Abstract
Differential protection of plants by Enterobacter cloacae was studied by investigating early sensing and response behavior of Pythium ultimum sporangia toward seeds in the presence or absence of E. cloacae. Ten percent of P. ultimum sporangia were activated within the first 30 min of exposure to cucumber seeds. In contrast, 44% of the sporangia were activated as early as 15 min after exposure to corn seeds with over 80% sporangial activation by 30 min. Germ tubes emerged from sporangia after 2.5 and 1.0 h in the cucumber and corn spermospheres, respectively. Seed application of the wild-type strain of E. cloacae (EcCT-501R3) reduced sporangial activation by 45% in the cucumber spermosphere, whereas no reduction was observed in the corn spermosphere. Fatty acid transport and degradation mutants of E. cloacae (strains EcL1 and Ec31, respectively) did not reduce sporangial activation in either of the spermospheres. Although wild-type or mutant strains of E. cloacae failed to reduce seed colonization incidence, pathogen biomass on cucumber seeds was reduced in the presence of E. cloacae strains EcCT-501R3 and Ec31 by 4 and 8 h after sowing, respectively. By 12 h, levels of P. ultimum on cucumber seeds treated with E. cloacae EcCT-501R3 did not differ from levels on noninoculated seeds. On corn seeds, P. ultimum biomass was not affected by the presence of any E. cloacae strain. When introduced after sporangial activation had occurred, E. cloacae failed to reduce P. ultimum biomass on cucumber seeds compared with that on nontreated seeds. Also, increasing numbers of sporangia used to inoculate seeds yielded increased pathogen biomass at each sampling time. This indicates a direct link between the level of seed-colonizing biomass of P. ultimum and the number of activated and germinated sporangia in the spermosphere, suggesting that E. cloacae suppresses P. ultimum seed infections by reducing sporangial activation and germination within the first 30 to 90 min after sowing.
Pythium ultimum is a widespread and destructive oomycete pathogen of plants, notorious for its rapid pathogenic development in response to germinating seeds. P. ultimum infects seeds and seedlings, leading to a disease known as damping-off (10). Disease development is generally initiated from sporangia, which together with oospores serve as quiescent survival structures in the P. ultimum life cycle, each requiring seed exudates to break dormancy and initiate plant infections (9). Sporangia can transition from quiescence to active growth in as little as 1.5 h after exposure to a seed (7, 19). Seed exudates from many plant species can induce these rapid responses (4, 21). Although the specific exudate elicitors from most plants are unknown, the elicitors from cottonseed exudates have been identified as long-chain unsaturated fatty acids (LCUFA), comprised largely of oleic and linoleic acids (9, 10, 18). Other exudate molecules such as saturated fatty acids, sugars, amino acids, and other organic acids do not elicit sporangial germination (13, 14, 18). Once sporangia have germinated, high levels of seed colonization and subsequent infection are observed within 24 to 48 h, leading to rapid seed and seedling death (1, 2, 8, 9, 24). However, if exudate stimulants are removed by physical or biological means, sporangia do not germinate and little or no seed colonization and disease development occur (3, 7, 12, 15).
Enterobacter cloacae is a common seed-associated bacterium that suppresses seed infections, protecting a number of plant species from P. ultimum-induced damping-off (3, 4, 8, 12). Plant protection by E. cloacae is achieved largely by degradation of LCUFA in seed exudates (20), which eliminates the stimulation of P. ultimum sporangia (4, 21). E. cloacae suppresses P. ultimum infections when applied as a coating onto seeds of plants such as carrot, cotton, cucumber, lettuce, radish, sunflower, tomato, and wheat. However, no disease suppression is observed when E. cloacae is applied as a coating onto seeds of corn or pea (4, 12).
Currently, it is not known how quickly sporangial germination and subsequent disease development are initiated on these different host species. Furthermore, it is not known whether reduced disease development that accompanies the removal of exudate stimulants is due solely to reductions in sporangial germination or whether E. cloacae may also suppress later stages of pathogenesis. Because most of the current knowledge of seed-induced stimulation of sporangial germination comes from work with in vitro-collected seed exudates (4, 18, 20, 21), little is known of the temporal dynamics of sporangial responses in the spermosphere and how these responses relate to the temporal exudation of stimulatory molecules.
