Abstract
Numerous secondary metabolites have been isolated from the insect pathogenic fungus Metarhizium anisopliae, but the roles of these compounds as virulence factors in disease development are poorly understood. We targeted for disruption by Agrobacterium tumefaciens-mediated transformation a putative nonribosomal peptide synthetase (NPS) gene, MaNPS1. Four of six gene disruption mutants identified were examined further. Chemical analyses showed the presence of serinocyclins, cyclic heptapeptides, in the extracts of conidia of control strains, whereas the compounds were undetectable in ΔManps1 mutants treated identically or in other developmental stages, suggesting that MaNPS1 encodes a serinocyclin synthetase. Production of the cyclic depsipeptide destruxins, M. anisopliae metabolites also predicted to be synthesized by an NPS, was similar in ΔManps1 mutant and control strains, indicating that MaNPS1 does not contribute to destruxin biosynthesis. Surprisingly, a MaNPS1 fragment detected DNA polymorphisms that correlated with relative destruxin levels produced in vitro, and MaNPS1 was expressed concurrently with in vitro destruxin production. ΔManps1 mutants exhibited in vitro development and responses to external stresses comparable to control strains. No detectable differences in pathogenicity of the ΔManps1 mutants were observed in bioassays against beet armyworm and Colorado potato beetle in comparison to control strains. This is the first report of targeted disruption of a secondary metabolite gene in M. anisopliae, which revealed a novel cyclic peptide spore factor.
Metarhizium anisopliae is at the forefront of efforts to develop entomopathogenic fungi as insect biocontrol agents and has been used worldwide for invertebrate pest control as an alternative to chemical pesticides (16, 31, 67). The best-known use of Metarhizium has been for the control of locust and grasshopper plagues in Africa, Australia, and South America (51, 54). Secondary metabolite production by M. anisopliae is predicted to be critical for disease development in host insects (10, 66, 75, 81). However, the roles of secondary metabolites as virulence factors for insect pathogenic fungi are poorly understood due to the lack of molecular genetic studies unequivocally demonstrating their contributions to the disease process (13, 31, 42). Furthermore, it is unclear what roles they play in the biology of these organisms. They might confer antimicrobial activity for niche establishment, serve as signaling molecules that modulate fungal or insect development, or act as self-defense compounds that protect the fungus from invertebrate-derived, microbe-derived, or environmentally derived stresses (e.g., light, heat, desiccation, or reactive oxygen species). Directly or indirectly, secondary metabolites could accordingly serve as insect toxins.
Only a few families of peptide metabolites have been described from Metarhizium spp. The destruxin toxins, a large family of cyclic depsipeptides, are the most prevalent of the secondary metabolites produced by M. anisopliae in fermentation and, by far, the most exhaustively researched toxins of the entomopathogenic fungi (60). They are produced by several taxonomically distinct fungi and are thought to be involved in the disease processes of Alternaria brassicae and M. anisopliae on their respective plant and invertebrate hosts (42, 57, 60). Destruxins A, B, and E are the primary constituents reported from fermentation broths of M. anisopliae (56), and their synthesis is likely encoded by a nonribosomal peptide synthetase (NPS) (39). Bailey et al. (4) characterized an NPS gene from M. anisopliae called pesA (GenBank accession no. CAA61605) but concluded it probably did not encode a destruxin synthetase, based on the absence of methylation domains in the predicted protein (12).
Generally, in vitro production of destruxins by M. anisopliae is positively correlated with virulence of the fungus against its hosts, i.e., strains that produce relatively high levels of destruxin in vitro tend to exhibit greater virulence against insects, although exceptions to this trend exist (3, 20, 42, 69). Furthermore, in vitro assays utilizing purified destruxins have demonstrated a wide range of biological activities against insects, including immunosuppression, activation of calcium channels in muscle, antifeedant activity, and acute toxicity (28, 57, 60, 66, 67, 75, 81). M. anisopliae also produces the myroridin antibiotics (44), which are linear heptapeptides that show antifungal and antiviral activities, as well as cell growth inhibition (32).
Recently, we provided the first report of mutagen production by M. anisopliae, which secretes the compounds NG-391 and NG-393 into culture media of the fungus (46). These compounds are structurally related to the fusarins produced by Fusarium spp. (25), which are formed as hybrid polyketide-nonribosomal peptides (25, 64, 73). Like the fusarins, they exhibited potent S9-dependent mutagenic activity in the Salmonella mutagenicity test but were inactive in antimicrobial and mosquitocidal assays (46).
In addition to nonribosomal peptides and hybrid molecules containing them, Metarhizium spp. produce secondary metabolites in other chemical classes. These include the cytochalasins (1, 24), helvolic acid (19), swainsonine (33, 59), 12-hydroxyovalicin (47), viridoxins (29), fungerins (78), and metacytofilin (37). Although most of the known metabolites of Metarhizium exhibit biological activities in vitro against mammalian, insect, microbial, or plant cells, none has yet been demonstrated conclusively by genetic means to play an essential role in invertebrate disease. Thus, their roles in insect-fungus interactions, as well as in the biology of Metarhizium as a saprophyte, remain unknown (28, 67, 75).
We report here the first targeted disruption of a secondary metabolite gene in M. anisopliae by Agrobacterium tumefaciens-mediated transformation (ATMT), which facilitated the discovery of a new class of nonribosomal peptides called serinocyclins (45) that were detected in conidia of control strains but not in gene disruption mutants of MaNPS1 (ΔManps1). We also report the in vitro and in vivo characterization of the resulting ΔManps1 mutants. Moon et al. (55) previously described ATMT of M. anisopliae and early aspects of the present study.
MATERIALS AND METHODS
Strains and general growth conditions.
Sixteen Metarhizium strains were chosen from the Agricultural Research Service Collection of Entomopathogenic Fungi (ARSEF) in Ithaca, NY, for evaluation of destruxin production in vitro and insect virulence (Table 1) . For routine culturing, Metarhizium strains were grown in one-quarter-strength Sabouraud dextrose agar plus yeast extract (¼SDAY; 10 g of dextrose, 2.5 g of yeast extract, 2.5 g of Bacto peptone, and 15 g of Bacto agar per liter; 20-ml/9-cm diameter petri dish) at 25°C in the dark.
TABLE 1.
M. anisopliae isolates vary in virulence against second-instar BAW and in destruxin production in vitro
| ARSEF isolate | Metarhizium species | Host origin | Destruxins A and B (SEM)a | Relative levels of Dtxb | Mean time (days) until deathc | MaNPS1 RFLPd |
|---|---|---|---|---|---|---|
| 324 | M. anisopliae var. acridum | Orthoptera | 0.38 (0.20) | L | 5.4a | A |
| 703 | M. anisopliae | Lepidoptera | 0.66 (0.21) | L | 3.6bc | B |
| 1015 | M. anisopliae var. majus | Lepidoptera | 0.32 (0.32) | L | 4.1b | B |
| 2421 | M. anisopliae | Homoptera | 2.97 (0.21) | L | NT | B |
| 297 | M. anisopliae var. majus | Coleoptera | 1.03 (0.22) | L | NT | B |
| 2353 | Metarhizium sp. | Homoptera | 27.91 (0.94) | I | 4.1b | C |
| 1094 | M. anisopliae | Lepidoptera | 10.35 (1.72) | I | NT | C |
| 1080 | M. anisopliae | Lepidoptera | 12.26 (1.45) | I | 3.9b | C |
| 3335 | M. anisopliae var. anisopliae | Coleoptera | 10.13 (1.13) | I | NT | C |
| 808 | M. anisopliae | Coleoptera | 9.95 (0.41) | I | NT | C |
| 2162 | M. anisopliae var. anisopliae | Coleoptera | 40.75 (2.17) | H | NT | C |
| 2575 | M. anisopliae var. anisopliae | Coleoptera | 110.41 (13.38) | H | 3.4c | C |
| 1095 | M. anisopliae var. anisopliae | Lepidoptera | 83.54 (3.37) | H | 3.5c | B |
| 549 | M. anisopliae | Unknown | 59.13 (7.91) | H | NT | C |
| 2547 | M. anisopliae | Coleoptera | 95.35 (8.47) | H | NT | C |
| 23 | M. anisopliae var. anisopliae | Coleoptera | 160.34 (14.28) | H | 3.4c | C |
Estimates are expressed in mg/liter of broth at 14 days postinoculation for destruxins A and B.
Arbitrarily assigned categories of destruxin (Dtx) A and B production. L, low (0 to 5 mg/liter of broth); I, intermediate (>5 to 30 mg/liter of broth); H, high (>30 to 170 mg/liter of broth).
Dosage of ca. 10 spores/mm2. Means followed by the same letter are not significantly different as determined by analysis of variance (F7,347 = 16.0; P < 0.0001; Tukey's test, P < 0.05). NT, not tested.
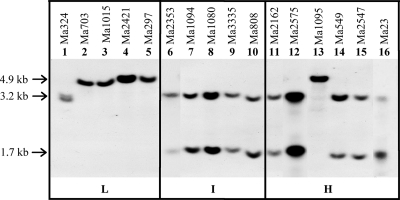
See Fig. 2. A, double band unique to ARSEF 324; B, single 4.9-kb band typical of most low destruxin producers; C, two bands at 3.2 and 1.7 kb typical of intermediate to high destruxin producers. ARSEF 1095 was an exception to this trend.
For synchronous production of conidia, spore suspensions were spread uniformly across the surface of one-quarter-strength Sabouraud starch agar plus yeast extract (¼SSAY) plates prepared identically as for ¼SDAY but with corn starch (no. S9679; Sigma-Aldrich Co., St. Louis, MO) replacing dextrose; cultures were incubated for at least 1 week at 25°C in the dark. To limit mycelium production, plates were incubated without Parafilm in a sealed plastic container, which allowed sufficient airflow to prevent moisture build-up inside the petri dish lids. Alternatively, conidia were produced under the same conditions on barley medium (7.5 g of organic barley flour per liter and 1.5% Bacto agar) with incubation for at least 12 days. Submerged blastospore-like propagules (21) were obtained in half-strength SDY supplemented with 50 mM citrate-phosphate buffer (pH 6.5) and inoculated with 107 spores/100 ml of medium in a 250-ml flask. Flasks were incubated at 27°C for 3 days in the dark with shaking (150 rpm). For the production of destruxins, conidia (106) were transferred to 100 ml of Difco or HiMedia Czapek-Dox broth (CDB; Becton Dickinson [BD], Franklin Lakes, NJ, or VWR, West Chester, PA, respectively) with 0.5% BD Bacto peptone (CDBP) in 250-ml Erlenmeyer flasks and grown at 25°C with shaking (150 rpm) for up to 2 weeks.
E. coli strain JM109 was used for the construction and amplification of the binary vector carrying the MaNPS1 gene disruption construct (pBD1-ma267KO) (Fig. 1). E. coli was grown at 37°C in Luria-Bertani (LB) broth or on LB agar with kanamycin (75 μg/ml). A. tumefaciens strain GV3101 was used for transformation of M. anisopliae and was grown at 29°C in yeast extract and peptone medium (YEP) (11) with gentamicin (25 μg/ml). All restriction endonucleases were purchased from New England Biolabs (Ipswich, MA), and the pGEM-T Easy vector and T4 DNA ligase were purchased from Promega (Madison, WI).
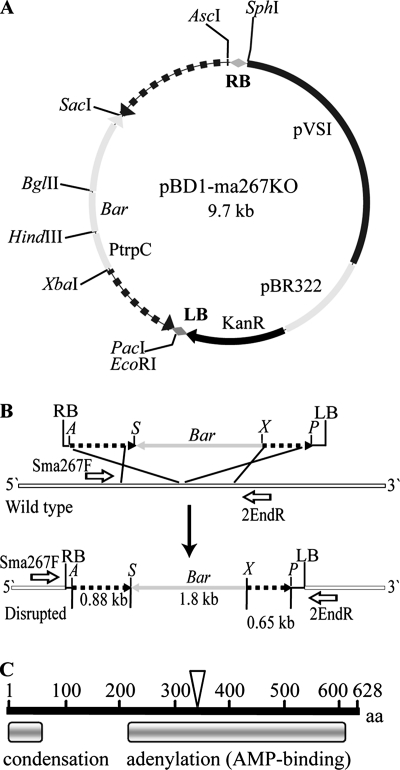
FIG. 1.
Strategy for targeted disruption of MaNPS1. (A) Binary vector pBD1-ma267KO, which encodes the Bar gene conferring resistance to GA whose expression is driven by the TrpC promoter (PtrpC) of Aspergillus nidulans. Hashed lines on either side of the PtrpC-Bar gene are the flanking regions of clone Ma#267 of MaNPS1. (B) Schematic of a homologous recombination event between the Ma#267 gene disruption construct and the wild-type MaNPS1 gene, which results in the insertion of a functional copy of the Bar gene into the MaNPS1 gene. Sma267F and 2EndR are forward and reverse primers, respectively, that hybridize to wild-type MaNPS1 DNA flanking the insertion point of the disruption construct. These primers were used to screen Bar-resistant transformants for a targeted gene disruption event. (C) Schematic of the conserved domains of the predicted protein of clone Ma#267 of MaNPS1, which contains part of a condensation domain and a full-length adenylation domain of an elongation module. The inverted triangle represents the approximate site at which the MaNPS1 gene was disrupted with the Bar gene.
Nucleic acid isolations.
