and influence gene expression in different zones within biofilms (4, 9, 13, 14, 52, 55).
Most studies of gene expression by bacteria in biofilms report levels that reflect the average gene expression across the entire population (2). Techniques often are insensitive to the diverse range of activities that occur among cells at different locations within the biofilm and therefore do not account for the contribution of specific parameters, such as the role of changing environmental conditions on the regulation of gene expression (2). However, due to the importance of understanding bacterial activities in situ, several approaches have been developed to evaluate local gene expression within biofilms. One approach uses fluorescent reporter genes fused to promoter regions of interest as well as epifluorescence microscopy (14, 33, 55, 65). Although useful, this technique requires the prior genetic manipulation of the bacteria and the reintroduction of the constructed strains back into the environment. The expression of the reporter gene may induce physiological changes in the cells of the constructed strains that do not occur in the wild-type cell. Thus, this approach precludes the ability to study gene expression in the wild-type strain in its native state. Fluorescent in situ hybridization (FISH) has been used to detect specific RNA sequences in bacterial cells in their native assemblages (1, 15, 32). This approach is most useful for studying high-abundance RNAs, such as rRNAs, but is less useful for detecting mRNA transcripts in low abundance. Measurements of cellular activity may be obtained by combining FISH with microautoradiography or Raman spectroscopy after the uptake of labeled substrates into cellular material (26, 56). To measure the gene expression of cells in biofilms, in situ reverse transcription-PCR has been developed to amplify mRNA present in low copy numbers (29). This approach uses fluorescently labeled probes to detect individual cells expressing a gene of interest, but it is limited by a lack of mRNA quantification.
Here, we developed a strategy based on advances in laser capture microdissection microscopy (LCMM) (7, 16-18, 39, 48, 49) and multiplex quantitative real-time reverse transcriptase PCR (qRT-PCR) (10, 23, 24, 34) for the quantitative assessment of any gene expressed by bacteria in their native environment. The approach allows the quantification of low-abundance mRNA transcripts without the need to genetically manipulate the cells or expose cells to labeled substrates. We used this strategy to study the vertical distribution of RNA transcripts in Pseudomonas aeruginosa biofilms. In particular, we characterized the distribution of a housekeeping gene, acpP; the 16S rRNA; and two genes regulated by quorum sensing (QS), phzA1 and aprA. The results demonstrate that RNA abundances for individual genes are not uniformly distributed throughout biofilms, and that cells in relatively close proximity to each other may have vast differences in abundances of individual RNA transcripts.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa PAO1 and P. aeruginosa PAO1(pAB1) were used for these studies. Plasmid pAB1 allows the isopropyl-β-d-thiogalactopyranoside (IPTG) induction of green fluorescent protein (GFP) (3). Inocula of P. aeruginosa containing pAB1 were supplemented with 150 μg/ml of carbenicillin. Two biofilm growth conditions were used: colony biofilms and biofilms cultivation in drip-flow reactors (61, 64). For colony biofilms, planktonic cultures of P. aeruginosa were incubated overnight at 37°C in Luria-Bertani (LB) broth (6). Cultures were diluted in sterile LB broth to an optical density at 600 nm of 0.4 (1-cm path length), and 25 μl was used to inoculate sterilized black, polycarbonate membrane filters (13-mm diameter, 0.2-μm pore size; GE Water & Process Technologies). The filters were placed on LB agar (Difco Laboratories) and incubated for a total of 52 h at 37°C. The membranes containing the colonies were transferred to fresh LB agar every 12 h. For the IPTG induction of GFP in strains containing pAB1, membranes were transferred to LB agar containing 1.0 mM IPTG and were incubated for an additional 4 h.
Biofilms also were cultivated on stainless steel coupons in continuous flow by using drip-flow reactors (64). The reactor consisted of a one-time flowthrough system containing a medium reservoir, pump, silicon tubing, drip-flow chamber, and waste container. Biofilm minimal medium (BMM) (42) contained (per liter) 9.0 mM sodium glutamate, 50 mM glycerol, 0.02 mM MgSO4, 0.15 mM NaH2PO4, 0.34 mM K2HPO4, 145 mM NaCl, 20 μl trace metals, and 1 ml vitamin solution. The trace metal solution contained (per liter of 0.83 M HCl) 5.0 g CuSO4·5H2O, 5.0 g ZnSO4·7H2O, 5.0 g FeSO4·7H2O, and 2.0 g MnCl2·4H2O. The vitamin solution contained (per liter) 0.5 g thiamine and 1 mg biotin. The pH of the medium was adjusted to 7.0. Prior to inoculation into the system, each strain was incubated in BMM for 18 h at 37°C. The cultures were transferred to fresh BMM for 4 h to reach an optical density at 600 nm of 0.2. The inoculum was diluted 20-fold in 0.85% NaCl, and 5 ml was used to inoculate steel coupons for 25 min. BMM then was pumped through the reactors at 1.2 ml/min for 72 h. Reactors were maintained at 37°C throughout the incubation. For biofilms for which the induction of the GFP was necessary, BMM was supplemented with 1.0 mM IPTG for the final 4 h of incubation.
Cryoprocessing of biofilms.
Following incubation, biofilms were cryoembedded by flash-freezing the stainless steel slides containing drip-flow-cultivated or colony biofilms on dry ice. Biofilms were immersed in Tissue-Tek O.C.T. compound (Sakura Finetechnical Co.). Vertical transects of the biofilms were obtained by sectioning the solidified O.C.T.-containing biofilms with a cryomicrotome. Thin sections (5 μm) of vertical transects of the biofilms were placed onto membrane-coated microscope slides (P.A.L.M. Microlaser Technologies). The microscope slides were maintained on dry ice until examination and sampling.
LCMM.
LCMM (Zeiss/P.A.L.M. Laser-MicroBeam system) was used to dissect and capture sections from different regions within the biofilms. Microscope slides containing the O.C.T.-embedded biofilms were thawed on the microscope stage for 5 s. The biofilms then were examined using lenses with objectives of ×10 to ×40 magnification. Areas of the biofilms ranging from 500 to 100,000 μm2 were obtained using the laser catapult parameter, which allows dissected samples to be catapulted into 40 μl of lysis buffer (0.3 M sucrose and 0.02 M sodium acetate at pH 4.5) contained in the caps of 0.5-ml microcentrifuge tubes.