To better understand the differential protection of plants by E. cloacae, this study was focused on the dynamics of P. ultimum during early stages of pathogenesis in the presence and absence of E. cloacae on two differentially protected hosts. The main goals of this work are to identify how quickly E. cloacae reduces P. ultimum sporangial activation and germ tube emergence in the spermosphere and to establish whether this relates directly to reductions in subsequent P. ultimum seed colonization. A further goal is to better understand the potential role of E. cloacae fatty acid degradation during early stages of disease development.
MATERIALS AND METHODS
Plant material.
To reduce experimental variability, corn (Zea mays cv. Northern X-tra Sweet) and cucumber (Cucumis sativus cv. Marketmore) seeds were sorted by discarding cracked, deformed, or discolored seeds. Seeds were then surface sterilized for 3 min in 0.05% sodium hypochlorite with 1 to 2 drops of Tween 20, rinsed with sterile deionized water (sdw), and blotted dry. Seeds weighing between 160 and 200 mg and between 24 and 30 mg were used for all corn and cucumber assays, respectively.
Production and germination of Pythium ultimum sporangia.
Pythium ultimum P4 was routinely grown on a mineral salts medium containing 0.1% α-phosphatidylcholine (SM+L). Sporangia produced on this medium mimic the behavior of sporangia produced on live plant tissue (13). Agar discs (4-mm diameter) were cut from 5-day-old cultures kept at 27°C. Discs were leached twice in darkness for 10 minutes each followed by a 3-h leaching by adding leaching buffer [0.01 M Ca(NO3)2·4H2O, 0.04 M MgSO4·7H2O, 0.05 M KNO3, pH 5.8] and replacing it with fresh buffer at the end of each leaching period. Discs were rinsed twice with sdw and kept at 24°C for 2 days in darkness.
In preparation of defined levels of sporangia to be used as inocula, entire SM+L plates (9-cm diameter) were leached as described above. Sporangia were harvested by adding 500 μl sdw per plate and scraping sporangia from the surface of the culture. Sporangia were filtered though sterile cheesecloth, centrifuged, and washed twice with sdw. Concentrations of sporangia were adjusted with a hemacytometer. Five microliters of sporangial suspension was then added to the surface of water agar discs (4-mm diameter) and allowed to be absorbed. Sporangial germination rates were determined by staining sporangia with 0.03% acid fuchsin in 85% lactic acid and viewing them microscopically at ×100 to ×250.
Production of E. cloacae cells.
E. cloacae strains used in this study were the wild type, EcCT-501R3, and two fatty acid metabolic mutants, EcL1 (fadLΔ) and Ec31 (fadBAΔ) (20). Strain EcL1 is unable to transport long-chain fatty acids into the cell but otherwise has a fully functional β-oxidation pathway, and strain Ec31 is a β-oxidation mutant unable to degrade fatty acids. Tryptic soy broth (BD Diagnostics, New Jersey) amended with rifampin (100 μg/ml) and kanamycin (50 μg/ml) when appropriate was inoculated with E. cloacae cells, and cells were grown for 18 h at 30°C and harvested by centrifugation. Cells were washed twice in Tris-buffered saline (pH 7.2) and then resuspended in Tris-buffered saline. Cell density was adjusted to 109 CFU/ml at an optical density at 600 nm (0.56) to make a stock cell suspension. The stock was diluted to make suspensions that would give final concentrations of cells of 104, 106, or 108 CFU/cm3 substrate.
P. ultimum sporangial activation and germination assays.
Activation experiments were designed to establish when sporangia first perceive exudate stimulants, whereas the germination experiments were designed to assess sporangial germ tube emergence. Glass beads (1-cm3 volume, 0.1-mm diameter) were added to wells of Corning 12-well tissue culture plates, and sdw was added to adjust the water content to 18%. A sporangial disc was added to each well, and a surface-sterilized seed was placed on the surface of the disc. Seeds were covered with an additional 3 cm3 of glass beads, and the final water content was brought up to 18%.