To prepare genomic DNA for Southern blot analysis, M. anisopliae strains were grown as for destruxin production above in CDBP for 8 days. Mycelia were harvested, rinsed thoroughly with sterile distilled water, and lyophilized. Genomic DNA was isolated by using modifications of the protocol from the Wizard genomic DNA purification kit (Promega) as described previously (86), except that the lysis buffer was 150 mM EDTA, 50 mM Tris-Cl (pH 8.0), with 1% Sarkosyl (49). In addition, the protocol was modified as follows. Lyophilized mycelium (0.7 to 1.0 ml) was ground to a fine powder without the use of glass beads; each tube was vortex mixed for 30 to 60 s after the addition of lysis buffer. The 30-min incubation step at 65°C was not included, and after the isopropanol precipitation step the DNA was pelleted with a 30- to 60-s spin at 12,000 × g instead of a 10-min spin. After the suspension of DNA in 1× TE (1 M Tris-HCl [pH 8] and 0.5 M EDTA [pH 8]) as described by Wu et al. (86), the DNA was treated with proteinase K at a final concentration of 200 μg/ml for 1 h at 37°C, followed by heat inactivation at 65°C for 5 min. The DNA was phenol-chloroform extracted once, chloroform extracted once, and then ethanol precipitated with a 0.1 volume of 3 M sodium acetate and 2 volumes of cold 100% ethanol (68). Finally, the DNA was suspended in 1× TE and treated with a 1/100 dilution of 10 mg of RNase A (Fermentas, Inc., Hanover, MD)/ml for 30 min at 37°C.
To screen transformants by PCR for disruption of the MaNPS1 gene, genomic DNA was extracted according to the small scale (miniprep) DNA preparation method of Irelan et al. (38) but modified as follows. A small piece of mycelium was transferred to 500 μl of 1× SDY with 1% Bacto peptone broth in a 1.5-ml Eppendorf tube and grown for 2 days at 25°C with shaking (150 rpm). The culture was transferred to a 2-ml screw-cap microcentrifuge vial, the mycelium was pelleted, and the supernatant was removed, followed by the addition of approximately 150 μl of sterile white quartz sand (Sigma no. S-9887) and 500 μl of isolation buffer (38). The mycelia were disrupted with a Mini-BeadBeater (Biospec Products, Inc., Bartlesville, OK) for 10 s at setting 5 (4,800 oscillations/min) and then incubated at 65°C for 10 min. After treatment with 7.5 M ammonium acetate as described previously (38), the tubes were spun for 10 min at 16,000 × g, and 700 μl of supernatant was transferred to a fresh tube. DNA was precipitated with isopropanol as described previously (38), pelleted by centrifugation as before, washed briefly with 75% ethanol, pelleted, air dried briefly, and resuspended in 300 μl of 1× TE. The DNA was treated with 3 μl of RNase A (10-mg/ml stock) for 1 h at 37°C, extracted with an equivalent volume of saturated phenol-chloroform-isoamyl alcohol (25:24:1), precipitated by using standard methods (68), pelleted and washed with 75% ethanol, and resuspended in 50 μl of 1× TE buffer after air drying briefly.
Total RNA for cDNA preparation and reverse transcription-PCR (RT-PCR) was isolated from lyophilized mycelia of 3- and 6-day-old cultures grown as described above for large-scale DNA isolation in CDBP. Lyophilized mycelium was ground with a mortar and pestle in liquid N2, and 50 mg of material was treated with 1 ml of TRIzol reagent (Invitrogen, Carlsbad, CA). Phase separation was obtained by adding a 0.2 volume of chloroform, followed by gentle inversion of the tube, while RNA precipitation was achieved by using a 0.5 volume of isopropanol plus a 0.5 volume of high-salt precipitation solution (Molecular Research Center, Inc., Cincinnati, OH). RNA to be used for RT-PCR was suspended in 53 μl of diethyl pyrocarbonate (DEPC)-treated water plus 6 μl of 10× buffer (Promega) and treated with 1 μl of RNase-free DNase (Promega) for 30 min at 37°C. RNA was purified again with TRIzol as described above but starting with 0.5 ml of TRIzol solution and followed finally by suspension in 30 μl of DEPC-treated water.
Preparation of cDNA and amplification conditions for RT-PCR.
First-strand cDNA was synthesized by using random hexamer primers, 1 μg of total RNA per sample, and 2 μl each of RNaseOUT (Invitrogen) and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's protocol. First-strand cDNA (2 μl) from each time point, and the fungus strain assayed was used as a template for PCRs using the primer pair DesF (5′-GTCCGGACGCATTTACACGAA-3′) and DesR (5′-GACAGCCAGCTTAGAAACA-3′) to detect the expression of MaNPS1. Each reaction contained final concentrations of 1 U of Taq polymerase (Promega), 1× reaction buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.5 μM concentrations of each primer, and 2 μl of cDNA in a total vol of 25 μl. The thermocycler parameters used were 94°C for 2 min (initial denaturation), followed by 35 cycles of 94°C for 30 s (denaturation), 55°C for 1 min (primer annealing), and 72°C for 1 min (primer extension), with a final extension at 72°C for 5 min. The β-tubulin gene was used as a constitutive internal control to confirm the presence of high-quality cDNA that was not contaminated with genomic DNA (primers β-tub 40F, 5′-CACCTTCAGACCGGTCAGTG-3′; β-tub 750R, 5′-GAAGCGGCCATCATGTTCTTAG-3′). Total RNA (50 ng) was used as a template in each experiment to confirm the removal of genomic DNA before cDNA preparation. ARSEF 2575 genomic DNA was used as a template with β-tubulin primers as a positive control, and for comparison of this intron-containing PCR product with the smaller product derived from cDNA.
Construction of the Ma#267 gene replacement cassette.
Clone Ma#267 is an ∼1.9-kb cDNA fragment kindly provided by S. Screen and R. St. Leger (University of Maryland, College Park) that includes the sequence of expressed sequence tag (EST) AJ272930 (623 bp), which was isolated after 24 h of growth of M. anisopliae ARSEF 2575 on cockroach cuticle-containing minimal medium (23). Clone Ma#267 contains part of an NPS condensation domain at the 5′ end of the sequence and a complete NPS adenylation (AMP-binding) domain downstream (Fig. 1C). A gene disruption cassette was constructed so that placement of the Bar gene disrupted the adenylation domain (Fig. 1). The first portion of the gene (fragment F1, 0.88 kbp) was amplified from clone Ma#267 with the primers 1PacIF (5′-TTAATTAAGAGACAGTTGCCTTTGGC-3′) and 1XbaIR (5′-TCTAGAGCAGAGGATTGAAAGTCA-3′) by PCR. A second fragment of clone Ma#267 (fragment F2, 0.65 kbp) was amplified with the primers 2SacIF (5′-GAGCTCGAGGAAGATTGTCATTTC-3′) and 2AscIR (5′-GGCGCGCCTATTGCGCCAATAGGCAT-3′). PacI, XbaI, SacI, and AscI restriction sites were added to the respective primers (underlined nucleotides). Clone Ma#267 template DNA was amplified by using the following thermocycler parameters: 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 57°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 5 min, with approximately 20 ng of DNA and the same components and concentrations as described above for the RT-PCRs. PCR-amplified F1 and F2 fragments were cloned into the pGEM-T Easy vector according to the manufacturer's recommendations (Promega) and then double-digested with PacI/XbaI or SacI/AscI to give sticky-ended fragments. These fragments were ligated with the 1.8-kb Bar gene expression cassette obtained from double digestion of pBARKS1 (53, 58; Fungal Genetics Stock Center, Kansas City, MO) with XbaI/SacI. The full 3.3-kb fragment was cloned into pUCAP (80) by insertion into the AscI and PacI restriction sites of the multiple cloning site and designated pUCAP-ma267KO. This vector was amplified in JM109, and the AscI/PacI fragment was then cloned into the AscI/PacI sites of binary vector pBD1 (B. G. G. Donzelli and A. C. L. Churchill, unpublished results), a derivative of the binary vector pPK2 (14), to give the vector pBD1-ma267KO (Fig. 1A).
ATMT.
MaNPS1 gene disruption vector pBD1-ma267KO was electroporated into A. tumefaciens GV3101 with a Gene Pulser II (Bio-Rad, Hercules, CA) using standard settings recommended by the manufacturer. ATMT was performed by using a modified method of Covert et al. (14). pBD1-ma267KO-transfected GV3101 cells were grown in 50 ml of YEP medium with kanamycin (50 μg/ml) and gentamicin (25 μg/ml) overnight at 29°C until reaching an optical density at 600 nm (OD600) of 0.6 to 0.8. Cells were washed twice with 4°C sterile water and then diluted with induction medium (7) containing 200 μM acetosyringone (3′,5′-dimethoxy-4′-hydroxyacetophenone; Aldrich, Milwaukee, WI) to an OD600 of 0.15 and grown at 29°C until reaching an OD600 of 0.5. ARSEF 2575 conidia (2 to 3 weeks old) were harvested from ¼SDAY in sterile water with 0.05% Silwet l-77 (Loveland Industries, Inc., Greeley, CO), filtered through either sterile nylon membrane (20-μm pore size) or three layers of sterile Kimwipes held in a funnel to remove hyphae, counted by using a hemacytometer, and used for ATMT.
pBD1-ma267KO-transfected GV3101 cells were mixed 1:1 with M. anisopliae spores (108 spores/ml), and 200 μl of this mixture was spread on the surface of sterile black filter paper (Thomas Scientific, Swedesboro, NJ), which had been overlaid onto each induction medium plate with 200 μM acetosyringone. Plates were incubated for 2 days in the dark at 25°C. Transformant selection was achieved by transferring each filter paper onto plates containing M-100 medium (74) amended with 6.9 ml of Finale (AgrEvo Environmental Health, Montvale, NJ)/liter, which corresponds to 400 μg of the active ingredient, glufosinate ammonium (GA), per ml. The medium also contained 300 μg of either carbenicillin (Research Products International, Mount Prospect, IL) or cefotaxime (Research Products International)/ml to kill bacteria. Each filter paper was then overlaid with 10 ml of M-100 medium containing the same antibiotics as in the basal medium and 0.9% agar. Plates were incubated for 7 to 10 days in the dark at 25°C until the transformants had grown to the surface of the top agar. Each primary transformant was transferred to a single well of a 24-well microtiter plate containing Aspergillus minimal medium (ATCC medium 687, Pontecorvo's Aspergillus medium), which was modified to contain 4.4 g of monobasic ammonium phosphate/liter in place of 6 g of sodium nitrate/liter (referred to here as modified Aspergillus minimal medium[mAMM]). mAMM was amended with 3.45 ml of Finale/liter (200 μg of GA/ml), and the cultures were incubated in the dark at 25°C.
Screening primary transformants for disruption of the MaNPS1 gene and mitotic stability.
Miniprep genomic DNA (prepared as described above) was screened by PCR to detect a homologous recombination event in which the gene disruption construct containing the Bar cassette was inserted into the wild-type copy of the gene. DNA was amplified with the primers Sma267F (5′-CCATTGTGCTTCAATGCT-3′) and 2EndR (5′-CCGGAGGTAAATAACGTCGA-3′), which anneal outside of the two flanking regions used to make the disruption construct of pBD1-ma267KO (Fig. 1B). PCR amplification was performed with a blend of DNA polymerases in the ratio of 0.2 U of Platinum Pfx DNA polymerase (Invitrogen) and 5 U of Taq DNA polymerase (Promega) (52) in the reaction mix. Miniprep genomic DNA was diluted 10-fold, and 1 μl (approximately 30 ng) was used as a template in reactions containing 0.1 μl of the Pfx/Taq polymerase mix and final concentrations of 1× Pfx amplification buffer, 0.2 mM dNTPs, 1 mM MgSO4, and 0.5 μM concentrations of each primer in a final volume of 25 μl. The following thermocycler parameters were used: 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 62°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 5 min. An amplicon of 1.9 kb is expected when an intact copy of the MaNPS1 gene is present in transformants with an ectopic integration event or from wild-type ARSEF 2575. In the case of a homologous recombination event at the Ma#267 locus, an amplicon of 3.7 kb is expected from the ΔManps1 mutants since this event would result in an amplicon that is larger in size than the intact, wild-type gene fragment by 1.8 kb, the size of the Bar cassette (Fig. 1B). Primary transformants could potentially exhibit both a wild-type fragment (1.9 kb) and a disrupted MaNPS1 gene (3.7 kb) before homokaryon purification by isolation of colonies derived from single, mononucleate spores.
To select monoconidial isolates, spores from six putative ΔManps1 mutants and one ectopic transformant identified in the PCR screen were diluted and transferred to Difco CDB with 1.5% agar (CDA; BD) containing 3.45 ml of Finale/liter (200 μg of GA/ml). A representative single spore-derived colony from each transformant was determined mitotically stable by demonstrating growth on GA-containing medium after three weekly serial transfers in the absence of selection.
Southern blot analyses.
M. anisopliae isolates were screened for potential restriction fragment length polymorphisms (RFLPs) after digestion of genomic DNA with restriction enzyme, Southern blotting, and hybridization with individual Metarhizium NPS fragments. Analyses were repeated independently at least twice for each isolate. Genomic DNA (10 μg) of 16 M. anisopliae isolates (Table 1) was digested with BamHI, which digests clone Ma#267 (1.897 kb) of MaNPS1 into two fragments, and separated by gel electrophoresis (1% agarose, 1× TAE buffer [40 mM Tris-acetate, 2 mM Na2EDTA-2H2O; pH 8.5). Digested DNA was transferred to positively charged nylon membrane by capillary transfer as described in the digoxigenin (DIG) application manual for filter hybridization (Roche Molecular Biochemicals, Indianapolis, IN) and independently hybridized with each of five distinct NPS gene fragments from Metarhizium spp., denoted Ma#267, ma1095-2.1, ma1095-3.3, ma1015-2KA2, and ma2082-2040 (unpublished data). Each DNA fragment, cloned into a pGEM-T Easy vector, was PCR labeled using the PCR DIG probe synthesis kit (Roche Molecular Biochemicals) and gene-specific primers (Sma267F and 2EndR for clone Ma#267; unpublished data for other clones).
Prehybridization and high-stringency hybridization conditions were as described in Roche's DIG application manual with the following exceptions. Prehybridization in DIG Easy Hyb solution was for at least 2 h; up to 25 μl of denatured PCR-labeled probe (depending on the relative labeling efficiency) was added to 2.5 ml of DIG Easy Hyb solution and incubated with the membrane overnight at 45°C in a hybridization oven. The Roche protocol for washing blots after hybridization was modified by washing in low-stringency buffer at room temperature for 10 min three times, followed by two washes in high-stringency buffer (0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate) at 68°C for 30 min. Hybridization probes were detected using CDP-Star as the chemiluminescent substrate and the recommended Roche protocol with the following exceptions. The first incubation in blocking solution was for 2 h and then the membrane was incubated in 10 ml of 1× blocking solution containing 0.5 μl of anti-DIG-AP antibody solution for 30 min, followed by three 10-min rinses in washing buffer and detection with CDP-Star using the transparency technique. Each probe was hybridized to the identical blot that had been stripped of the previous probe using Roche's recommended stripping protocol.