RNA extraction.
RNA extraction followed the phenol-chloroform-based protocol of Chomczynski and Sacchi (11) as modified by Barton et al. (5). The samples, obtained by being laser catapulted into 40 μl lysis buffer, were mixed by inversion with additional buffer (160 μl of lysis buffer and 200 μl of 2% sodium dodecyl sulfate). An aliquot of luciferase (lucI) RNA (1.7 × 104 transcript copies) (Promega) was added as a spike-in control to assay for RNA loss during sample preparation. Samples were centrifuged for 10 s and then transferred to a 1.5-ml microcentrifuge tube containing water-saturated phenol (400 μl), and then they were heated in a water bath at 65°C for 5 min with frequent vortexing. The aqueous layer, formed after 25 min of centrifugation, was extracted with an equal volume of phenol:chloroform:isoamyl-alcohol (25:24:1) in a heavy Phase-Lock gel tube (Eppendorf). RNA was precipitated by adding 3 μl of PolyAcryl carrier (Molecular Research Center, Inc.), 32 μl of 0.25 μM sodium acetate, and 1,200 μl of absolute ethanol. Resuspended RNA was treated with DNase by using the Turbo DNA-free kit (Ambion, Inc.) in 10-μl reaction mixtures for 20 min. RNA was diluted to a final volume of 25 μl to reduce interference from possible PCR inhibitors. Total RNA was quantified using a NanoDrop fluorescence spectrophotometer (NanoDrop Technologies, Inc.) with Molecular Probes Ribogreen (Invitrogen). All samples were stored at −80°C until analysis by qRT-PCR.
Primers and dual-labeled probes.
PCR primer and dual-labeled probe sequences are shown in Table 1. The primers were designed using the Primer3 program (41) or with Primer Express v2.0 software (Applied Biosystems). Primers were assayed for melting temperature, primer dimers, and secondary structure using Mfold (30, 67) and Oligo Analyzer, version 3.0 (Integrated DNA Technologies, Inc.). Primers were purchased from Integrated DNA Technologies.
TABLE 1.
qRT-PCR primer and probe sequences and labels
| Primer | Sequence | Labela | Concn (nM) | Tempb (°C) | Product size (bp) |
|---|---|---|---|---|---|
| Primers and probes for qRT-PCR | |||||
| gfp-For | TTTCACTGGAGTTGTCCCAATTC | 400 | 60 | 80 | |
| gfp-Rev | CACCCTCTCCACTGACAGAAAAT | 400 | |||
| gfp-Probe | TGTGCCCATTAACATCACCATCTAATTCAACA | 5′ FAM, 3′ BHQ-1 | 300 | ||
| acpP-For | ACTCGGCGTGAAGGAAGAAG | 400c/50d | 60 | 80 | |
| acpP-Rev | CGACGGTGTCAAGGGAGT | 400c/900d | |||
| acpP-Probe | AAGTCACCAACAGCGCTTC | 5′ JOE, 3′ BHQ-1 | 200 | ||
| lucI-For | GTGTTGGGCGCGTTATTTATC | 200 | 60 | 78 | |
| lucI-Rev | ACTGTTGAGCAATTCACGTTCA | 200 | |||
| lucI-Probe | CGCCCGCGAACGACATTTAT | 5′ Cy5, 3′ BHQ-2 | 200 | ||
| aprA-For | GCTTCAGCCAGAACCAGAAGAT | 200 | 60 | 78 | |
| aprA-Rev | TCGACACATTGCCCTTCAAC | 200 | |||
| aprA-Probe | ACATCGGACAGCGCCTTCTCGTTG | 5′ FAM, 3′ BHQ-1 | 100 | ||
| phzA1-For | TAAAACGTAATCGCGAGTTCATG | 900 | 60 | 74 | |
| phzA1-Rev | TTTTATTTGCGGAACGGCTATT | 900 | |||
| phzA1-Probe | CCAATGCACGCAGTTTCTGTATCGGGT | 5′ ROX, 3′ BHQ-2 | 150 | ||
| 16S rRNA-Fore | CAAAACTACTGAGCTAGAGTACG | 300 | 59 | 215 | |
| 16S rRNA-Reve | TAAGATCTCAAGGATCCCAACGGCT | 300 | |||
| Primers used for IVT | |||||
| gfp-T7-for | TAATACGACTCACTATAGGGGGAGAAGAACTTTTCACTGG | ||||
| gfp-T7-rev | GAAAGGGCAGATTGTGTGGAC | ||||
| acpP-T7-for | TAATACGACTCACTATAGGGCCATCGAAGAACGCGTTAAG | ||||
| acpP-T7-rev | CCTGAACGGTGGTGATCTTT | ||||
| aprA-T7-for | TAATACGACTCACTATAGGGCTTGCATTGAAAGGTCGTAGC | ||||
| aprA-T7-rev | GATATCGCCGTAGACGAAGGT | ||||
| phzA1-T7-for | TAATACGACTCACTATAGGGGTCAGCGGTACAGGGAAACAC | ||||
| phzA1-T7-rev | GTGGGAATACCGTCACGTTT |
FAM, 6-carboxyfluorescein; BHQ-1, black hole quencher 1; JOE, carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein; and ROX, carboxy-X-rhodamine.
Annealing temperature for PCR.
Primer concentrations used in acpP, gfp, and lucI triplex reactions.
Primer concentrations used in acpP, aprA, and phzA1 triplex reactions.
Primers sequences used by Matsuda et al. (31). The acquisition step of the 16S rRNA assay was performed at 72°C.
IVT.
RNA standards were generated by in vitro transcription (IVT) (19). The gene of interest was amplified from P. aeruginosa PAO1 genomic DNA by using gene-specific primers containing a T7 promoter (Table 1). The PCR products were purified and used for IVT with the MEGAscript T7 kit (Ambion), which included a DNase treatment (Turbo DNase for 15 min at 37°C). IVT RNA products were purified using the RNeasy mini kit (Qiagen) and analyzed by electrophoresis on an RNA nanochip using a Bioanalyzer 2100 (Agilent Technologies). RNA standards were quantified using UV absorption at 260 nm, aliquoted, and stored at −80°C.