For activation assays, sporangial discs were harvested at 30-min intervals up to 3 h and at hourly intervals up to 6 h after exposure to cucumber seeds or at 15-min intervals up to 1 h and at 30-min intervals up to 3 h and with an additional sampling at 4 h after exposure to corn seeds. After retrieval from the glass bead matrix, sporangial discs were further incubated so that the total incubation times (activation times in the spermosphere plus subsequent incubation for germ tube emergence) were 6 and 4 h for sporangia exposed to cucumber and corn, respectively. Sporangial discs in wells containing substrate and water but no seeds served as negative controls. Five replicates were prepared for each treatment, and each experiment was repeated three times.
For germ tube emergence assays, sporangial discs were retrieved, rinsed, and immediately assessed for germination. Sporangial discs in wells containing substrate and water but no seeds served as negative controls. Five replicates were prepared for each treatment, and each experiment was repeated three times. At each sampling time, seeds were removed and blotted dry and the seed weight was recorded.
Activation and germ tube emergence assays were also performed with E. cloacae EcCT-501R3 cells added at 104, 106, or 108 CFU/cm3 substrate. A second set of experiments evaluated E. cloacae strain EcCT-501R3, EcL1, or Ec31 at 108 CFU/cm3 substrate. Sporangial discs in wells containing no seeds but with E. cloacae EcCT-501R3 added at a rate of 108 CFU/cm3 substrate served as negative controls. Positive controls consisted of sporangial discs exposed to seeds, with E. cloacae EcCT-501R3 cells added after disc harvest to match the approximate number of cells from the maximum concentration used in the study. This permitted a check on the level of sporangial activation but also demonstrated that E. cloacae does not significantly reduce germ tube emergence during the incubation period. Three replicates were prepared for each treatment, and each experiment was repeated three times.
Seed colonization incidence.
Seeds were placed in Corning 12-well tissue culture plates as described above. E. cloacae strains EcCT-501R3, EcL1, and Ec31 were added at 108 CFU/cm3 substrate. Seeds in wells containing no sporangia but with E. cloacae EcCT-501R3 added at a rate of 108 CFU/cm3 substrate served as negative controls. Seeds inoculated with P. ultimum sporangia served as positive controls. Seeds were removed at 0, 1, 2, 4, 6, and 8 h after sowing; rinsed; and placed on a medium containing 1% water agar amended with 15 μg/ml each of rifampin and penicillin G. Plates were incubated at 27°C, and the percentage of colonized seeds was assessed after 48 h by observing characteristic colony growth emerging from seeds. Six replicates were prepared for each treatment, and the experiment was repeated three times.
Quantification of P. ultimum seed colonization by qPCR.
Quantitative PCR (qPCR) was used to quantitatively assess seed colonization by determining the amount of P. ultimum biomass on seed surfaces. To equate DNA levels with mycelial biomass equivalents, P. ultimum was grown for 6 days at 27°C on potato dextrose agar (Difco, United States) plates overlaid with cellophane. Mycelium was harvested, weighed, frozen, and lyophilized. After dry weights were determined, lyophilized mycelia were stored at −20°C prior to DNA extraction. DNA was extracted using the Qiagen DNeasy plant minikit (Qiagen Sciences, Maryland) according to the manufacturer's instructions for optimized yield. DNA was quantified using the Quant-iT PicoGreen double-stranded DNA assay kit (Molecular Probes, Oregon). Since increasing amounts of P. ultimum mycelia were linearly correlated with increasing amounts of DNA {r2 = 0.920, P < 0.0001, DNA (ng) = [26.721 × mycelium fresh weight (mg)] + 57.865}, DNA levels were subsequently used as a biomass estimator.