To characterize the ΔManps1 mutants and control strains for Bar gene location and copy number, genomic DNA (10 μg) was digested with XbaI, SacI, BamHI, or SacI/BamHI; separated by gel electrophoresis (0.8% agarose, 1× TAE buffer); and transferred to positively charged nylon membrane by capillary transfer as previously described. The blot was hybridized sequentially with either digoxigenin-labeled Ma#267 or a Bar gene fragment as described above.
Insect bioassays.
Eight Metarhizium isolates were chosen for analysis in insect bioassays to evaluate the range of virulence levels present in this subset (Table 1). Because of their varied host and culture history, all isolates were first passed through beet armyworm (BAW), Spodoptera exigua, and reisolated from cadavers before initiating virulence bioassays. BAW eggs were obtained from the U.S. Department of Agriculture-Agricultural Research Service (USDA-ARS) Southern Insect Management Unit, Stoneville, MS. For the narrow-host-range, orthopteran isolate ARSEF 324, initial infection was accomplished only by inoculating larvae with a very high dose of spores. Single-spore isolates of all BAW-derived strains, stored in 10% glycerol at −80°C, were used in all further analyses.
To screen ARSEF isolates for virulence, BAW were inoculated as second instars using an enclosed Plexiglas spray tower with a calibrated nozzle (79) at a dosage of 10 spores mm−2, including treatment with the suspending medium (0.04% Silwet l-77) as a negative control. One dish of 1% yeast extract agar was included inside the spray tower during inoculation to quantify spore germination and estimate dosage. Larvae were monitored daily for 7 days for signs and symptoms of disease and mortality, and dead larvae were removed from dishes, surface disinfected, and incubated on sterile 1.5% water agar to confirm mycosis. Average survival times and percentage mortality among fungus-inoculated S. exigua larvae were determined for the range of isolates assayed.
To screen ΔManps1 mutants and control strains for virulence against BAW, fungal isolates were cultured on ¼SSAY (9-cm diameter petri dishes) for 10 days at 25°C with a 15-h light/9-h dark photoperiod. Plates were uncovered and dried for 8 h at 20% relative humidity within a laminar flow cabinet and then covered, sealed with Parafilm, and stored at −20°C for 1 to 21 days before use in BAW bioassays. Spores were flooded into suspension on the culture surface using 0.01% Tween 80. Suspensions were transferred to sterile 50-ml screw-cap centrifuge tubes and shaken for 15 min on a wrist-action shaker. Concentrations were adjusted by using a hemacytometer to 3.5 × 107 conidia/ml, using an additional 0.01% Tween 80 as a diluent, to a total volume of 15 ml. Diluent alone was used to inoculate controls.
BAW were obtained as eggs from Benzon Research (Carlisle, PA). Newly hatched larvae were reared on diet containing chlortetracycline, methyl paraben, and potassium sorbate (product no. F9220B; Bio-Serv, Frenchtown, NJ) at 25°C and for a 15-h/9-h light/dark cycle until reaching the second instar. Second instars were dipped in spore suspensions (or diluent for controls) three times in quick succession and placed individually on a 1-cm3 cube of BAW diet containing chlortetracycline as the only antibiotic (Southland Products, Inc., Lake Village, AR) in a 3.5-cm-diameter petri dish. Methyl paraben and potassium sorbate were excluded from the postinoculation diet since they inhibit growth of Metarhizium. Larval mortality was tabulated daily for 7 days at 25°C with a 15-h/9-h light/dark cycle, changing the diet daily. Aliquots of each spore suspension used for inoculations were spread on one-quarter-strength SDA and incubated for 20 h before scoring at least 100 spores/isolate for germination.
Six assays were conducted using four isolates in each assay. Wild-type and ectopic strains were included in each of the six assays. Two of three ΔManps1 mutants were selected randomly for each assay so that each isolate was tested in four assays. Sixty second instars were inoculated with each isolate in each assay. Larvae dying within 24 h of inoculation were discarded, resulting in 47 to 60 viable larvae per isolate per assay. One-way analyses of variance were done to test for strain differences in arcsin square root-transformed percentage mortality and germination. Pearson correlation analysis was conducted between transformed percentage mortality and arcsine square root-transformed percentage germination.
To screen ΔManps1 mutants and control strains for virulence against Colorado potato beetle (CPB; Leptotarsa decemlineata), insect eggs were obtained from the USDA Phillip Alampi Beneficial Insect Laboratory (West Trenton, NJ) and hatched on potato foliage in a 24°C incubator with a 12-h/12-h light/dark cycle. Insects were inoculated as third instars, using an enclosed Plexiglas spray tower with a calibrated nozzle (79), at four spore dosages (range, ca. 100 to 1,500 spores/mm2, 12-day-old fresh conidia from ¼SDAY) and with a negative control (i.e., the suspending medium, 0.04% Silwet l-77). One dish of 1% yeast extract agar was included to quantify spore germination and estimate dosage. After spraying, each insect was transferred to a sterile petri dish (60 by 15 mm2) lined with moist filter paper and containing potato foliage. Each treatment consisted of two replicates of 30 larvae each, and the experiment was independently replicated at least twice. Larvae were monitored daily for up to 7 days for signs of disease and mortality; dead larvae were removed and checked as described above for confirmation of mycosis.
Mitotic stability of ΔManps1 mutants isolated from infected insects.
Second-instar BAW larvae were inoculated and maintained as described above. Dead, infected larvae that had begun to sporulate were collected and transferred to water agar containing a cocktail of antibiotics (25 μg of gentamicin/ml, 100 μg of mefoxin/ml, 100 μg of streptomycin/ml, 100 μg of carbenicillin/ml, 100 μg of chloramphenicol/ml, 30 μg of hygromycin B/ml) to inhibit the growth of microbes other than M. anisopliae, which was allowed to grow for approximately 1 week before transfer to ¼SSAY without Bar selection. A small amount of mycelium was scraped from each culture on ¼SSAY, and DNA was extracted by using 100 μl of DNAzol (Molecular Research Center, Inc.) according to the manufacturer's instructions. DNA (1 μl) was used as a template for PCR using the primer pair DesF and DesR (see above), which differentiates between ΔManps1 mutants (single band, 0.6 kb), the Ect A-18 strain (two PCR products, 0.6 and 0.7 kb), and ARSEF 2575 (single band, 0.7 kb). The ∼0.7-kb band represents the genomic DNA copy of the gene, whereas the ∼0.6-kb band represents the cDNA copy of the transgene at the site of MaNPS1 gene disruption in the mutants or the vector integration site in the ectopic transformant. Reactions contained 0.5 U of Taq polymerase mix, 1× Taq amplification buffer, 0.2 mM dNTPs, 2 mM MgCl2, and 0.5 μM concentrations of each primer in a final volume of 15 μl. The thermocycler parameters used were 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, with a final extension at 72°C for 10 min.
High-pressure liquid chromatography (HPLC) analyses of extracts of broth and mycelia of M. anisopliae.
For quantitative analysis of destruxins from broth and mycelia, 100-ml aliquots of CDBP were inoculated with 1 ml of 106 spores/ml in triplicate in 250-ml Erlenmeyer flasks and grown for up to 14 days at 160 rpm at 25°C ± 2°C with ambient light. Mycelia were harvested from broth by vacuum filtration through two sheets of Whatman no.1 filter paper in a Buchner funnel, and each mycelial mat was oven dried for 48 h to obtain the dry weight.
For initial comparisons of destruxin production in vitro by Metarhizium isolates and for gene expression analyses, broth from fungal cultures (10 ml) was passed through a preconditioned C18 EV SPE column (200 mg; no. 405150; Alltech Associates, Inc., Deerfield, IL), rinsed with water, eluted with 2 ml of methanol, and dried. For destruxin analyses of ΔManps1 mutants and control strains, the entire broth fraction was extracted twice with a 0.5 volume of dichloromethane, and then the organic layers were combined and dried over anhydrous Na2SO4, and the solvent was removed in vacuo. Mycelial mats were blended in approximately 100 ml of ethanol and stirred for 24 h at 25°C ± 2°C in ambient light. The ethanolic extract was collected via filtration through two sheets of Whatman no. 1 paper and concentrated to dryness in vacuo, and the residue was partitioned between dichloromethane and water. The organic layer was dried over anhydrous Na2SO4 and concentrated in vacuo. The mycelial mats were oven dried for 48 h to obtain the dry weights. All extracts were dissolved in methanol before HPLC analysis.
For HPLC analyses comparing destruxin production by Metarhizium spp. and during MaNPS1 gene expression, destruxins A and B were estimated by using a 10-point calibration curve and integrated areas for each peak in extract chromatograms that matched the retention time of authentic destruxin A (Sigma-Aldrich Co.) and destruxin B (HPLC purified). HPLC used a reversed-phase C18-A column (Varian 5 μm, 250 by 4.6 mm) with a premixed isocratic mobile phase (acetonitrile-water, 50:50), a flow rate of 1 ml/min, primary detection by UV at 220 nm, and a wavelength scan of 200 to 350 nm. The conditions for HPLC analysis of destruxin production in Δmanps1 mutants and control strains were as described by Krasnoff et al. (46) except that detection was at 215 nm. The limits of detection for destruxins A and B were 5.9 and 8.8 μg, respectively. The limits of quantification for destruxins A and B were 3.0 and 5.7 μg, respectively.
ESIMS analyses of conidial extracts.
Cultures were grown on barley medium in 9-cm petri dishes for 8 days at 25°C. Conidia were harvested by scraping the surface of cultures with a sterile loop in 10 ml of water, followed by a second rinse of the culture with 5 ml of water. The combined conidial suspension (∼15 ml) was filtered through cheesecloth and pelleted by centrifugation at 600 × g for 6 min. After removal of the supernatant, the pellet was resuspended in 1 ml of MeOH by vortex mixing and then sonicated for 5 min. The suspension was then filtered through a 0.2-μm-pore-size syringe filter and dried under N2. Samples were diluted in MeOH to a concentration of 0.1 mg/ml and infused by a syringe pump (Harvard apparatus) at 5 μl/min into a Micromass ZMD-4000 spectrometer for electrospray ionization mass spectrometric (ESIMS) analysis in positive-ion mode using capillary and cone voltages of 3.25 kV and 50 V, respectively. The data were acquired by scanning four times through the range m/z 2 to 1,200 at 400 amu/s (3.0 s/scan) with a 0.1-s interscan delay. Serinocyclin A, purified as described previously (45), was used as a calibration standard. Standard curves were generated, using 0.01-, 0.1-, 1.0-, 5.0-, and 10.0-ng/ml solutions in methanol, by summing the ion counts of the pseudomolecular ions [M+H]+, [M+Na]+, and [M+K]+ of serinocyclin A at m/z 673, 695, and 711, respectively. Detector response measured in this manner was linear through the range from 0.1 to 10 ng/μl. The limit of quantification was established at 0.1 ng/μl (signal-to-noise ratio, 5). The limit of detection for the method was established at 0.06 ng/μl (signal-to-noise ratio, 3).
In vitro phenotype assays of ΔManps1 mutants.
Methodologies for assessing asexual sporulation, colony hydrophobicity, radial growth with or without hydrogen peroxide (H2O2) treatment, germination of fresh or aged conidia on agar medium or one supplemented with H2O2 or KCl, conidial germination on cockroach wings, conidial adhesion, and conidial surface morphology as evaluated by scanning electron microscopy are described in the supplemental material.
RESULTS
Identification of an M. anisopliae target isolate for MaNPS1 gene disruption.
A representative selection of isolates from the ARSEF collection was screened for virulence against BAW and in vitro destruxin production to identify a single strain to be used for MaNPS1 gene disruption and further experimentation. All eight isolates assayed caused mycosis among second-instar BAW larvae (Table 1). Generally, and as reported by others in the literature (3, 42, 69), isolates that produced relatively high levels of destruxin in vitro were more virulent (i.e., killed insects faster) than those that produced relatively low levels of destruxin in culture. ARSEF 703 was an exception to this trend in that it produced low destruxin titers in vitro but was highly virulent against BAW. ARSEF 324, which is an M. anisopliae var. acridum isolate with a narrow host range generally limited to orthoptera, was the least virulent of the strains assayed and killed larvae most slowly, as well as producing low levels of destruxins in vitro. It is notable that lepidopteran isolates of M. anisopliae exhibited a range of virulence levels against the lepidopteran S. exigua and were not always the most virulent. In fact, two of the four most virulent isolates against BAW (ARSEF 2575 and ARSEF 23) were originally isolated from coleopteran hosts. Ultimately, ARSEF 2575 (also referred to here as Ma2575; formerly known as ME1) was chosen as the primary isolate for further study because of its high virulence against BAW and relatively high levels of destruxin production in vitro (Table 1). In addition, this strain is a broad-host-range entomopathogen, one of the most commonly referenced M. anisopliae strains in the literature, and has been utilized for a wide range of molecular biology studies.
Sequence analysis of clone Ma#267 of MaNPS1.