The copy number of RNA standards was calculated using the following equation: number of molecules per microliters = (number of grams of RNA per microliter/molecular weight) × Avogadro's constant. Copy numbers per reaction were assigned to each standard using Rotor Gene software, version 6.0 (Corbett Research). For each standard, the concentration was plotted against the cycle number at which the fluorescence exceeded the background (i.e., the threshold cycle [CT]). The slope of the calibration curve was used to determine the reaction efficiency (E) of the CT according to the equation E = [10(−1/slope)] − 1, where an E of 1 indicates an exponential amplification of the product. The efficiency of serially diluted samples was evaluated to determine the assay's sensitivity and to ensure the assay's compatibility with different probes in the multiplex reactions.
qRT-PCR.
One-step qRT-PCR was used for all mRNA quantifications. qRT-PCR mixtures (25 μl) containing 2 to 3 μl of template RNA were used according to the manufacturer's instructions. Three kits were used depending on the type of reaction. QuantiTect probe RT-PCR and QuantiTect multiplex RT-PCR NR kits (Qiagen) were used for dual-labeled probe analysis and multiplex dual-labeled probe analysis. A QuantiTect Sybr green RT-PCR kit was used for Sybr green labeling analysis.
Primer and probe concentrations were determined by performing the optimization protocol recommended for the Rotor Gene system (real-time summary, version 1.7). Every assay contained negative controls (samples lacking reverse transcriptase or template) and positive controls (samples with appropriate standards added). Standard RNA, which was derived from IVT, and the external spike-in control RNA of lucI were serially diluted in 8 μl/ml of PolyAcryl carrier (Molecular Research Center, Inc.) to accommodate 101 to 106 molecules per reaction. Cycling parameters were established according to the kit's instructions for single-probe assays or for multiplex assays (Corbett Research). Three replicates were used to generate standard curves. Each sample was assayed in duplicate. Primer specificity (for the single PCR product) was confirmed by electrophoresis using a Bioanalyzer 2100 and a DNA 500 chip (Agilent Technologies).
Transcript amounts for acpP, gfp, aprA, phzA1, and lucI were calculated from calibration curves, with a normalization factor of calculated lucI per sample/1.7 × 104, since 1.7 × 104 lucI RNA transcripts were added to each sample immediately after laser capture as a spike-in control. To quantify the relative amounts of 16S rRNA, CT values were log transformed with the formula amplification−CT × 1010, where CT and amplification values were derived from the comparative quantitation analysis that used Rotor Gene software (40).
Validation of qRT-PCR efficiency, linearity, and reproducibility.
qRT-PCR efficiencies were calculated from the slope of each calibration curve run in multiplex reactions. A qRT-PCR efficiency (E) of 1 indicates an exponential amplification of the product. We observed high efficiency for acpP (0.91), gfp (0.93), and lucI (1.02) when run in multiplex reactions using dual-labeled probes. Standard curves for these transcripts exhibited linear responses from 50 to 500,000 transcripts (r2 > 0.98 for each). The efficiencies for multiplex reactions were the following: aprA, 1.04; phzA1, 1.04; acpP, 0.86; and lucI, 0.94. These transcripts exhibited linear responses from approximately 300 to 30,000 transcript copies (r2 > 0.96 for each).
Sybr green-based qRT-PCR was used to measure 16S rRNA. This assay had an efficiency of 0.95 and was linear from 10 to 100,000 transcripts (r2 > 0.99). The interexperimental precision was approximately 16%, and the intrasample precision was 11%. These calculations were based on the calculated number of copies per reaction and their variations from the means. We chose to obtain the reproducible measure from the calculated number of copies per reaction and not from CT values to avoid underestimating the true variability (28).
Statistical analysis.
Since data sets obtained from qRT-PCR do not have uniform distributions, differences between means were calculated using the two-tailed Mann-Whitney test.
RESULTS
GFP fluorescence correlates with gfp mRNA amounts.
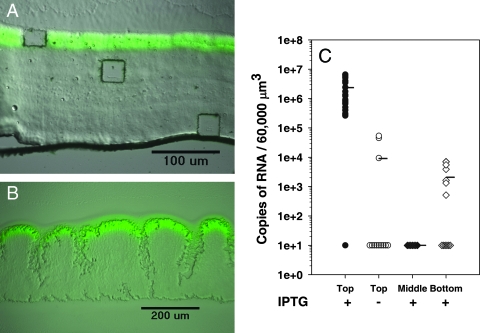
To quantify RNA levels from small groups of cells within a larger surrounding biofilm population, we first developed a system by which mRNA transcript numbers could be determined and compared to those of a spatially defined phenotypic trait. For this we used a strain of P. aeruginosa with an IPTG-inducible gene for GFP (gfp). GFP fluorescence was assayed by microscopy, and gfp mRNA abundance was assayed by LCMM/qRT-PCR. When biofilms of this strain are incubated for 52 h in the absence of IPTG and then induced with IPTG for the final 4 h of incubation, a band of green fluorescence is observed in the top 30-μm zone of the biofilm (Fig. 1A and B) (8, 61). This fluorescent band is observed whether IPTG is applied from the bottom of the biofilm in the colony biofilm format (Fig. 1B) or from the liquid medium in drip-flow-cultivated biofilms (Fig. 1A). The LCMM was used to dissect and capture cells from the top fluorescent layer in the IPTG-induced biofilms and from the equivalent top zone of control biofilms not amended with IPTG. The captured dissects ranged from 500 to 48,000 μm2 (Fig. 1B). Figure 1c shows the gfp mRNA copy abundances for 26 samples after normalizing the results to the 60,000-μm3 biofilm volume. On average, in IPTG-induced biofilms the copy number of gfp mRNA was 250-fold greater than that from the biofilm samples not induced with IPTG (P < 0.001), for which no fluorescent zone was observed. Of the 12 samples taken from biofilms to which no IPTG was added, 9 had gfp mRNA levels below the detection limit, indicating little or no expression of this gene in the absence of the inducing agent. The other three samples had transcript levels above the detection limits but approximately 100-fold less than that of the IPTG-induced biofilms. Of the 26 samples from the IPTG-induced biofilms, one sample showed no gfp mRNA transcripts. The lack of gfp mRNA in this sample was not due to sampling error or mRNA degradation, since the lucI spike-in control was not anomalous and since the sample contained 16S rRNA (described below).