To prepare standards for qPCR analysis, P. ultimum was grown as described above. Mycelial mass was harvested, ground in liquid nitrogen, and transferred to 4 ml extraction buffer (0.05 M EDTA, 0.1 M Tris, 0.5 M NaCl, 0.7% β-mercaptoethanol, 0.25% sodium dodecyl sulfate). Mycelia were lysed at 65°C for 1 h, after which 1.3 ml 5 M potassium acetate was added, and samples were incubated on ice for 20 min. Samples were centrifuged (3,000 rpm, 10 min), 3.2 ml isopropanol was added to supernatants, and they were kept on ice for 30 min. Samples were centrifuged, and pellets containing DNA were air dried and resuspended in TE buffer (0.01 M Tris, 1 mM EDTA). The suspension was treated with 10 μg RNase A at 65°C for 10 min. DNA was then extracted using phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) followed by chloroform-isoamyl alcohol (24:1, vol/vol) and ethanol precipitation. DNA was redissolved in 10 mM Tris (pH 8.0), and the quality and integrity of the DNA were evaluated by gel electrophoresis. DNA was quantified using the Quant-iT PicoGreen double-stranded DNA assay kit and split into aliquots that were stored at −80°C.
To determine seed-colonizing biomass of P. ultimum, seeds were prepared in glass beads and Corning 12-well tissue culture plates as described above. Treatment mixtures consisted of inoculated seeds with either E. cloacae strain EcCT-501R3 or strain Ec31 (108 CFU/cm3) added at the time of sowing for the first experiment and at 2 h after sowing for the second experiment. Nontreated, inoculated seeds served as positive controls, and noninoculated seeds were used as negative controls for all experiments. P. ultimum biomass was measured on both corn and cucumber seeds in the first experiment but only on cucumber in the second experiment. In a third series of seed colonization experiments, cucumber seeds were inoculated with 0, 100, 300, or 600 sporangia per seed (sporangia were obtained as described above). Seeds were removed at 4, 8, and 12 h after sowing; rinsed; cut into six to eight pieces, transferred to cryo-vials; and frozen in liquid nitrogen. Seeds were kept at −80°C until extracted. Seed pieces were ground in a mortar and pestle and with glass beads in liquid nitrogen. DNA was then extracted using the Qiagen DNeasy plant minikit according to the manufacturer's instructions for optimized yield. DNA samples were kept at −20°C until analyzed. Three replicates were prepared for each treatment, and each experiment was repeated twice.
The amount of P. ultimum DNA on and in seeds was measured in triplicate in 96-well plates (Phenix, North Carolina) using qPCR. qPCR mixtures were prepared by mixing 7 μl water, 0.5 μl 10 μM primer AFP276 (5′-TGTATGGAGACGCTGCATT-3′) (5), 0.5 μl 10 μM primer ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (22), 10 μl Platinum Sybr green qPCR SuperMix-uracil DNA glycosylase with 6-carboxyl-X-rhodamine (Invitrogen, California), and 2 μl template. Samples were amplified using the ABI Prism 7000 sequence detection system (Applied Biosystems, California) with the following conditions: initial denaturation at 95°C for 3 min followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 60 s, and a final extension at 72°C for 10 min. A dissociation protocol starting at 60°C was used to assess the presence of primer-dimer formation and other artifacts. Quantification was achieved by comparing cycle threshold (CT) values in samples to CT values of exogenous P. ultimum DNA standards ranging from 1.5 fg to 75 ng. Standards were established for every 96-well plate analyzed, and the relationship between CT and DNA amount was used to calculate the amount of Pythium DNA in seed samples. All regression coefficients were ≥0.98. Slopes from the standard curves were used to calculate the efficiency (E) of the PCR using the formula E = 10[−1/slope] − 1. The average efficiency was 106% (±3%).
Statistical analysis.