Clone Ma#267 (1.897 kb) is a cDNA fragment of a predicted NPS gene identified initially as AJ272930 (623 bp) from an EST library of M. anisopliae ARSEF 2575 grown for 24 h in a minimal medium containing 1% cockroach cuticle after a 30 h preculture period in a rich Sabouraud-glucose broth with 0.5% yeast extract (23). Using the NCBI Basic Local Alignment Search Tool (BLAST) (2) and the discontinuous Megablast or Blastn program against the nonredundant database, clone Ma#267 showed greatest identities over a 311-nucleotide (nt) region (69%, 8e-24) and over an 858-nt region (64%, 6e-38) within the locus XM_001227983, which encodes hypothetical protein CHGG_10057 from the fungus Chaetomium globosum, a saprophyte and human nail and skin pathogen (analyses were conducted im May 2008 [data not shown]). Protein CHGG_10057 has the highest similarity to the predicted protein of clone Ma#267 (Table 2) and contains several domains typical of NPSs, including seven predicted AMP-binding domains. Ma#267 showed the greatest similarity to the first predicted amino acid activation domain of the predicted C. globosum protein, with an E-value of 3e-141 and identities and positive amino acids over an 873-nt region of 59 and 75%, respectively. Identities of 25 to 65% were seen over other regions of the protein. From genome sequence data, the CHGG_10057 protein is encoded by a gene and mRNA of 27,672 nt. The C. globosum gene is associated with other predicted genes in an apparent gene cluster. However, relatively few of the associated genes have been annotated as containing domains common to secondary metabolite biosynthetic genes. Blastx analyses of clone Ma#267 at the Joint Genome Institute eukaryotic genomics site (http://genome.jgi-psf.org/euk_home.html) revealed high similarity (E-value of 1e-120) to a predicted NPS gene (fgenesh1_pg.C_scaffold_19000036) with six adenylation domains on scaffold 19 of the genome of the mycoparasite Trichoderma virens. The T. virens hypothetical protein was highly similar to the predicted protein CIMG_09750 of the human pathogen Coccidioides immitis, which has the second highest similarity to the predicted protein of clone Ma#267 of MaNPS1 (Table 2). The T. virens protein also showed similarity to the majority of other proteins in Table 2, except C. globosum CHGG_10057. Support Vector Machine-based software (63) was unable to predict the specificity of the adenylation domain of clone Ma#267 of M. anisopliae MaNPS1, which has GenBank accession no. EU275151.
TABLE 2.
Top 11 NCBI sequences producing significant Blastx alignments with clone Ma#267 of MaNPS1
| Accession no. | Sequence description | E value | Gene (reference)a |
|---|---|---|---|
| XP_001227984.1 | Hypothetical protein CHGG_10057 (Chaetomium globosum) | 3e-141 | |
| XP_001240129.1 | Hypothetical protein CIMG_09750 (Coccidioides immitis) | 4e-108 | |
| BAE64605.1 | Unnamed protein product (Aspergillus oryzae) | 2e-104 | |
| XP_390720.1 | Hypothetical protein FG10544.1 (Gibberella zeae) | 5e-104 | NPS18 (77) |
| XP_001213501.1 | Predicted protein (Aspergillus terreus) | 6e-102 | |
| XP_001222884.1 | Hypothetical protein CHGG_06789 (Chaetomium globosum) | 1e-98 | |
| XP_753380.1 | Nonribosomal peptide synthase, putative (Aspergillus fumigatus) | 8e-98 | NPS8 (15) |
| XP_001267502.1 | Nonribosomal peptide synthase, putative (Neosartorya fischeri) | 1e-96 | |
| EDP52010.1 | Nonribosomal peptide synthase, putative (Aspergillus fumigatus) | 3e-96 | |
| XP_382491.1 | Hypothetical protein FG02315.1 (Gibberella zeae) | 2e-95 | NPS4 (77) |
| AAP78735.1 | Nonribosomal peptide synthase (Alternaria brassicae) | 1e-93 | AbNPS2 homologue (43) |
References are indicated in cases where gene expression data could be found or inferred for the respective gene.
Using Blastn against the GenBank EST database, a 59-nt region of clone Ma#267 showed 76% identity to three ESTs (GenBank accession numbers CO035825, CO032944, and CO021837) from both saprobic and pathogenic spherule cDNA libraries of Coccidioides posadasii (17, 41). These ESTs form a contig of 1,558 nt that is 98% identical over the same length to the mRNA of GenBank accession no. XM_001240128, which encodes hypothetical protein CIMG_09750 from the closely related fungus C. immitis (Table 2). Further Blastx analyses of the C. posadasii EST sequences, as well as other fungal ESTs with short regions (32 to 35 nt) of high similarity to Ma#267 (Corynascus heterothallicus EB062651; Neurospora crassa AI399499; and Ophiostoma piliferum EB044438) suggested that these genes likely encode NPSs or other proteins containing AMP-binding domains that are distinct from MaNPS1 and the C. globosum CHGG_10057 predicted protein.
Although clone Ma#267 showed significant similarity to many proteins predicted to encode NPSs, most have not been characterized functionally and are not linked to known natural products. It is notable that of the proteins with greatest similarity to the M. anisopliae MaNPS1 sequence, most were from human fungal pathogens (Chaetomium, Coccidioides, Aspergillus terreus, A. fumigatus, and Neosartorya fischeri) or predicted human allergens (A. oryzae, Gibberella, and Alternaria brassicae), of which Gibberella and Alternaria were the only plant pathogens represented. Of all the fungi with genes most similar to clone Ma#267, Gibberella and Trichoderma are the most closely related taxonomically, both being in the same order, Hypocreales, as M. anisopliae.
Southern blot analyses to screen M. anisopliae strains for RFLPs.
Southern analysis was conducted to determine the hybridization pattern and copy number of MaNPS1 by hybridizing clone Ma#267 with isolates of M. anisopliae that vary in virulence and destruxin production in vitro (Table 1). Most isolates appeared to contain a single copy of clone Ma#267, suggesting a single copy of the gene. The exception was ARSEF 324, which in multiple analyses showed a doublet suggesting the possibility of two slightly different sized copies of MaNPS1.
Clone Ma#267 detected RFLPs that correlated, albeit imperfectly, with relative levels of destruxin production in vitro (Fig. 2 and Table 1). The majority of isolates that produced intermediate and high levels of destruxins exhibited a hybridization pattern identical to that of ARSEF 2575, which had two hybridizing bands of ca. 3.2 and 1.7 kb in size that were expected since clone Ma#267 contains a single BamHI site. ARSEF 1095 was a clear exception to the pattern in that it exhibited the polymorphism typical of most low producers of destruxin, i.e., a single band of ca. 4.9 kb, and yet produced high levels of destruxins in vitro. Since the two bands present in most intermediate or high producers totaled 4.9 kb, the results suggested that the single 4.9-kb band might have arisen by loss of the BamHI site via a mutational event.
FIG. 2.
Southern blot of genomic DNA of M. anisopliae strains digested with BamHI and hybridized with DIG-labeled Ma#267 (1.897 kb). Clone Ma#267 of the MaNPS1 gene detects DNA polymorphisms that correlate with levels of in vitro destruxin production by M. anisopliae. L, I, and H refer to relative levels of destruxin A and B (low, intermediate, and high, respectively) produced by the selected isolates in vitro as shown in Table 1.
The correlative results shown in Fig. 2 are in distinct contrast to those we observed using four additional unique NPS gene fragments from M. anisopliae isolates ARSEF 1095, ARSEF 1015, or M. album isolate ARSEF 2082 as probes (data not shown). These fragments were cloned from genomic DNA of the respective strains using a PCR-based approach to amplify regions containing conserved domains within NPSs (unpublished data). Sequential hybridization of the blot shown in Fig. 2 with each of these clones showed no discernible pattern that correlated with fungal virulence or in vitro destruxin levels, unlike the Ma#267 clone from ARSEF 2575.
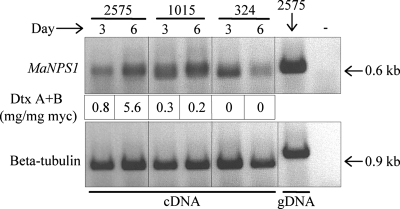
In vitro expression of MaNPS1.
RT-PCR experiments demonstrated that MaNPS1 is expressed concurrently with destruxin production in two of three isolates assayed 3 and 6 days after inoculation of CDBP with conidia (Fig. 3). The relative expression levels increased as destruxin production increased for M. anisopliae strain ARSEF 2575, which exhibited high levels of virulence against BAW and high levels of destruxin production in vitro (Table 1). However, for ARSEF 1015, which exhibited intermediate virulence against BAW and low in vitro production of destruxins, increased expression of MaNPS1 over time was not correlated with an increase in destruxin production relative to mycelial mass. Furthermore, MaNPS1 expression did not increase with time in ARSEF 324, the narrow-host-range M. anisopliae var. acridum isolate that exhibited low virulence against BAW and low in vitro production of destruxins. In addition, the relative levels of gene expression in ARSEF 2575 and 1015 did not correlate well with the levels of destruxin measured in the same cultures, i.e., gene expression was relatively high in ARSEF 1015, but only low amounts of destruxin were detected compared to the amounts present in ARSEF 2575 cultures.
FIG. 3.
In vitro MaNPS1 gene expression by M. anisopliae. Relative expression levels of MaNPS1, detected by RT-PCR, correlated with an increase in destruxin production over time for M. anisopliae isolate ARSEF 2575 (M. anisopliae var. anisopliae; high producer) but not for isolates ARSEF 1015 (M. anisopliae var. majus; low producer) or ARSEF 324 (M. anisopliae var. acridum; low producer). The numbers under each lane of the top gel represent the average destruxin A+B production relative to the average dry weight of the mycelium (mg of destruxin A+B/mg of mycelium [myc]) per replicate culture at 3 days (six replicates/isolate) or 6 days (three replicates/isolate) postinoculation. The “+ control” was genomic DNA used as a template for the PCR; the “− control” was the PCR lacking template DNA. No signals were detected when DNase-treated RNA from each sample was used as a template (data not shown).
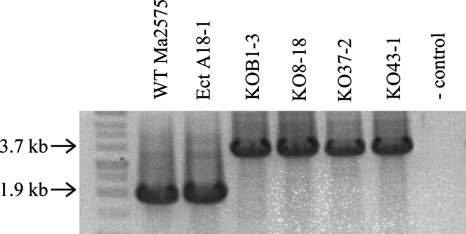
MaNPS1 gene disruption.
GA-resistant transformants were screened by PCR to detect the amplification products indicative of an intact, wild-type MaNPS1 gene versus those in which the Bar expression cassette was inserted within the MaNPS1 coding region. An amplicon of 1.9 kb, representing an intact copy of MaNPS1, was detected in wild-type ARSEF 2575 and an ectopic, GA-resistant transformant, Ect A-18 (Fig. 4). Transformants in which a homologous recombination event caused insertion of the Bar expression cassette (1.8 kb) into the wild-type Ma#267 fragment (1.9 kb) produced an amplicon of 3.7 kb (Fig. 1B and 4). Four ΔManps1 mutants (KO B1-3, KO 8-18, KO 37-2, and KO 43-1) and Ect A18-1 were chosen for further evaluations of potential phenotype changes. All strains were mitotically stable in vitro in the absence of drug selection for more than 3 weeks (data not shown). Single spore isolates of the ARSEF 2575 strain passed through BAW, and all transformants described here were deposited into the ARSEF culture collection and given the accession numbers ARSEF 2575 (ARSEF 8322), Ect A-18 (ARSEF 8323), KO B1-3 (ARSEF 8324), KO 8-18 (ARSEF 8325), KO 37-2 (ARSEF 8326), and KO 43-1 (ARSEF 8327).
FIG. 4.
GA-resistant transformants in which the MaNPS1 gene was disrupted (KO B1-3, KO 8-18, KO 37-2, and KO 43-1) exhibited a 3.7-kb amplicon after PCR with primer pair Sma267F and 2EndR, whereas the wild-type strain ARSEF 2575 and the ectopic transformant (A-18-1) had a 1.9-kb amplicon representative of the intact gene without the Bar expression cassette (1.8 kb in size) (Fig. 1B).
Southern analyses of ΔManps1 mutants and control strains.
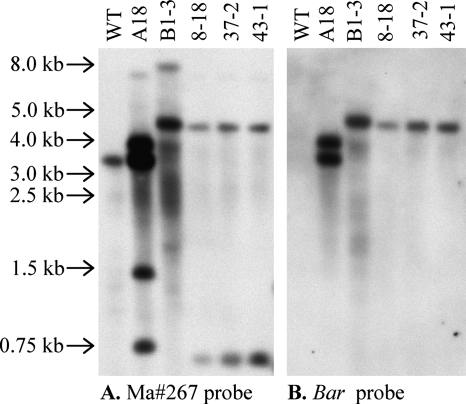
All four ΔManps1 mutants contained a single copy of the Bar gene (Fig. 5B, data not shown). In contrast, Ect A18-1, a control transformant in which the MaNPS1 disruption cassette integrated into the genome by nonhomologous recombination, contained two copies of the Bar gene inserted at sites external to the Ma#267 locus (Fig. 5 and data not shown). KO B1-3 contained at least one additional mutation not tagged by Bar that revealed polymorphisms when genomic DNA was digested by SacI (Fig. 5A), XbaI (data not shown), BamHI (data not shown), or SacI/BamHI (data not shown). In comparison to the other ΔManps1 mutants, SacI-digested KO B1-3 was missing a band ∼0.6 kb in size and contained an ∼8-kb band not present in the other strains (Fig. 5A). The bands and background signals were more intense in the Ect A18-1 and KO B1-3 lanes in comparison to other strains due to DNA overloading of these samples to confirm the absence of the 0.6-kb band in KO B1-3 and the presence of additional bands in Ect A18-1 and KO B1-3 (Fig. 5A; data not shown).
FIG. 5.
Southern analysis of SacI-digested genomic DNA of ΔManps1 mutants and control strains. The membrane was probed first with a DIG-labeled fragment of clone Ma#267 and then stripped and probed with a DIG-labeled fragment of the Bar gene. WT, ARSEF 2575; A18, Ect A18-1; B1-3, KO-B1-3; 8-18, KO 8-18; 37-2, KO 37-2; 43-1, KO 43-1. Select size markers from a 1-kb DNA ladder (Promega) are indicated on the left side of the figure.
Chemical analyses of ΔManps1 mutants and control strains.