FIG. 1.
Localized GFP fluorescence of (A) biofilm cultivated in a drip-flow reactor and (B) colony biofilm. Biofilms were incubated without an inducing agent for 72 and 52 h, respectively, and then were induced with 1 mM IPTG for the final 4 h. (B) LCMM was used to obtain microdissected samples from regions within the biofilm ranging from 500 to 60,000 μm2. (C) qRT-PCR of gfp mRNA obtained from the top fluorescent zone of the IPTG-induced biofilms (n = 26) and from the equivalent top zone of the noninduced biofilms (n = 12). Sections were normalized to 60,000 μm3 for comparison. Each point represents an individual measurement. The points plotted at 101 are below detection limits (+IPTG, 1 sample; −IPTG, 9 samples). A two-tailed Mann-Whitney test to calculate differences in means between IPTG-induced and noninduced biofilms was used (P < 0.01). The LCMM also was used to obtain biofilm sections from the middle (n = 7) and bottom (n = 13) of the biofilm. A value of 101 indicates amounts below the level of detection (middle, 7 samples; bottom, 6 samples).
Stratified gfp expression in IPTG-induced biofilms.
Since GFP requires oxygen for activity, there is the possibility that it is expressed throughout the biofilm but only shows fluorescence in the oxygen-exposed top layer. To determine if gfp expression, as opposed to GFP activity, is greatest at the top of the biofilms, LCMM was used to sample the nonfluorescent zones of the IPTG-induced biofilms. The results show little gfp mRNA in the underlying portions of the biofilm compared to that in the top 30-μm fluorescent zone (P < 0.001 for values for the top layer compared to those for the middle or bottom layer) (Fig. 1C). No gfp expression was observed for any of the samples in the middle zone, and six samples from the bottom zone had no gfp expression. Some of the samples from the bottom zones had small amounts of gfp expression that were approximately 1,000-fold lower than those in the top fluorescent zone. Therefore, in this system, the deeper biofilm zones are not fluorescent due to low levels of gfp expression rather than to a lack of GFP activation with oxygen. These results demonstrate that it is possible to use LCMM and qRT-PCR to quantify the RNA amounts of a regulated gene that is not homogenously distributed throughout a stratified biofilm.
Localized expression of the housekeeping gene acpP.
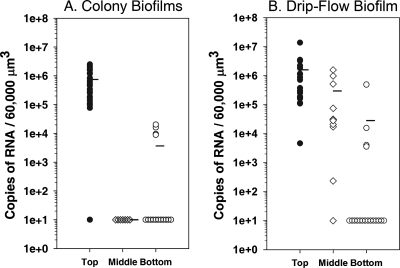
By using multiplex qRT-PCR, it is possible to assay the RNA amounts of several genes simultaneously. In addition to gfp and the lucI spike-in control, we chose to assay the gene for the acyl carrier protein, acpP, as an internal housekeeping gene control. AcpP is required for fatty acid biosynthesis and therefore is required for the production of new cell membrane material. Interestingly, acpP mRNA amounts are not uniform throughout these biofilms. In the colony biofilms (Fig. 2A), acpP mRNA abundances reflected gfp amounts, with the greatest amounts in the top 30-μm zone of the biofilm (P < 0.001 for values for the top layer compared to those for the middle or bottom layer). None of the samples isolated from the middle of the colony biofilms had acpP mRNA amounts that were above the detection limit by qRT-PCR. Similarly, most samples from the bottom of the biofilm had acpP mRNA levels that were below the level of detection. Several of the samples from the bottom of the biofilm had acpP levels that were above the detection level, but each of these amounts was approximately 100-fold less than the average amounts from samples from the top 30 μm of the biofilm. The results indicate that in these biofilms, which averaged 200 μm thick, the top 30 μm contains cells with a high abundance of acpP mRNA, suggesting that these cells are synthesizing new membrane material; the deeper portions of the biofilms had much less expression of this gene.
FIG. 2.
Stratified expression of acpP mRNA in biofilms. qRT-PCR was performed on acpP obtained from the top (n = 26 and 21), middle (n = 8 and 11), or bottom (n = 15 and 18) of colony biofilms and drip-flow biofilms, respectively. Two-tailed Mann-Whitney test results demonstrated significant differences in acpP expression (P < 0.01 for values for the top layer compared to those for the middle or bottom layer). The number of samples below the level of detection are shown as 101 for colony biofilms (top, 1 sample; middle, 8 samples; bottom, 11 samples) and drip-flow biofilms (top, 0 samples; middle, 1 sample; bottom, 14 samples).
The drip-flow-cultivated biofilms also showed the greatest acpP abundance in the top 30-μm zone of the biofilm. However, these biofilms had greater variability in mRNA amounts than the colony biofilms (Fig. 2B). Most samples from the bottom of the drip-flow biofilm had acpP mRNA levels that were below detection (14 of 18 samples), and the average abundance of acpP was 100-fold less at the base of the biofilm than at the top. The middle of the biofilm showed a large range of variability in acpP mRNA amounts, with an average that was approximately 10-fold less than that at the top of the biofilm. The drip-flow biofilms have more structural features than the colony biofilms (Fig. 1A and B), with increased amounts of water channels and mushroom-like features. The increased variability observed in Fig. 2B for the middle of the biofilms likely reflects the proximity of the dissected sample to adjacent water channels, which affects nutrient and oxygen availability. The results for acpP for both the colony biofilms and for the drip-flow biofilms indicate that there is significant variability in the expression of this housekeeping gene that is dependent on the location of the cells within a biofilm. The results also demonstrate that acpP may not be a good internal housekeeping gene control for normalizing the activity of other genes, since its expression is not uniform throughout these biofilms.
Uniform amounts of 16S rRNA throughout biofilms.