HPLC analyses of extracts of culture broth and mycelia (Table 3) showed that the amounts of destruxin A in broth or from mycelium did not vary significantly with strain (broth, F = 0.2; df = 3, 8; P < 0.9; mycelium, F = 0.7; df = 3, 8; P < 0.6). Average destruxin A production for all strains was 69.0 mg/liter in broth (standard error of the mean [SEM] = 6.5) and 4.9 mg/liter in mycelium (SEM = 1.5). Destruxin B production in broth varied significantly with strain (Table 3; F = 4.8; df = 3, 8; P < 0.03). Strain KO B1-3 was the only strain to produce significantly less destruxin B than control strain Ect A-18 (Dunnett's test, P < 0.04). Mycelial dry weights did not differ significantly among strains (F = 1.9; df = 3, 8; P < 0.2) with an average dry weight for all strains of 898.1 g (SEM = 55.6). Destruxin A and B production in broth by the ΔManps1 mutants KO 37-2 and KO 43-1 (data not shown) was comparable to that of ARSEF 2575, Ect A-18, and KO 8-18.
TABLE 3.
Quantitative estimates of destruxins A and B in crude broth and mycelial extracts of M. anisopliae ΔManps1 mutant (KO 8-18 and KO B1-3) and control (ARSEF 2575 and Ect A-18) strainsa
| Strain | Mycelial dry wt (mean mg/culture ± SEM) | Amt in broth (mean mg/liter ± SEM)
|
Amt in mycelium (mean mg/liter ± SEM)
|
||
|---|---|---|---|---|---|
| Destruxin A | Destruxin B | Destruxin A | Destruxin B | ||
| ARSEF 2575 | 742 ± 15 a | 72.7 ± 0.7 a | 37.7 ± 0.6 a | 4.5 ± 0.6 a | 2.8 ± 0.4 a |
| Ect A-18 | 983 ± 63 a | 75.3 ± 16.4 a | 36.8 ± 3.1 a | 2.7 ± 1.0 a | 2.8 ± 0.3 a |
| KO 8-18 | 1038 ± 167 a | 62.5 ± 22.6 a | 36.6 ± 8.6 a | 8.5 ± 6.0 a | 4.7 ± 2.5 a |
| KO B1-3 | 829 ± 87 a | 65.6 ± 9.6 a | 16.7 ± 1.5 b | 3.8 ± 0.9 a | 1.1 ± 0.2 ab |
Estimates are expressed as the mean of three replicated extractions from 11-day-old 100-ml cultures. Means within a column followed by the same letter are not significantly different at P < 0.05 in Tukey's HSD test.
The amount of destruxin B produced by KO B1-3 in mycelium was determined to be significantly less than that of the two control strains only when KO 8-18, which was highly variable in this assay, was removed from the statistical analysis. See the text discussing the chemical analyses of the ΔManps1 mutants and control strains in Results for more details.
In mycelium, destruxin B production did not vary significantly with strain (Table 3; F = 1.3; df = 3, 8; P < 0.3). The average production for all strains was 2.9 mg/liter (SEM = 0.7). However, strain KO 8-18 gave particularly variable results in this assay, which swamped differences that were evident when it was not included in the statistical analyses. For example, it became apparent that strain KO B1-3 produced significantly less destruxin B in mycelium when it was compared only to the control strains Ect A-18 and ARSEF 2575 (F = 9.7; df = 2, 6; P < 0.01). Besides producing less destruxin B in broth and mycelium, the ΔManps1 mutant KO B1-3 was distinct in that its culture fluid was more yellow than the others due to overproduction of the mutagens NG-391 and NG-393 by ∼10-fold relative to ARSEF 2575 (46). This increase in NG-39X production by KO B1-3 was presumably caused by a nonspecific mutation that occurred during ATMT, which was distinct from the MaNPS1 targeted gene disruption event. Evidence for this explanation was suggested by the RFLP patterns unique to KO B1-3 compared to the other three ΔManps1 mutants (Fig. 5A and data not shown).
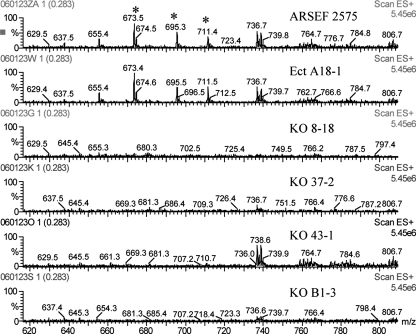
Extracts of conidia, culture fluids, mycelia, and submerged blastospore-like propagules were analyzed by ESIMS. Conidial extracts of control strains ARSEF 2575 and Ect A18-1, but not of the four ΔManps1 mutants, produced mass spectra with peaks at m/z 673, 695, and 711 Da (Fig. 6) that are consistent with the pseudomolecular ions [M+H]+, [M+Na]+, and [M+K]+, respectively, of the novel cyclic heptapeptide serinocyclin A (45). Serinocyclin A was estimated in conidiating cultures of ARSEF 2575 and Ect A18-1 at 24.0 ± 6.3 and 19.1 ± 12.9 μg/9-cm plate, respectively (average of three replicates ± the standard deviation). Neither serinocyclin A nor serinocyclin B was detected in the extracts of other developmental stages of the control strains, including mycelia, culture fluids, or submerged blastospore-like propagules (data not shown).
FIG. 6.
ESIMS analyses of conidial extracts from Δmanps1 mutants and control strains. Spectra represent the average of four scans, 3 s in duration, through the mass range m/z 2 to 1,202. The peaks denoted with asterisks at m/z 673, 695, and 711 Da are the pseudomolecular ions [M+H]+, [M+Na]+, and [M+K]+, respectively, of serinocyclin A.
Insect bioassays with ΔManps1 mutants and control strains.
No differences were detected in the ability of ΔManps1 mutants to kill hosts from two orders of insects in comparison to the control strains. Using BAW (Lepidoptera) as the host, the average mortality did not differ significantly among strains in single-dosage bioassays (Table 4). Mortality ranged from 86 to 95% among fungus-treated larvae and 0 to 4% among control larvae in all assays. The survival time for infected second instars ranged from 3.0 to 3.2 days across all strains. Conidial germination (i.e., the viability of the inoculum used for bioassays) did not vary significantly among isolates (F = 0.9, df = 4, 19, P < 0.5) and ranged from 82 to 97%. There was no correlation between the spore viability of each strain and larval mortality (Pearson r = 0.02). There were no differences in virulence between conidia stored at −20°C for 1 day versus aged spores stored under the same conditions for 21 days before use in bioassays. Each transformant, grown as mycelia originating from the surface of sporulating cadavers of infected BAW, was demonstrated by PCR to be mitotically stable in the absence of Bar selection for up to several weeks after passage through the insect host. The PCR products amplified from recovered mycelium of each strain were of the sizes expected for the ΔManps1 mutants containing a disrupted copy of the MaNPS1 gene, as well as control strains containing a wild-type copy (data not shown).
TABLE 4.
Mortality and survival times for larvae of the BAW, Spodoptera exigua, and CPB, Leptinotarsa decemlineata, inoculated with spores from the ΔManps1 mutant and control strains of M. anisopliaea
| Strainb |
Spodoptera exigua
|
Leptinotarsa decemlineata
|
||||
|---|---|---|---|---|---|---|
| Avg % mortality (SEM) | Mean survival time (SEM) | No. of insects | Avg % mortality (SEM) | Mean survival time (SEM) | No. of insects | |
| ARSEF 2575 | 86 (7) | 3.1 (0.05) | 282 | 44 (9) | 4.5 (0.13) | 63 |
| Ect A-18 | 89 (7) | 3.1 (0.04) | 292 | 43 (11) | 4.5 (0.23) | 32 |
| KO 8-18 | 89 (8) | 3.1 (0.05) | 193 | 44 (9) | 4.3 (0.13) | 54 |
| KO 37-2 | 89 (11) | 3.2 (0.06) | 220 | 48 (8) | 4.6 (0.15) | 62 |
| KO 43-1 | 95 (2) | 3.0 (0.04) | 224 | 44 (11) | 4.5 (0.14) | 40 |
| KO B1-3 | NT | NT | NT | 38 (9) | 4.8 (0.22) | 30 |
For the percent mortality, no significant difference was observed among strains by analysis of variance. Analysis was done on arcsine-square root-transformed percentages. For S. exigua, F = 0.8, df = 4, 19, and P < 0.5; for L. decemlineata, F = 0.13. df = 5, 37, and P < 0.9). For the survival time (in days), no significant difference was observed in insect survival times among isolates. For S. exigua, F = 1.4, df = 4, 1,206, and P < 0.2; for L. decemlineata, F = 0.9, df = 5, 275, and P < 0.5. The number of insects refers to the number of insects dead from fungal infection within 7 days of inoculation. NT, not tested.
Four (L. decemlineata) to six (S. exigua) assays were conducted with each strain, using 47 to 60 insects per strain per assay.
Using CPB (Coleoptera) as the host, the average mortality did not differ significantly among strains in multiple-dosage bioassays (Table 4). Mortality ranged from 38 to 48% among fungus-treated larvae and 0 to 8% among control larvae in all assays. Survival times for infected third instars ranged from 4.3 to 4.8 days across all strains. Conidial germination ranged from 92 to 99% among strains and did not vary significantly among isolates (F = 0.6; df = 5, 24; P < 0.7). It was notable that KO B1-3, the overproducer of the mutagenic NG-39X compounds in vitro (46), was no more virulent against CPB than any of the other strains.
Colony hydrophobicity and growth rate of ΔManps1 mutants and control strains in the absence or presence of H2O2.
No differences in the hydrophobicity of conidiating colonies on ¼SDAY could be detected when water droplets were placed on the surface of cultures of ΔManps1 mutants and control strains. No significant differences were detected in the radial growth of colonies of the four ΔManps1 mutants and two control strains on ¼SDAY at 12 days of growth (37 to 39 mm in diameter) or on CDA at 12 days (35 to 37 mm in diameter; experiment 1) or 11 days (30 to 33 mm in diameter; experiment 2) of growth. When H2O2 (10 mM or 16 mM) was assayed in CDA medium against all six strains for an effect on radial growth in two independent experiments, there was a significant dose and strain interaction in each experiment. Since the variation by experiment was significant (due to the 1-day difference in the age of colonies measured at either 12 or 11 days after inoculation), each experiment was analyzed separately (experiment 1, interaction F = 33.0, df = 10, 36, P < 0.0001; experiment 2, interaction F = 8.0, df = 10, 36, P < 0.0001). KO B1-3, the overproducer of the NG-39X mutagens, was the only transformant that grew significantly less than the other ΔManps1 mutants and control strains at both concentrations of H2O2, as determined by Tukey's HSD test (experiment 1, dose 10 mM, effect of strain F = 47.4, df = 5, 12, P < 0.0001; experiment 1, dose 16 mM, effect of strain F = 71.2, df = 5, 12, P < 0.0001; experiment 2, dose 10 mM, effect of strain F = 19.0, df = 5, 12, P < 0.0001; experiment 2, dose 16 mM, effect of strain F = 9.4, df = 5, 12, P < 0.001). In experiment 1, the average diameter of colonies after 12 days of growth was 27 mm (SEM = 0.3) on 10 mM H2O2 for all isolates except KO B1-3, which averaged 20 mm in diameter (SEM = 0.8); at 16 mM H2O2, all isolates had an average colony diameter of 17 mm (SEM = 0.3) except KO B1-3, which averaged 1 mM in diameter (SEM = 1.3). For experiment 2, the average diameter of colonies after 11 days of growth was 17 mm (SEM = 0.4) on 10 mM H2O2 for all isolates except KO B1-3, which averaged 10 mm (SEM = 0.7) in diameter; at 16 mM H2O2, all isolates had an average colony diameter of 6 mm (SEM = 0.6) except KO B1-3, which showed no measurable growth.
Sporulation and spore size of ΔManps1 mutants and control strains.
No significant differences were detected between ΔManps1 mutants (KO B1-3 and KO 8-18) and control strains (ARSEF 2575 and Ect A-18) in ability to sporulate on ¼SDAY. Average numbers of conidia per plate ranged from 4.4 × 109 to 5.4 × 109, with three replicates per isolate. The average lengths of hydrated, 1-week-old spores of ΔManps1 mutants (KO 8-18, KO 37-2, and KO 43-1) and control strains (ARSEF 2575 and Ect A-18), produced on ¼SSAY and measured at 9 h postplating, were comparable and ranged from 6.1 to 6.3 μm (100 spores per isolate measured), which is within the size range of ≤9 μm previously reported for M. anisopliae var. anisopliae (35). Similarly, spore areas (where the total numbers of pixels were converted to microns) for these conidia were comparable, with average area measurements for ΔManps1 mutants ranging from 126 to 142 μm and the two control strains at 134 μm.
Conidial germination of ΔManps1 mutants and control strains on cockroach wings and in response to ageing, H2O2, or KCl treatment.
Spore germination, extensive hyphal growth, and appressorium formation were evident on the surface of cockroach wings inoculated with ¼SSAY-produced conidia from three ΔManps1 mutants (KO 8-18, KO 37-2, and KO 43-1) or two control strains (ARSEF 2575 and Ect A-18). Images of each isolate were recorded at 25 h postinoculation. There were no discernible differences among the isolates in their abilities to germinate, spread across the cuticle surface, or develop appressoria. Significant variability in germination was evident on each wing, with some areas supporting greater germination and growth than others.
No significant differences in viability of fresh (1-week-old) or aged (3-week-old) ¼SSAY-produced conidia germinated on agar medium were detected between ΔManps1 mutants (KO 8-18, KO 37-2, and KO 43-1) and control strains (ARSEF 2575 and Ect A-18). The average percent germination levels for 1- and 3-week-old conidia at 9 h postplating and 12 h postplating, respectively, were 80.4% (SEM = 3.9) and 84.0% (SEM = 3.1), respectively. Similar results were obtained in two additional experiments assaying germination of 8- or 12-day-old conidia at 12 or 10 h postplating, respectively (data not shown). Control strains and ΔManps1 mutants were comparable in their rates of germination tube elongation, with an average germ tube length for 1-week-old spores of 5.7 μm (SEM = 0.7) at 9 h after plating.
No significant differences were detected between 10-day-old spores of three ΔManps1 mutants (KO 8-18, KO 37-2, and KO 43-1) and two control strains (ARSEF 2575 and Ect A-18) in their abilities to germinate in the presence of increasing concentrations of H2O2 (0, 5, 7.5, and 10 mM). The percent germination decreased in a linear fashion (germination = 92 − 9.3 [concentration], t = 13.6, df = 1, r2 = 0.91) with increasing H2O2 concentration, demonstrating a significant effect of H2O2 on germination (F = 148, df = 1, 14, P < 0.0001), but all strains behaved similarly (F = 0.1, df = 4, 14, P < 0.97). The average percent germination levels at each concentration of H2O2 were 98.2% (0 mM), 35.6% (5 mM), 20.6% (7.5 mM), and 5.6% (10 mM) 19 h after plating.
No significant differences were detected between 6-day-old conidia of three ΔManps1 mutants (KO 8-18, KO 37-2, and KO 43-1) and two control strains (ARSEF 2575 and Ect A-18) in the germination of conidia on 0.8 M KCl. The average percentage germination at 17 h postplating without exposure to KCl was 67.4% (SEM = 3.1), whereas germination in the presence of 0.8 M KCl averaged 49.6% (SEM = 3.8). The biggest effect of 0.8 M KCl across all strains, besides delaying germination, was a noticeable inhibition of hyphal growth and elongation by 36 h postplating (data not shown).
Adhesion of conidia of ΔManps1 mutants and control strains to different surfaces with various levels of polarity.
Attachment of conidia from ΔManps1 mutants (KO B1-3, KO 8-18, KO 37-2, and KO 43-1) and control strains (ARSEF 2575 and Ect A-18) to untreated polystyrene (uncharged, hydrophobic) was comparable for all of the strains (F = 1.1, df = 5, 12, P < 0.431) and ranged from 32 to 45%, with an overall average of attachment of 38% (SEM = 2.0). The average numbers of CFU counted on the unwashed and washed plates were 271 (SEM = 8.0) and 103 (SEM = 5.0), respectively.
Cell surface hydrophobicity (CSH), as measured with a BATH (bacterial adhesion to hydrocarbon) assay, did not vary significantly among conidia of ΔManps1 mutants and control strains (F = 0.5, df = 4, 71, P < 0.741). Furthermore, CSH did not vary with age of conidia (F = 0.3, df = 1, 71, P < 0.584) or with the medium (Barley or ¼SSAY) from which conidia were produced (F = 1.1, df = 1, 71, P < 0.295). The average CSH index for 7- to 30-day-old conidia was 90.4% (SEM = 0.6). Conidia of ΔManps1 mutants (KO 8-18 and KO 43-1) and control strains (ARSEF 2575 and Ect A-18) bound equally poorly to weakly polar (untreated Permanox plastic slides), highly hydrophobic (Sigmacote-coated glass slides), and charged, hydrophilic (poly-d-lysine-treated Permanox plastic slides) surfaces, with average binding efficiencies of 0.04, 0.10, and 0.16%, respectively, over a 30-min period; relatively few conidia remained attached after the first of two washes. Conidia of both the ΔManps1 mutants and control strains bound equally well to DEAE-Sephadex with a range of recovery of 7.2 to 18.0% and an overall average of 12.9% recovery (SEM = 2.0). Similarly, no significant differences were observed across strains in the binding of conidia to CM-Sephadex, although binding occurred at a much lower frequency, with a range of recovery of 76.5 to 88.5% and an average of 84.0% (SEM = 2.1). Boucias et al. (6) reported similar results for the Sephadex-binding experiments.
Surface morphology of M. anisopliae var. anisopliae conidia.
Germination assays of dried conidia stored at −20°C for 2 months before scanning electron microscopic analyses showed no major differences in viability between conidia of control strains (ARSEF 2575, 90%; Ect A-18, 79%) and the ΔManps1 mutants (KO 8-18, 82%; KO 37-2, 73%; KO 43-1, 73%). Scanning electron microscopy of these conidia at magnifications as high as ×50,000 revealed no clear differences between the surface morphologies of control and ΔManps1 mutants (data not shown). The surfaces of aerial conidia of M. anisopliae var. anisopliae were minutely cerebriform, with a resemblance to the gyri and sulci (bumps and grooves, respectively) of the cerebral cortex. The sizes and shapes of the conidia and superficial substructure were similar, except for minor differences in orientation and contour lengths of the 100-nm-wide folds and thin crevices comprising the cerebriform texture of the entire surface of each conidium. Boucias et al. (6) demonstrated by transmission electron microscopy that high-resolution surface replicas of M. anisopliae conidia exhibited a similar topography, with linear arrays of short rodlets and a regular cross-striation, similar to the organization and dimensions observed here.
DISCUSSION
Initial RFLP analyses suggested a correlation, albeit imperfect, between MaNPS1 and destruxin production by M. anisopliae. In addition, MaNPS1 was expressed in liquid culture conditions supporting mycelial growth and destruxin production. Because these results suggested that MaNPS1 might encode the destruxin NPS, this gene was chosen as a target for disruption to evaluate gene function. Gene disruption results showed clearly that MaNPS1 does not encode a destruxin synthetase since the ΔManps1 mutants produced amounts of destruxin comparable to the wild-type and ectopic control strains. The only exception to this result was KO B1-3, which produced less destruxin B than all other strains. The altered metabolism of this strain was probably caused by an independent, undefined mutation(s) that concurrently resulted in the overproduction of the NG-39X mutagens (46) and the underproduction of destruxin B. The presence of pseudomolecular ions representing the novel cyclic heptapeptide serinocyclin A in conidia of two control strains but not in four ΔManps1 mutants indicated that MaNPS1 encodes a serinocyclin synthetase and not a destruxin synthetase.
Although MaNPS1 was expressed in mycelia during vegetative growth in liquid CDBP, serinocyclins were not detected in these cultures. It is conceivable that the medium used for gene expression assays supported a preconidial developmental stage that could be associated with expression of the serinocyclin synthetase gene in the absence of conidia production. The absence of a natural product in the presence of gene expression could occur if the precursors necessary for synthesis of serinocyclins were unavailable in the culture medium or if posttranscriptional processes regulating serinocyclin production were developmentally or environmentally regulated and not supported by the culture conditions used. Under specific medium and environmental conditions, M. anisopliae has the potential to make at least three distinct single-cell types in vitro: blastospores, which have single-layered cell walls that bud from hyphae, and phialide-derived submerged conidia or aerial conidia, which have double-layered cell walls (40, 50). Thus far, serinocyclins have been detected only in aerial conidia of M. anisopliae var. anisopliae and M. anisopliae var. acridum produced in plate cultures (45). They were not detected in mycelial cultures of ARSEF 2575 grown in half-strength SDY (pH 6.5) to produce submerged blastospore-like propagules. Submerged conidium production has been reported in M. anisopliae var. acridum strains but appears to be rare in other species (40, 67), including M. anisopliae var. anisopliae.
It is unclear why clone Ma#267 of the serinocyclin synthetase gene MaNPS1 detected RFLP patterns that correlate with the relative levels of destruxin production, which the present study demonstrates occurs via a distinct biosynthetic pathway. The presence of the MaNPS1 gene in all 16 M. anisopliae strains assayed indicates that it is widespread among diverse strains and likely plays a significant role in the biology of the organism as a saprophyte or invertebrate pathogen. The presence of the gene, as well as the production of the serinocyclins, in both narrow- and broad-host-range pathogens is further evidence of its likely importance to the fungus. In contrast, two of the other four NPS genes evaluated for RFLPs were absent in a subset of these same strains (data not shown). Nevertheless, we were unable to identify a phenotype linked to the loss of metabolite production by the ΔManps1 mutants that would suggest function.
Only a single study to date has used molecular genetic approaches to directly address the role of secondary metabolites in invertebrate pathogenesis. Eley et al. (18) demonstrated that tenellin is not involved in the pathogenesis of Beauveria bassiana against wax moth larvae when they disrupted the hybrid NPS-polyketide synthase gene responsible for its biosynthesis. Similarly, in the present study, ΔManps1 mutants of M. anisopliae killed lepidopteran and coleopteran insect hosts as well as control strains, demonstrating that the serinocyclins do not function as virulence factors in these two insect systems. Disruption of the MaNPS1 gene had no measurable effect on hydrophobicity of sporulating colonies, conidial production, or colony growth rate in the presence of H2O2. The reduced colony growth rate of KO B1-3 after H2O2 treatment was likely related to other genetic and metabolic changes detected in this strain. Further studies will be necessary to characterize the phenotypic change in resistance to H2O2 unique to KO B1-3. The spore size after hydration, viability of aged conidia, germination tube length, spore germination on cockroach wings or agar medium after exposure to stressors, adherence to a variety of surfaces, and conidial surface morphology were unaffected by disruption of the MaNPS1 gene. It may be that MaNPS1 plays an early developmental role at a stage of the fungal life cycle not evaluated in our insect bioassays or under a unique set of environmental conditions not replicated in the laboratory. The present study provides a chemical phenotype by which to differentiate M. anisopliae strains and other fungal species encoding orthologues of MaNPS1 for the production of serinocyclin in conidia or other developmental stages.
Our results differ from what others have reported after disruption of spore-associated NPS genes from fungal pathogens of plants and humans. Reeves et al. (65) reported that Pes1 mutants of the human pathogen Aspergillus fumigatus exhibited increased conidial hydrophobicity, increased sensitivity to oxidative stress, altered conidial surface morphology, and reduced virulence against a lepidopteran model host. Furthermore, Pes1 expression, which is constitutive in vitro, increased in 3- and 4-day-old larvae that had been inoculated with conidia by injection. In contrast, MaNPS1 was expressed only weakly in 4-day-old BAW larvae infected through the cuticle by M. anisopliae (unpublished data). Kim et al. (43) reported that disruption of AbNPS2 from the Brassica pathogen Alternaria brassicicola resulted in decreased hydrophobicity of sporulating colonies, abnormal spore cell wall morphology, decreased spore germination rates in vitro and in vivo, reduced sporulation in vivo, and reduced virulence of aged spores. Furthermore, AbNPS1 expression was almost exclusively limited to conidia and conidiophores. Blastx and functional analyses further suggest that MaNPS1 is distinct from Pes1 of A. fumigatus and AbNPS2 of A. brassicicola, although all three pathways appear to encode spore-associated factors. The predicted proteins of Pes1 and AbNPS2 contain just four adenylation domains (43, 65) while, based on the cyclic heptapeptide structure of the serinocyclins, the MaNPS1 protein could contain as many as seven adenylation domains (48, 82). That the C. globosum protein CHGG_10057, to which clone Ma#267 of MaNPS1 is most similar, contains seven predicted AMP-binding domains makes it intriguing to consider that this fungus may make a family of metabolites similar to the serinocyclins.
It is notable that serinocyclins were detected at levels 5 to 10 times higher in conidia from a derivative isolate of the thermotolerant Green Muscle strain of M. anisopliae var. acridum (IMI 330189 from Niger, Africa) than in conidia of South Carolina isolate ARSEF 2575 (45). Green Muscle is a commercial biocontrol agent used against plagues of grasshoppers and locusts in arid and semi-arid areas of Africa. Furthermore, the predicted protein most similar to Ma#267 is from a fungal species, C. globosum, that comprises diverse strains adapted to grow under a wide range of environmental conditions, including as human pathogens or as rhizosphere-competent saprophytes in association with desert plants (85). Accordingly, such a range of growth conditions requires a capability of thermotolerance for at least some strains of Chaetomium. The documented thermotolerance of M. anisopliae conidia (62) and its plant-associated rhizosphere competence (34) are of interest in this regard. Whether the serinocyclins contribute to thermotolerance or rhizosphere competence of M. anisopliae under specific environmental or host conditions remains to be determined. Further studies also will be necessary to establish whether relatively high serinocyclin production is a specific characteristic of Green Muscle conidia or whether heat-tolerant M. anisopliae var. acridum strains generally produce more serinocyclins on average than other subspecies.
In contrast to Pes1 and AbNPS2, we have little knowledge of MaNPS1 function, but we have characterized the structures of the natural products produced by the enzyme and have some evidence indicating biological activity of serinocyclin A (45). Assays with pure serinocyclin A demonstrated that it was not cytotoxic but had a marked negative effect on the ability of mosquito larvae to swim directionally. Given this effect, it is appealing to consider that serinocyclins may play a protective role for the fungus in aquatic or highly humid environments since the compounds themselves are hydrophilic. For example, M. anisopliae ARSEF 2575 conidia have been reported to cause the death of embryos of inland silverside fish and grass shrimp (26, 27) in safety assessments evaluating effects on nontarget organisms. How the behavioral effect of serinocyclin A on mosquito larvae relates to its role in the biology of M. anisopliae as a soil and plant-associated saprophyte and insect pathogen is still unclear. Since M. anisopliae is being evaluated as a biological control agent for several disease-vectoring mosquito species (5, 70-72), it would be prudent to assess whether serinocyclins contribute a sublethal effect against mosquitoes (76) that could be beneficial to their control with the fungus.
Although Pes1 of A. fumigatus is constitutively expressed and AbNPS2 of A. brassicicola is expressed primarily in spores and conidiophores, the gene expression profile of MaNPS1 has been evaluated only partially under a variety of culture conditions with a few strains. Freimoser et al. (23) first reported the isolation of EST AJ272930, a fragment of clone Ma#267 of MaNPS1, from M. anisopliae var. anisopliae ARSEF 2575 after growth for 24 h in a minimal medium containing 1% cockroach cuticle. In microarray analyses, expression of AJ272930 by ARSEF 2575 was reported as either downregulated (22) or upregulated (83) in minimal medium with 1% cockroach cuticle, upregulated in minimal medium with either 1% beetle or 1% gypsy moth cuticle (22), downregulated in minimal medium containing insect hemolymph (22, 83), and nonresponsive to root exudate (83). We measured only weak expression of MaNPS1 in larvae of BAW infected for 4 days with ARSEF 2575 (unpublished data) but strong expression in mycelial cultures of M. anisopliae var. anisopliae (ARSEF 2575), M. anisopliae var. majus (ARSEF 1015), and M. anisopliae var. acridum (ARSEF 324) grown in CDBP.