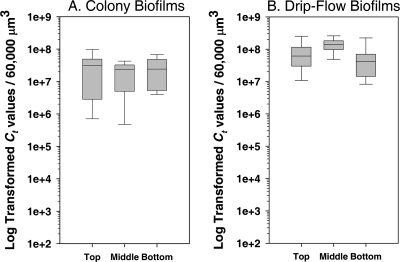
In contrast to the heterogeneous pattern observed for acpP and gfp, the 16S rRNA amounts were relatively constant in the different biofilm layers (Fig. 3). No statistically significant difference was observed for the 16S rRNA gene product at the top, middle, or bottom of the biofilms that had been cultivated in either the colony biofilm or the drip-flow biofilm format (P > 0.1 for each analysis). In addition, the average amounts of 16S rRNA were fairly consistent among all of the biofilms and biofilm zones. Although the expression of rRNA varies with cell growth (35, 43), the homogenous distribution of 16S rRNA likely is due to the higher stability of rRNA contained within ribosomes. The abundance of individual RNAs measured here by qRT-PCR is dependent on both the expression and half-life of the transcript. Whereas the half-life of the acpP and gfp mRNAs is short, the rRNA contained within ribosomes is more stable. These results are consistent with the visual observations of the 16S rRNA using FISH probing, which showed relatively constant amounts of 16S rRNA throughout a biofilm (63). The results also suggest that the 16S rRNA is used as an internal control for the comparison of the induction of genes that are differentially regulated through the biofilms. However, rRNA must be used with caution as an internal control, since the abundance of rRNA often is much higher than that of any mRNA species.
FIG. 3.
Box plot of 16S rRNA in biofilms. Sections from the top (n = 10 and 12), middle (n = 11 and 8), and bottom (n = 12 and 17) biofilm layers from colony biofilms or drip-flow biofilms, respectively, were analyzed by SYBR green-based qRT-PCR. CTs were log transformed and adjusted to 60,000 μm3. The line indicates the median; 50% of observations are within the box. Bars indicate samples with the minimum and maximum values. A two-tailed Mann-Whitney test for comparisons between top and bottom levels of 16S rRNA in colony biofilms showed no significant differences (P > 0.5). In addition, no significant difference was observed for the different layers of the drip-flow biofilms.
Stratified expression of QS-regulated genes.
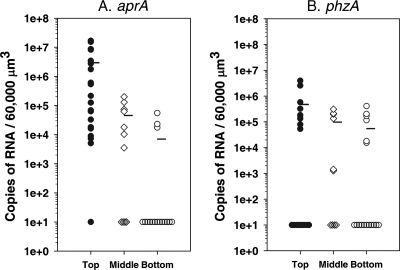
Bacterial cells within biofilms communicate through small diffusible chemical signals by QS (20, 21, 36, 37). We assayed the localized expression of two genes, aprA (encoding alkaline protease) and phzA1 (necessary for pyocyanin biosynthesis), that are known to be regulated by QS (46, 57) and that have been shown by proteomics to be upregulated in P. aeruginosa biofilms (42). P. aeruginosa PAO1 biofilms that had been cultivated in drip-flow reactors were cryoembedded and thinly sectioned for LCMM and then assayed for aprA and phzA1 expression along vertical transects of the biofilms. The amounts of aprA mRNA showed a trend similar to that observed for gfp and acpP, with the greatest expression in the outermost 30-μm portion of the biofilm (P < 0.001) (Fig. 4A). The expression of aprA averaged 1,000-fold higher than that of samples from the bottom of the biofilms, where most samples had expression that was below the detection limit. The middle of the biofilms had intermediate aprA expression that averaged approximately 100-fold less than that of samples from the top of the biofilms. In contrast, phzA1 amounts, although greater at the top of the biofilm, were more uniformly distributed at the different levels (P > 0.1) (Fig. 4B), possibly due to the lower overall expression of phzA1 compared to that of aprA. The 16S rRNA gene product was used as an internal control and showed uniform amounts throughout these biofilms (P > 0.5). The differences observed between the expression patterns of aprA and phzA1 demonstrate that factors other than QS, such as the physiological status of the cells, also may influence the expression of genes that are regulated by QS.
FIG. 4.
Expression of QS-regulated genes aprA and phzA1 in P. aeruginosa PAO1 biofilms that were cultivated in drip-flow reactors. (A) aprA mRNA obtained from the top (n = 20), middle (n = 11), and bottom (n = 17) of biofilms. The mRNA levels in some samples were below the level of detection (top, 1 sample; middle, 4 samples; bottom, 13 samples). (B) phzA1 mRNA from the same samples. The mRNA levels in some samples were below the level of detection (top, 8 samples; middle, 4 samples; bottom, 11 samples). Two-tailed Mann-Whitney test results demonstrated significant differences in aprA expression (P < 0.01 for values for the top layer compared to those for the middle or bottom layer). Two-tailed Mann-Whitney test results demonstrated no significant differences in phzA expression (P > 0.1 for values for the top layer compared to those for the middle or bottom layer).
Limits of detection for localized mRNA and rRNA amounts in biofilms.
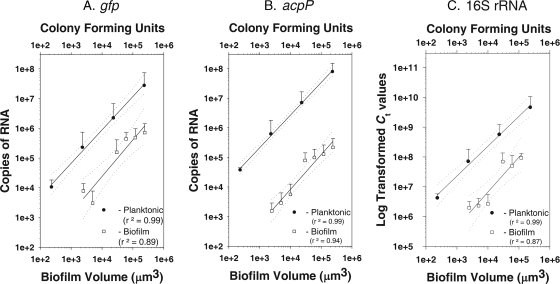
To determine the range of detection limits for LCMM/qRT-PCR, biofilm samples were dissected from the top zone of IPTG-induced colony biofilms. The amounts of gfp, acpP, and 16S rRNA were quantified and plotted against the sample volume. Since samples were taken from cryoembedded biofilms, viable cell counts could not be determined. However, assuming a cell volume of approximately 1 μm3 (51), a 500-μm2 area of a 5-μm-thick section contains approximately 2,500 cells. For acpP and gfp, a linear relationship was observed between the sample volume and the number of mRNA transcripts that ranged from 1 × 103 to 1 × 105 μm3 (r2 = 0.94 for acpP; r2 = 0.90 for gfp) (Fig. 5A and B). Therefore, this technique is useful for quantifying mRNA transcripts from approximately 2,000 cells. Similarly, log-transformed CT values of 16S rRNA also correlated with sample size (r2 = 0.90) (Fig. 5C). Due to the higher cellular amounts of 16S rRNA, the detection limit for rRNA is much less than 1,000 cells.
FIG. 5.
Limits of detection for microdissected biofilm samples and for planktonic cultures for gfp (A), acpP (B), and 16S rRNA (C). Detection limits were based on biofilm volume using 5-μm-thick sections of the top zones of the biofilms. Detection limits for planktonic cells were based on the numbers of CFU of serially diluted cultures. Dual-labeled probes were used in multiplex reactions for gfp (r2 = 0.89 for biofilm cells and r2 = 0.99 for planktonic cells) and for acpP (r2 = 0.94 for biofilm cells and r2 = 0.99 for planktonic cells). The levels of 16S rRNA were measured using the SYBR green-based chemistry of qRT-PCR (r2 = 0.87 for biofilm and r2 = 0.99 for planktonic).
The detection limits of qRT-PCR for gfp, acpP, and 16S rRNA also were determined for exponentially growing planktonic cells, allowing for the comparison of the mRNA transcript counts to the viable cell counts. RNA was extracted from serially diluted planktonic P. aeruginosa(pAB1) cultures containing from 1 to 109 cells (determined as the number of CFU) (Fig. 5). Linear responses were observed for cell numbers and mRNA transcripts (r2 = 0.97 for gfp; r2 = 0.99 for acpP). A linear response also was observed for the 16S rRNA (r2 = 0.99). According to the regression analysis, there are approximately 40 gfp and 900 acpP mRNA transcripts per cell growing in planktonic culture.
These results allowed the comparison of the gene expression levels of exponentially growing planktonic cells and of cells growing in the top zone of the IPTG-induced colony biofilms (Fig. 5). Planktonic cells had mRNA levels of gfp and acpP per cell that were higher than the expression in an equivalent volume of the biofilm. RNA amounts were approximately 40-, 250-, and 33-fold greater in planktonic cells for gfp, acpP, and 16S rRNA, respectively, than those from the top biofilm zone. Therefore, although mRNA amounts are highest at the top zone of these biofilms, the amounts in biofilms are less than those for exponentially growing planktonic cultures.
DISCUSSION
The goals of this research were (i) to develop methodology to quantify bacterial gene expression from small groups of bacterial cells isolated from biofilms without the prior genetic manipulation of the cells, and (ii) to use the method to determine if local conditions found within biofilms result in the spatial variation of bacterial gene expression. By using the cryosectioning of biofilms and LCMM, we were able to isolate and extract RNA from small groups of bacterial cells that were isolated from different regions within biofilms. Multiplex qRT-PCR allowed the quantification of gene expression of up to four genes simultaneously from these small groups of cells.
The approach was validated using an artificially induced tracer gene (gfp), a housekeeping gene (acpP), an RNA spike-in control (lucI), an internal control (16S rRNA), and two genes regulated by QS. For gfp and acpP, the qRT-PCR approach using dual-labeled probes was sensitive for detecting less than 1,000 mRNA transcripts. For exponentially growing planktonic cells, for which transcripts ranged from 100 to 400 copies per cell (Fig. 5), the qRT-PCR approach may be useful for estimating mRNA abundances from less than 10 cells. At the oxygen interface of these biofilms, we estimate that the number of transcripts for gfp and acpP is approximately 40- to 250-fold less per cell than for exponentially growing planktonic cells (Fig. 5). We measured the expression of these genes for approximately 2,000 biofilm-associated cells. However, based on the detection limits obtained for planktonic cultures, it should be possible to investigate gene expression from even smaller numbers of biofilm cells by using this approach.
The GFP fluorescent zone of the biofilm correlated with gfp mRNA amounts (Fig. 1). GFP amounts were significantly greater for IPTG-induced biofilm cultures than for noninduced samples, and they were greater for the GFP fluorescent zone than the nonfluorescent deeper areas of the biofilms. Variation in the amount gfp mRNA at different regions of the biofilms was expected, given the localized nature of the fluorescent zone. Interestingly, an active zone of acpP mRNA of similar size also was observed (Fig. 2). Housekeeping genes, such as acpP, often serve as internal controls in qRT-PCR experiments to normalize the expression of regulated genes (45). In biofilms, gradients of pH, oxygen, nutrients, and waste products are established at different depths in biofilms (53, 54). These variable environmental factors contribute to various metabolic activities of the bacteria. Since the AcpP analyzed in this study is required for the synthesis of fatty acids and new membrane material, the results suggest that the cells in the upper 30 μm of these biofilms likely are the most active in membrane synthesis. The cells in the deeper portions of the biofilm likely are in the slow-growth state. These results are consistent with those of another study that indicates that P. aeruginosa cells in deeper portions of the biofilm are less metabolically active (60). In the prior study, transmission electron microscopy was used to demonstrate that P. aeruginosa cells located in the top layer of a biofilm, at the air/biofilm interface, are more susceptible to antibiotic killing than those in the deeper portions (60), even though the antibiotics penetrate through biofilms (3, 60, 66). The prior study suggested that the cells in the deeper portions of the biofilm are more tolerant to the antibiotics primarily due to their slow growth rate. The results here, showing the localized expression of acpP, further support that cells in deeper portions of these P. aeruginosa biofilms are not as metabolically active as those at the top of the biofilm.
Due to the differential expression of acpP throughout the biofilms, acpP may not be used as an internal control for normalizing the activity of genes that are differentially expressed in biofilms. Using FISH probing, 16S rRNA showed a fairly uniform distribution in P. aeruginosa biofilms (63). The qRT-PCR results here confirm those results by demonstrating little variability in 16S rRNA amounts along the vertical layers of the biofilm (Fig. 3). These results were similar in all conditions tested, including colony biofilms, drip-flow-cultivated biofilms, and biofilms supplemented with IPTG. The expression of ribosomal RNAs is highly regulated in bacteria (35). The level of 16S rRNA has been shown to vary according to cellular activity, with slower metabolisms having lower levels of 16S rRNA (43). Although the top portion of these biofilms may be most active in membrane biogenesis, the cells at the top of the biofilm are not equivalent in growth to exponentially growing planktonic cells (Fig. 5). The final abundance of rRNA per cell, as measured here, was approximately 33-fold less for cells at the top of the biofilm than for the exponentially growing planktonic cells. The reduced growth of these cells (as indicated by the lower acpP and rRNA amounts), combined with the stability of rRNA within ribosomes of cells in the deeper portion of the biofilms, may explain the relative uniformity in rRNA abundance throughout the biofilm layers observed here and in a previous study (63).