Freimoser et al. (23) did not detect expression of AJ272930 in an EST library of M. anisopliae var. acridum ARSEF 324 after growth on minimal medium containing both 1% chitin and 1% cockroach cuticle. However, Wang and St. Leger (84) reported the gene is upregulated in ARSEF 324 in the presence of host (i.e., locust) dichloromethane cuticle extracts but is nonresponsive or downregulated in non-host cuticular extracts or minimal medium alone. In the same study, expression of three other NPS genes from the library was either nonresponsive to minimal medium or a variety of insect cuticular extracts, or downregulated, suggesting specificity of the response of the ARSEF 324 MaNPS1 gene to locust dichloromethane cuticular extract. Gene expression results reported to date suggest that regulation of expression of the serinocyclin synthetase gene MaNPS1 is complex and requires further study.
Not surprisingly, the expression information for genes similar to MaNPS1, but for which the natural product chemistries they produce are unknown, suggests a diversity of strategies under which such peptide synthetase genes are regulated. The gene encoding the hypothetical protein CIMG_09750 of C. immitis (with five AMP-binding domains), and its close relative C. posadasii, is expressed in both saprobic and spherule (infective) stages of development (41). Coccidioides pathogens grow asexually as filamentous saprobes in the soil and as multicellular, parasitic, endosporulating spherules in the human host (36). Similarly, Metarhizium spp. exhibit host infection-related dimorphism, growing as filamentous, conidiating saprobes in the soil and as hyphal bodies in the insect hemolymph. This latter stage is conducive to evasion of the host immune system but not adapted for survival outside the host (30, 61). NPS18, which encodes hypothetical protein FG10544.1 of G. zeae with seven predicted adenylation domains, is constitutively expressed (77). Prediction server results reported by Tobiasen et al. (77), which suggested amino acids potentially activated by the NPS18 enzyme, did not reveal any predicted similarity in function with the MaNPS1 enzyme, given the known amino acids composing the serinocyclin cyclic peptides (45). NRPS8 of A. fumigatus (XP_753380.1) encodes a predicted protein with six adenylation domains. It is considered a unique NPS gene in A. fumigatus since it is the only 1 of 14 whose transcripts are highly abundant in ungerminated conidia compared to other conditions, suggesting its involvement in the synthesis of small peptides that could be important in early stages of fungal development (15). NPS4 of G. zeae (FG02315.1) was expressed constitutively in some, but not all, Fusarium spp. assayed and is predicted to contain five adenylation domains (77). Tobiasen et al. (77) suggest that NPS4 may be involved in the synthesis of acuminatum, a cyclodepsipeptide from Fusarium spp. (9) that causes conidia of Penicillium spp. and Aspergillus spp. to swell, inhibiting germination in the process (8). If AbrePsy1 of A. brassicae (AAP78735.1) functions like AbNPS2 of A. brassicicola, expression of a four-module enzyme would be predicted to be unique to conidia and conidiophore production (43).
The present study represents the first demonstration of targeted disruption of a secondary metabolite gene in M. anisopliae, which revealed a novel pathway for the synthesis of a class of spore factors, the serinocyclins, of yet unknown function. The serinocyclins are cyclic heptapeptides that were not described previously from any other organism, and they have been detected to date only in relatively small quantities from M. anisopliae (45). Without the use of gene disruption techniques, it is probable that this new peptide class would have remained unknown since the compounds are not evident in mycelial liquid cultures of the fungus, from which most M. anisopliae metabolites have been characterized previously. Since this is a newly discovered group of compounds, the full spectrum of biological activity of the serinocyclins is unknown. The serinocyclins do not provide any measurable contribution to the virulence of M. anisopliae against BAW and CPB, but it is unknown whether they are involved in causing disease in other invertebrates, including mosquitoes, against which serinocyclin A alters larval swimming behavior. Further analyses are necessary to reveal the role of the serinocyclins as spore factors in the biology of the fungus. The serinocyclins are an example that demonstrates that not all secondary metabolites of M. anisopliae necessarily serve roles as broad-host-range toxins of insects, a premise that often has been inferred in the literature.
This research demonstrates the value of using targeted gene disruption methodologies in entomopathogens to reveal novel, biologically active metabolites, which may prove useful in agricultural or other systems. For biocontrol purposes, such approaches will allow a determination of which secondary metabolite pathways likely contribute to survival of the fungus as a saprophyte and which are necessary for fungal virulence against specific insect hosts. In this way, it will be possible to determine which pathways contribute to effective insect biocontrol and which are unnecessary, knowledge of which is especially valuable if a class of metabolites, such as the NG-39X mutagens (46), exhibits nonspecific toxicity to other organisms. Selection of strains lacking unnecessary, potentially toxic pathways will contribute to increased safety of biocontrol organisms, as will a greater understanding of when and if individual metabolites are produced during specific stages of fungal development and/or during the insect infection process. Ultimately, a more thorough understanding of how fungal biocontrol agents of insects cause disease should contribute to their broader use in agricultural and other systems and to a safer food supply, higher water quality, and a more sustainable environment.
Supplementary Material
Acknowledgments
We thank Steven Screen and Raymond St. Leger (University of Maryland) for providing cDNA clone Ma#267 and its extended DNA sequence before publication, as well as for helpful discussions. Jonathan Leehr, Rick Vaughan, and Jen Williams (USDA-ARS) provided excellent technical support. Pia Gavino (Boyce Thompson Institute; currently Illinois Central College) isolated three of the NPS fragments used as probes against genomic DNA in Southern blots for RFLP analysis. Karen Hansen and Richard Humber (USDA-ARS) provided cultures from the ARSEF Collection of Entomopathogenic Fungi. Jeffrey Scott (Cornell University) provided German cockroaches.
This project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant numbers 2002-35316-12207 and 2005-35607-15283.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aldridge, D. C., and W. B. Turner. 1969. Structures of cytochalasin C and cytochalasin D from Metarhizium anisopliae. J. Chem. Soc. C 6:923-928. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amiri-Besheli, B., B. Khambay, S. Cameron, M. L. Deadman, and T. M. Butt. 2000. Inter- and intra-specific variation in destruxin production by insect pathogenic Metarhizium spp. and its significance to pathogenesis. Mycol. Res. 104:447-452. [Google Scholar]
- 4.Bailey, A. M., M. J. Kershaw, B. A. Hunt, I. C. Paterson, A. K. Charnley, S. E. Reynolds, and J. M. Clarkson. 1996. Cloning and sequence analysis of an intron-containing domain from a peptide synthetase-encoding gene of the entomopathogenic fungus Metarhizium anisopliae. Gene 173:195-197. [DOI] [PubMed] [Google Scholar]
- 5.Blanford, S., B. H. K. Chan, N. Jenkins, D. Sim, R. J. Turner, A. F. Read, and M. B. Thomas. 2005. Fungal pathogen reduces potential for malaria transmission. Science 308:1638-1641. [DOI] [PubMed] [Google Scholar]
- 6.Boucias, D. G., J. C. Pendland, and J. P. Latge. 1988. Nonspecific factors involved in attachment of entomopathogenic Deuteromycetes to host insect cuticle. Appl. Environ. Microbiol. 54:1795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bundock, P., A. Den Dulk Ras, A. Beijersbergen, and P. J. J. Hooykaas. 1995. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14:3206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burmeister, H. R., J. J. Ellis, and R. F. Vesonder. 1981. Survey for Fusaria that produce an antibiotic that causes conidia of Penicillium digitatum to swell. Mycopathologia 74:29-33. [Google Scholar]
- 9.Carr, S. A., E. Block, and C. E. Costello. 1985. Structure determination of a new cyclodepsipeptide antibiotic from Fusaria fungi. J. Org. Chem. 50:2854-2858. [Google Scholar]
- 10.Charnley, A. K. 1989. Mechanisms of fungal pathogenesis in insects, p. 85-125. In J. M. Whipps and R. D. Lumsden (ed.), Biotechnology of fungi for improving plant growth. Cambridge University Press, London, England.
- 11.Chilton, M.-D., T. C. Currier, S. K. Farrand, A. J. Bendich, M. P. Gordon, and E. W. Nester. 1974. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. USA 71:3672-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarkson, J., S. Screen, A. Bailey, B. Cobb, and K. Charnley. 1998. Fungal pathogenesis in insects, p. 83-94. In P. Bridge, Y. Couteaudier, and J. Clarkson (ed.), Molecular variability of fungal pathogens. CAB International, Oxfordshire, United Kingdom.
- 13.Clarkson, J. M., and A. K. Charnley. 1996. New insights into the mechanisms of fungal pathogenesis in insects. Trends Microbiol. 4:197-203. [DOI] [PubMed] [Google Scholar]
- 14.Covert, S. F., P. Kapoor, M.-H. Lee, A. Briley, and C. J. Nairn. 2001. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. Res. 105:259-264. [Google Scholar]
- 15.Cramer, R. A., J. E. Stajich, Y. Yamanaka, F. S. Dietrich, W. J. Steinbach, and J. R. Perfect. 2006. Phylogenomic analysis of non-ribosomal peptide synthetases in the genus Aspergillus. Gene 383:24-32. [DOI] [PubMed] [Google Scholar]
- 16.de Faria, M. R., and S. P. Wraight. 2007. Mycoinsecticides and mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 43:237-256. [Google Scholar]
- 17.Delgado, N., C.-Y. Hung, E. Tarcha, M. J. Gardner, and G. T. Cole. 2004. Profiling gene expression in Coccidioides posadasii. Med. Mycol. 42:59-71. [DOI] [PubMed] [Google Scholar]
- 18.Eley, K. L., L. M. Halo, Z. Song, H. Powles, R. J. Cox, A. M. Bailey, C. M. Lazarus, and T. J. Simpson. 2007. Biosynthesis of the 2-pyridone tenellin in the insect pathogenic fungus Beauveria bassiana. ChemBioChem 8:289-297. [DOI] [PubMed] [Google Scholar]
- 19.Espada, A., and M. M. Dreyfuss. 1997. Effect of the cyclopeptolide 90-215 on the production of destruxins and helvolic acid by Metarhizium anisopliae. J. Ind. Microbiol. Biotechnol. 19:7-11. [Google Scholar]
- 20.Fargues, J., P. H. Robert, and A. Vey. 1985. Effet des destruxines A, B, et E dans la pathogénèse de Metarhizium anisopliae chez les larves de Coléoptères Scarabaeidae. Entomophaga 30:353-364. [Google Scholar]
- 21.Fargues, J., N. Smits, C. Vidal, A. Vey, F. Vega, G. Mercadier, and P. Quimby. 2002. Effect of liquid culture media on morphology, growth, propagule production, and pathogenic activity of the hyphomycete, Metarhizium flavoviride. Mycopathologia 154:127-138. [DOI] [PubMed] [Google Scholar]
- 22.Freimoser, F. M., G. Hu, and R. J. St. Leger. 2005. Variation in gene expression patterns as the insect pathogen Metarhizium anisopliae adapts to different host cuticles or nutrient deprivation in vitro. Microbiology 151:361-371. [DOI] [PubMed] [Google Scholar]
- 23.Freimoser, F. M., S. Screen, S. Bagga, G. Hu, and R. J. St. Leger. 2003. Expressed sequence tag (EST) analysis of two subspecies of Metarhizium anisopliae reveals a plethora of secreted proteins with potential activity in insect hosts. Microbiology 149:239-247. [DOI] [PubMed] [Google Scholar]
- 24.Fujii, Y., H. Tani, M. Ichinoe, and H. Nakajima. 2000. Zygosporin D and two new cytochalasins produced by the fungus Metarrhizium anisopliae. J. Nat. Prod. 63:132-135. [DOI] [PubMed] [Google Scholar]
- 25.Gelderblom, W. C. A., P. G. Thiel, K. J. V. D. Merwe, W. F. O. Marasas, and H. S. C. Spies. 1983. A mutagen produced by Fusarium moniliforme. Toxicon 21:467-473. [DOI] [PubMed] [Google Scholar]
- 26.Genthner, F. J., C. A. Chancy, J. A. Couch, S. S. Foss, D. P. Middaugh, S. E. George, M. A. Warren, and J. A. Bantle. 1998. Toxicity and pathogenicity testing of the insect pest control fungus Metarhizium anisopliae. Arch. Environ. Contam. Toxicol. 35:317-324. [DOI] [PubMed] [Google Scholar]
- 27.Genthner, F. J., and D. P. Middaugh. 1995. Nontarget testing of an insect control fungus: effects of Metarhizium anisopliae on developing embryos of the inland silverside fish Menidia beryllina. Dis. Aquat. Org. 22:163-171. [Google Scholar]
- 28.Gillespie, A. T., and N. Claydon. 1989. The use of entomogenous fungi for pest control and the role of toxins in pathogenesis. Pest. Sci. 27:203-215. [Google Scholar]
- 29.Gupta, S., S. B. Krasnoff, J. A. A. Renwick, D. W. Roberts, J. R. Steiner, and J. Clardy. 1993. Viridoxins A and B: novel toxins from the fungus Metarhizium flavoviride. J. Org. Chem. 58:1062-1067. [Google Scholar]
- 30.Hajek, A. E. 1997. Ecology of terrestrial fungal entomopathogens. Adv. Microb. Ecol. 15:193-249. [Google Scholar]
- 31.Hajek, A. E., S. P. Wraight, and J. D. Vandenberg. 2001. Control of arthropods using pathogenic fungi, p. 309-347. In S. B. Pointing and K. D. Hyde (ed.), Bio-exploitation of filamentous fungi, vol. 6. Fungal Diversity Press, Hong Kong, SAR, China. [Google Scholar]
- 32.Hill, S. R., R. Bonjouklian, G. Powis, R. T. Abraham, C. L. Ashendel, and L. H. Zalkow. 1994. A multisample assay for inhibitors of phosphatidylinositol phospholipase C: identification of naturally occurring peptide inhibitors with antiproliferative activity. Anti-Cancer Drug Des. 9:353-361. [PubMed] [Google Scholar]
- 33.Hino, M., O. Nakaayama, Y. Tsurumi, K. Adachi, T. Shibata, H. Terano, M. Kohsaka, H. Aoki, and H. Imanaka. 1985. Studies of an immunomodulator swainsonine. I. Enhancement of immune response by swainsonine in vitro. J. Antibiot. 38:926-935. [DOI] [PubMed] [Google Scholar]
- 34.Hu, G., and R. J. St. Leger. 2002. Field studies using a recombinant mycoinsecticide (Metarhizium anisopliae) reveal that it is rhizosphere competent. Appl. Environ. Microbiol. 68:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humber, R. A. 1997. Fungi: identification, p. 153-185. In L. A. Lacey (ed.), Manual of techniques in insect pathology. Harcourt Brace, Chatham, Kent, United Kingdom.