The results indicate that rRNA can be used as a technical control for determining if cell material is captured by the laser catapult method and retained during RNA sample preparation. However, the abundance of 16S rRNA (which is much higher than that of any of the mRNA species tested) is a drawback to using it as an internal control for quantifying the relative expression of other genes. In order to identify an appropriate alternative transcript that combines an average level of abundance with a uniform distribution along the biofilm layers, it may be necessary to develop genome-wide approaches that look at global distributions of gene expression throughout subpopulations within the biofilm.
Genes regulated by QS are responsive to small diffusible molecules, such as acyl homoserine lactones, the concentrations of which increase with increasing cell density (20, 21, 36, 37). Since cells in biofilms are at high density, QS is one factor that plays a role during biofilm development (12, 47). Here, we assayed the expression of two genes regulated by QS and found differences in their expression patterns. The amounts of aprA mRNA showed stratification and were 100 to 1,000-fold greater at the top of the drip-flow biofilms thab those of the middle or bottom layer (Fig. 4). In contrast, phzA1 amounts, although slightly higher at the top of the biofilm, had more equivalent average amounts throughout the biofilms compared to those of aprA. The results demonstrate that genes that are regulated by high cell density show the stratification and heterogeneity of expression in response to growth in biofilm microenvironments. The results for aprA are in contrast to those of a previous study that used reporter genes to characterize the local expression of two other QS-regulated gene, lasI and rhlI (13). The prior studies demonstrated that most of the cells expressing lasI and rhlI were near the biofilm substratum. The differences between the QS-regulated genes shown here and those in the prior study likely are due to the differences in the biofilm cultivation systems. Whereas the lasI/rhlI study used flowthrough biofilms, we used drip-flow biofilms that resulted in much thicker biofilms. The thicker biofilms likely resulted in steeper chemical gradients of oxygen and nutrients (53, 54), thereby possibly reducing the metabolic activity of bacteria in the deeper portions of the biofilms. QS may be affected by cell density as well as other factors, such as the metabolic status of the cells.
Global proteomic and transcriptomic studies have been performed to characterize the physiological differences between P. aeruginosa growing in biofilms and cells growing in planktonic cultures (44, 50, 58, 59, 62). Studies also have compared changes in protein profiles and gene expression during biofilm developmental processes (44). Those studies have been useful for identifying genes uniquely expressed in biofilms. Here, we demonstrate that gene expression differences occur not only between planktonic and biofilm-associated cells but also for cells at different locations within biofilms. The results may help explain some of the differences observed between global transcriptional and proteomic data. Since proteins in general have a longer half-life than mRNA, proteomic data would represent the accumulation of protein and cell material in biofilms up to the time of sampling (i.e., from all layers of the biofilms). However, due to the short half-life of the mRNA, the transcriptional studies would assay only genes that are actively expressed at the time of sampling. The results here suggest that the transcription of individual genes varies dramatically in different regions of the biofilms. Combining LCMM and qRT-PCR will be useful for characterizing the spatial and temporal aspects of these localized gene expression events.
Acknowledgments
We thank Kathleen McInnerney and Paula J. Wilderman for their assistance with experimental protocols and Jill Finkel for her assistance with statistical analyses.
This work was supported by Public Health Service grant AI-065906 (M.J.F.) from the National Institute of Allergy and Infectious Diseases and by the American Society for Microbiology Robert D. Watkins Graduate Research Fellowship (A.P.L.). We acknowledge the Montana State University INBRE Functional Genomics Core Facility supported by NIH grant P20-RR16455.
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Amann, R. I. 1995. Fluorescently labelled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol. Ecol. 4:543-554. [Google Scholar]
- 2.An, D., and M. R. Parsek. 2007. The promise and peril of transcriptional profiling in biofilm communities. Curr. Opin. Microbiol. 10:292-296. [DOI] [PubMed] [Google Scholar]
- 3.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagge, N., M. Schuster, M. Hentzer, O. Ciofu, M. Givskov, E. P. Greenberg, and N. Hoiby. 2004. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob. Agents Chemother. 48:1175-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, H. A., Z. Johnson, C. D. Cox, A. I. Vasil, and M. L. Vasil. 1996. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol. Microbiol. 21:1001-1017. [DOI] [PubMed] [Google Scholar]
- 6.Bertani, L., and G. Bertani. 1970. Preparation and characterization of temperate, non-inducible bacteriophage P2 (host: Escherichia coli). J. Gen. Virol. 2:201-212. [DOI] [PubMed] [Google Scholar]
- 7.Bonner, R. F., M. Emmert-Buck, K. Cole, T. Pohida, R. Chuaqui, S. Goldstein, and L. A. Liotta. 1997. Laser capture microdissection: molecular analysis of tissue. Science 278:1481-1483. [DOI] [PubMed] [Google Scholar]
- 8.Borriello, G., E. Werner, F. Roe, A. M. Kim, G. D. Ehrlich, and P. S. Stewart. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48:2659-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustin, S. A., V. Benes, T. Nolan, and M. W. Pfaffl. 2005. Quantitative real-time RT-PCR—a perspective. J. Mol. Endocrinol. 34:597-601. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 12.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 13.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Kievit, T. R., M. D. Parkins, R. J. Gillis, R. Srikumar, H. Ceri, K. Poole, B. H. Iglewski, and D. G. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 45:1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 16.Emmert-Buck, M. R., R. F. Bonner, P. D. Smith, R. F. Chuaqui, Z. Zhuang, S. R. Goldstein, R. A. Weiss, and L. A. Liotta. 1996. Laser capture microdissection. Science 274:998-1001. [DOI] [PubMed] [Google Scholar]
- 17.Espina, V., J. D. Wulfkuhle, V. S. Calvert, A. VanMeter, W. Zhou, G. Coukos, D. H. Geho, E. F. Petricoin III, and L. A. Liotta. 2006. Laser-capture microdissection. Nat. Protoc. 1:586-603. [DOI] [PubMed] [Google Scholar]
- 18.Fend, F., M. R. Emmert-Buck, R. Chuaqui, K. Cole, J. Lee, L. A. Liotta, and M. Raffeld. 1999. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am. J. Pathol. 154:61-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fey, S. J., A. Nawrocki, M. R. Larsen, A. Gorg, P. Roepstorff, G. N. Skews, R. Williams, and P. M. Larsen. 1997. Proteome analysis of Saccharomyces cerevisiae: a methodological outline. Electrophoresis 18:1361-1372. [DOI] [PubMed] [Google Scholar]
- 20.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 21.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 22.Geesey, G. G., R. J. Gillis, R. Avci, D. Daly, M. Hamilton, P. Shope, and G. Harkin. 1996. Influence of surface features on bacterial colonization and subsequent substratum chemical changes on 316L stainless steel. Corr. Sci. 38:73-95. [Google Scholar]
- 23.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 24.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 25.Huang, C. T., K. D. Xu, G. A. McFeters, and P. S. Stewart. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 64:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, W. E., K. Stoecker, R. Griffiths, L. Newbold, H. Daims, A. S. Whiteley, and M. Wagner. 2007. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ. Microbiol. 9:1878-1889. [DOI] [PubMed] [Google Scholar]
- 27.Lewandowski, Z., and H. Beyenal. 2001. Limiting-current-type microelectrodes for quantifying mass transport dynamics in biofilms. Methods Enzymol. 337:339-359. [DOI] [PubMed] [Google Scholar]
- 28.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 29.Magnuson, T. S., A. L. Neal, and G. G. Geesey. 2004. Combining in situ reverse transcriptase polymerase chain reaction, optical microscopy, and X-ray photoelectron spectroscopy to investigate mineral surface-associated microbial activities. Microb. Ecol. 48:578-588. [DOI] [PubMed] [Google Scholar]
- 30.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 31.Matsuda, K., H. Tsuji, T. Asahara, Y. Kado, and K. Nomoto. 2007. Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription-PCR. Appl. Environ. Microbiol. 73:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Møller, S., A. R. Pedersen, L. K. Poulsen, E. Arvin, and S. Molin. 1996. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl. Environ. Microbiol. 62:4632-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Møller, S., C. Sternberg, J. B. Anderson, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolan, T., R. E. Hands, and S. A. Bustin. 2006. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1:1559-1582. [DOI] [PubMed] [Google Scholar]
- 35.Nomura, M., R. Gourse, and G. Baughman. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 53:75-117. [DOI] [PubMed] [Google Scholar]
- 36.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for the expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen, K., and Z. Lewandowski. 1998. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol. Bioeng. 59:302-309. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, E., L. Charboneau, V. Espina, L. Liotta, E. Petricoin, and K. Dreher. 2004. Application of laser capture microdissection and protein microarray technologies in the molecular analysis of airway injury following pollution particle exposure. J. Toxicol. Environ. Health A 67:851-861. [DOI] [PubMed] [Google Scholar]
- 40.Roussel, Y., A. Harris, M. H. Lee, and M. Wilks. 2007. Novel methods of quantitative real-time PCR data analysis in a murine Helicobacter pylori vaccine model. Vaccine 25:2919-2929. [DOI] [PubMed] [Google Scholar]
- 41.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 42.Sarkisova, S., M. A. Patrauchan, D. Berglund, D. E. Nivens, and M. J. Franklin. 2005. Calcium-induced virulence factors associated with the extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:4327-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarmientos, P., and M. Cashel. 1983. Carbon starvation and growth rate-dependent regulation of the Escherichia coli ribosomal RNA promoters: differential control of dual promoters. Proc. Natl. Acad. Sci. USA 80:7010-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savli, H., A. Karadenizli, F. Kolayli, S. Gundes, U. Ozbek, and H. Vahaboglu. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52:403-408. [DOI] [PubMed] [Google Scholar]
- 46.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264-1277. [DOI] [PubMed] [Google Scholar]
- 48.Simone, N. L., R. F. Bonner, J. W. Gillespie, M. R. Emmert-Buck, and L. A. Liotta. 1998. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 14:272-276. [DOI] [PubMed] [Google Scholar]
- 49.Simone, N. L., C. P. Paweletz, L. Charboneau, E. F. Petricoin III, and L. A. Liotta. 2000. Laser capture microdissection: beyond functional genomics to proteomics. Mol. Diagn. 5:301-307. [DOI] [PubMed] [Google Scholar]
- 50.Southey-Pillig, C. J., D. G. Davies, and K. Sauer. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:8114-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinberger, R. E., A. R. Allen, H. G. Hansma, and P. A. Holden. 2002. Elongation correlates with nutrient deprivation in Pseudomonas aeruginosa unsaturated biofilms. Microb. Ecol. 43:416-423. [DOI] [PubMed] [Google Scholar]
- 52.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart, P. S. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart, P. S., and M. J. Franklin. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199-210. [DOI] [PubMed] [Google Scholar]
- 55.Teal, T. K., D. P. Lies, B. J. Wold, and D. K. Newman. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl. Environ. Microbiol. 72:7324-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner, M., P. H. Nielsen, A. Loy, J. L. Nielsen, and H. Daims. 2006. Linking microbial community structure with function: fluorescence in situ hybridization-microautoradiography and isotope arrays. Curr. Opin. Biotechnol. 17:83-91. [DOI] [PubMed] [Google Scholar]
- 57.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waite, R. D., A. Paccanaro, A. Papakonstantinopoulou, J. M. Hurst, M. Saqi, E. Littler, and M. A. Curtis. 2006. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waite, R. D., A. Papakonstantinopoulou, E. Littler, and M. A. Curtis. 2005. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J. Bacteriol. 187:6571-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Werner, E., F. Roe, A. Bugnicourt, M. J. Franklin, A. Heydorn, S. Molin, B. Pitts, and P. S. Stewart. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 63.Xu, K. D., G. A. McFeters, and P. S. Stewart. 2000. Biofilm resistance to antimicrobial agents. Microbiology 146:547-549. [DOI] [PubMed] [Google Scholar]
- 64.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng, Z., and P. S. Stewart. 2002. Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 46:900-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]