- 36.Hung, C.-Y., J.-J. Yu, K. R. Seshan, U. Reichard, and G. T. Cole. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iijima, M., T. Masuda, H. Nakamura, H. Naganawa, S. Kurosawa, Y. Okami, M. Ishizuka, T. Takeuchi, and Y. Iitaka. 1992. Metacytofilin, a novel immunomodulator produced by Metarhizium sp. TA2759. J. Antibiot. 45:1553-1556. [DOI] [PubMed] [Google Scholar]
- 38.Irelan, J., V. Miao, and E. U. Selker. 1993. Small scale DNA preps for Neurospora crassa. Fungal Genet. Newsl. 40:24. [Google Scholar]
- 39.Jegorov, A., P. Sedmera, and V. Matha. 1993. Biosynthesis of destruxins. Phytochemistry 33:1403-1405. [DOI] [PubMed] [Google Scholar]
- 40.Jenkins, N. E., and C. Prior. 1993. Growth and formation of true conidia by Metarhizium flavoviride in a simple liquid medium. Mycol. Res. 97:1489-1494. [Google Scholar]
- 41.Johannesson, H., T. Kasuga, R. A. Schaller, B. Good, M. J. Gardner, J. P. Townsend, G. T. Cole, and J. W. Taylor. 2006. Phase-specific gene expression underlying morphological adaptations of the dimorphic human pathogenic fungus, Coccidioides posadasii. Fungal Genet. Biol. 43:545-559. [DOI] [PubMed] [Google Scholar]
- 42.Kershaw, M. J., E. R. Moorehouse, R. Bateman, S. E. Reynolds, and A. K. Charnley. 1999. The role of destruxins in the pathogenicity of Metarhizium anisopliae for three species of insect. J. Invert. Pathol. 74:213-223. [DOI] [PubMed] [Google Scholar]
- 43.Kim, K.-H., Y. Cho, M. L. Rota, R. A. Cramer, and C. B. Lawrence. 2007. Functional analysis of the Alternaria brassicicola non-ribosomal peptide synthetase gene AbNPS2 reveals a role in conidial cell wall construction. Mol. Plant Pathol. 8:23-39. [DOI] [PubMed] [Google Scholar]
- 44.Kondo, S., N. Meguriya, H. Mogi, T. Aota, K. Miura, T. Fujii, I. Hayashi, K. Makino, M. Yamamota, and N. Nakajima. 1980. K-582, a new peptide antibiotic. I. J. Antibiot. 33:533-542. [DOI] [PubMed] [Google Scholar]
- 45.Krasnoff, S. B., I. Keresztes, R. E. Gillilan, D. M. E. Szebenyi, B. G. G. Donzelli, A. C. L. Churchill, and D. M. Gibson. 2007. Serinocyclins A and B, cyclic heptapeptides from Metarhizium anisopliae. J. Nat. Prod. 70:1919-1924. [DOI] [PubMed] [Google Scholar]
- 46.Krasnoff, S. B., C. H. Sommers, Y.-S. Moon, B. G. G. Donzelli, J. D. Vandenberg, A. C. L. Churchill, and D. M. Gibson. 2006. Production of mutagenic metabolites by Metarhizium anisopliae. J. Agric. Food Chem. 54:7083-7088. (Erratum, 56:1158, 2008.) [DOI] [PubMed] [Google Scholar]
- 47.Kuboki, H., T. Tsuchida, K. Wakazono, K. Isshiki, H. Kumagai, and T. Yoshioka. 1999. Mer-f3, 12-hydroxy-ovalicin, produced by Metarhizium sp. f3. J. Antibiot. 52:590-593. [DOI] [PubMed] [Google Scholar]
- 48.Lautru, S., and G. L. Challis. 2004. Substrate recognition by nonribosomal peptide synthetase multi-enzymes. Microbiology 150:1629-1636. [DOI] [PubMed] [Google Scholar]
- 49.Lee, T., S.-H. Yun, K. T. Hodge, R. A. Humber, S. B. Krasnoff, G. B. Turgeon, O. C. Yoder, and D. M. Gibson. 2001. Polyketide synthase genes in insect- and nematode-associated fungi. Appl. Microbiol. Biotechnol. 56:181-187. [DOI] [PubMed] [Google Scholar]
- 50.Leland, J. E., D. E. Mullins, L. J. Vaughan, and H. L. Warren. 2005. Effects of media composition on submerged culture spores of the entomopathogenic fungus, Metarhizium anisopliae var. acridum, part 1: comparison of cell wall characteristics and drying stability among three spore types. Biocontrol Sci. Technol. 15:379-392. [Google Scholar]
- 51.Lomer, C. J., R. P. Bateman, D. L. Johnson, J. Langewald, and M. Thomas. 2001. Biological control of locusts and grasshoppers. Annu. Rev. Entomol. 46:667-702. [DOI] [PubMed] [Google Scholar]
- 52.Martens, J. 2004. Long PCR amplification of p300 cDNA using a blend of Taq DNA polymerase and proofreading Pfu DNA polymerase. Promega eNotes. Promega, Madison, WI.
- 53.McCluskey, K. 2003. The Fungal Genetics Stock Center: from molds to molecules, p. 245-262. In A. I. Laskin, J. W. Bennett, and G. M. Gadd (ed.), Advances in applied microbiology, vol. 52. Elsevier, Inc., St. Louis, MO. [DOI] [PubMed] [Google Scholar]
- 54.Milner, R. J. 2000. Current status of Metarhizium as a mycoinsecticide in Australia. Biocontrol News Info. 21:47N-50N. [Google Scholar]
- 55.Moon, Y.-S., S. B. Krasnoff, J. D. Vandenberg, D. M. Gibson, and A. C. L. Churchill. 2005. Genetic analyses of a peptide synthetase gene from the insect pathogen Metarhizium anisopliae, abstr. Fungal Genet. Newsl. 52(Suppl.):517. [Google Scholar]
- 56.Païs, M., B. C. Das, and P. Ferron. 1981. Depsipeptides from Metarhizium anisopliae. Phytochemistry 20:715-723. [Google Scholar]
- 57.Pal, S., R. J. St. Leger, and L. P. Wu. 2007. Fungal peptide destruxin A plays a specific role in suppressing the innate immune response in Drosophila melanogaster. J. Biol. Chem. 282:8969-8977. [DOI] [PubMed] [Google Scholar]
- 58.Pall, M. L., and J. P. Brunelli. 1993. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. News 40:59-62. [Google Scholar]
- 59.Patrick, M., M. W. Adlard, and T. Keshavarz. 1993. Production of an indolizidine alkaloid, swainsonine by the filamentous fungus, Metarhizium anisopliae. Biotechnol. Lett. 15:997-1000. [Google Scholar]
- 60.Pedras, M. S. C., L. I. Zaharia, and D. E. Ward. 2002. The destruxins: synthesis, biosynthesis, biotransformation, and biological activity. Phytochemistry 59:579-596. [DOI] [PubMed] [Google Scholar]
- 61.Pendland, J. C., S.-Y. Hung, and D. G. Boucias. 1993. Evasion of host defense by in vivo-produced protoplast-like cells of the insect mycopathogen Beauveria bassiana. J. Bacteriol. 175:5962-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rangel, D. E. N., G. U. L. Braga, A. J. Anderson, and D. W. Roberts. 2005. Variability in conidial thermotolerance of Metarhizium anisopliae isolates from different geographic origins. J. Invert. Pathol. 88:116-125. [DOI] [PubMed] [Google Scholar]
- 63.Rausch, C., T. Weber, O. Kohlbacher, W. Wohlleben, and D. H. Huson. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rees, D. O., N. Bushby, R. J. Cox, J. R. Harding, T. J. Simpson, and C. L. Willis. 2007. Synthesis of [1,2-13C2,15N]-l-homoserine and its incorporation by the PKS-NRPS system of Fusarium moniliforme into the mycotoxin fusarin C. ChemBioChem 8:46-50. [DOI] [PubMed] [Google Scholar]
- 65.Reeves, E. P., K. Reiber, C. Neville, O. Scheibner, K. Kavanagh, and S. Doyle. 2006. A nonribosomal peptide synthetase (Pes1) confers protection against oxidative stress in Aspergillus fumigatus. FEBS J. 273:3038-3053. [DOI] [PubMed] [Google Scholar]
- 66.Roberts, D. W., S. Gupta, and R. J. St. Leger. 1992. Metabolite production by entomopathogenic fungus. Pesqui. Agropec. Bras. 27:325-347. [Google Scholar]
- 67.Roberts, D. W., and R. J. St. Leger. 2004. Metarhizium spp., cosmopolitan insect-pathogenic fungi: mycological aspects, p. 1-70. In A. I. Laskin, J. W. Bennett, and G. Gadd (ed.), Advances in applied microbiology, vol. 54. Elsevier, Inc., London, United Kingdom. [DOI] [PubMed] [Google Scholar]
- 68.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 69.Samuels, R. I., A. K. Charnley, and S. E. Reynolds. 1988. The role of destruxins in the pathogenicity of three strains of Metarhizium anisopliae for the tobacco hornworm Manduca sexta. Mycopathologia 104:51-58. [Google Scholar]
- 70.Scholte, E.-J., B. G. J. Knols, and W. Takken. 2006. Infection of the malaria mosquito Anopheles gambiae with the entomopathogenic fungus Metarhizium anisopliae reduces blood feeding and fecundity. J. Invert. Pathol. 91:43-49. [DOI] [PubMed] [Google Scholar]
- 71.Scholte, E.-J., K. Ng'habi, J. Kihonda, W. Takken, K. Paaijmans, S. Abdulla, G. F. Killeen, and B. G. J. Knols. 2005. An entomopathogenic fungus for control of adult African malaria mosquitoes. Science 308:1641-1642. [DOI] [PubMed] [Google Scholar]
- 72.Scholte, E.-J., W. Takken, and B. G. J. Knols. 2007. Infection of adult Aedes aegypti and A. albopictus mosquitoes with the entomopathogenic fungus Metarhizium anisopliae. Acta Trop. 102:151-158. [DOI] [PubMed] [Google Scholar]
- 73.Song, Z., R. J. Cox, C. M. Lazarus, and T. J. Simpson. 2004. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. ChemBioChem 5:1196-1203. [DOI] [PubMed] [Google Scholar]
- 74.Stevens, R. 1974. Mycology guidebook. University of Washington Press, Seattle.
- 75.Strasser, H., A. Vey, and T. M. Butt. 2000. Are there risks in using entomopathogenic fungi for pest control, with particular reference to the bioactive metabolites of Metarhizium, Tolypocladium, and Beauveria species? Biocontrol Sci. Technol. 10:717-735. [Google Scholar]
- 76.Thomas, M. B., and A. F. Read. 2007. Can fungal biopesticides control malaria? Nat. Rev. Microbiol. 5:377-383. [DOI] [PubMed] [Google Scholar]
- 77.Tobiasen, C., J. Aahman, K. S. Ravnholt, M. J. Bjerrum, M. N. Grell, and H. Giese. 2007. Nonribosomal peptide synthetase (NPS) genes in Fusarium graminearum, F. culmorum, and F. pseudograminearium and identification of NPS2 as the producer of ferricrocin. Curr. Genet. 51:43-58. [DOI] [PubMed] [Google Scholar]
- 78.Uchida, R., R. Imasoto, Y. Yamaguchi, R. Masuma, K. Shiomi, H. Tomoda, and S. Ōmura. 2005. New insecticidal antibiotics, hydroxyfungerins A and B, produced by Metarhizium sp. FKI-1079. J. Antibiot. 58:804-809. [DOI] [PubMed] [Google Scholar]
- 79.Vandenberg, J. D. 1996. Standardized bioassay and screening of Beauveria bassiana and Paecilomyces fumosoroseus against the Russian wheat aphid (Homoptera: Aphididae). J. Econ. Entomol. 89:1418-1423. [Google Scholar]
- 80.van Engelen, F. A., J. W. Molthoff, A. J. Conner, J.-P. Nap, A. Pereira, and W. J. Stiekema. 1995. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4:288-290. [DOI] [PubMed] [Google Scholar]
- 81.Vey, A., R. E. Hoagland, and T. M. Butt. 2001. Toxic metabolites of fungal biocontrol agents, p. 311-346. In T. M. Butt, C. Jackson, and N. Magan (ed.), Fungi as biocontrol agents. CAB International, New York, NY.
- 82.von Döhren, H. 2004. Biochemistry and general genetics of nonribosomal peptide synthetases in fungi. Adv. Biochem. Eng./Biotechnol. 88:217-264. [DOI] [PubMed] [Google Scholar]
- 83.Wang, C., G. Hu, and R. J. S. Leger. 2005. Differential gene expression by Metarhizium anisopliae growing in root exudate and host (Manduca sexta) cuticle or hemolymph reveals mechanisms of physiological adaptation. Fungal Genet. Biol. 42:704-718. [DOI] [PubMed] [Google Scholar]
- 84.Wang, C., and R. J. St. Leger. 2005. Developmental and transcriptional responses to host and nonhost cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot. Cell 4:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wijeratne, E. M. K., T. J. Turbyville, A. Fritz, L. Whitesell, and A. A. L. Gunatilaka. 2006. A new dihydroxanthenone from a plant-associated strain of the fungus Chaetomium globosum demonstrates anticancer activity. Bioorg. Med. Chem. 14:7917-7923. [DOI] [PubMed] [Google Scholar]
- 86.Wu, Q., T. M. Sandrock, B. G. Turgeon, O. C. Yoder, S. G. Wirsel, and J. R. Aist. 1998. A fungal kinesin required for organelle motility, hyphal growth, and morphogenesis. Mol. Biol. Cell 9:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.