The PROC GLM procedure in SAS v9.1 (SAS Institute, Cary, NC) was used for statistical analyses. All assays were analyzed individually and thereafter pooled and reanalyzed. Data sets for inoculated corn and cucumber seeds were also pooled and analyzed for comparison. Seed imbibition, sporangial activation, germ tube emergence, Pythium seed colonization incidence, and Pythium seed colonization biomass data were analyzed using two-way analysis of variance with “water uptake,” “activation percent,” “germination percent,” “colonization percent,” and “amount DNA” as response variables, respectively. “Time” and “treatment” were used as continuous and categorical predictors, respectively. Additionally, simple linear regression (SLR) was performed individually on all response variables with the exception of the “water uptake” variable (pooled data sets) and treatments using “time” as a continuous variable. This was followed by multiple regressions with “treatment” as an additional predictor variable and analysis of covariance (ANCOVA) for comparison of treatment slopes. SLR, multiple regression, and ANCOVA for activation and germination data were performed for data within the linear response so that time points with no activation/germination and full activation/germination were omitted. The relationship between DNA level and Pythium mycelial weight was analyzed using SLR with “amount DNA” as the response variable and “mycelial weight” as the predictor. qPCR standard calibrations of Pythium DNA versus CT values were also analyzed using SLR for estimation of the curve formula. Percentage data were transformed {arcsine [sqrt(p)] where p is the proportion of sporangia activated or with germ tubes and seed colonization incidence} to stabilize the normality and variance when necessary. The response variable “amount DNA” in the Pythium biomass data sets was transformed [−log (pg DNA + 1)] to achieve normality. Appropriate diagnostic plots were established to ensure that all assumptions of the tests were fulfilled and to check for influential points. Insignificant terms (α = 0.05) were dropped in all tested models. Results considered to be significant are comparisons with P < 0.05.
RESULTS
Seed characterizations.
Seed imbibition and germination percentages were used to gauge seed exudation rates and ensure seed viability and seedling quality. The germination rates of both cucumber and corn seeds were >99%. Both cucumber and corn seeds rapidly imbibed a significant amount of water within the first 30 min and continued to take up water over the 12-h sampling period (data not shown). When imbibition rates were expressed as mg H2O/mg seed, no differences were observed between corn and cucumber seeds during the first 4 h of imbibition. Beyond 4 h, significantly greater amounts of water were taken up by corn seeds than by cucumber seeds. Corn seeds weighed an average of 163 (±2.5) mg, which is about seven times greater than the weight of cucumber seeds (25 [±0.6] mg). Therefore, expressing imbibition rates as mg H2O/seed revealed that the overall amount of water imbibed by corn seeds greatly exceeded that of cucumber as early as 30 min after sowing.
P. ultimum sporangial activation and germ tube emergence in the spermosphere.
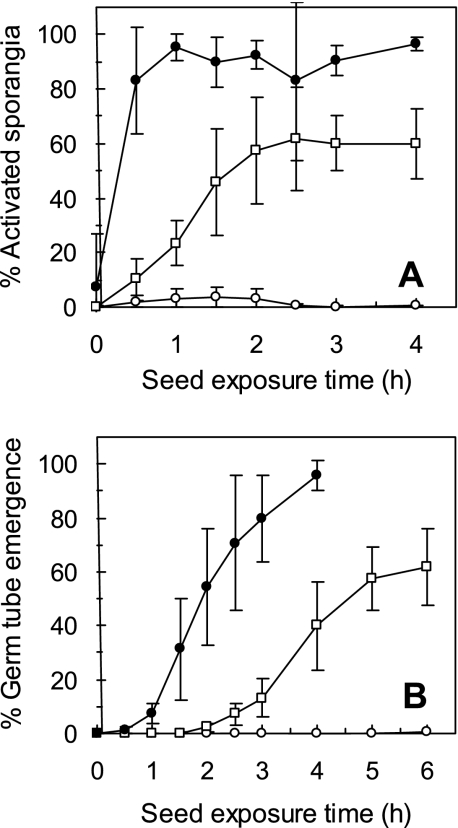
A low but significant percentage of P. ultimum sporangia were activated for germination in the cucumber spermosphere within the first 30 min of exposure to the seed (Fig. 1A). Maximum levels of sporangial activation (60 to 70%) were observed by 1.5 to 2.5 h of exposure to seeds. In contrast, 44% of the sporangia were activated in the corn spermosphere as early as 15 min after exposure to seeds. Maximum activation levels (80 to 100%) were observed as early as 30 min after seed exposure.
FIG. 1.
Activation (A) and germ tube emergence (B) of Pythium sporangia in the corn (•) and cucumber (□) spermospheres and in the absence of seeds (○) during the first 4 and 6 h of seed germination, respectively. Activation was assessed as percent sporangia with visible germ tubes after sporangia were exposed to seeds for various periods of time and then incubated in the absence of seeds, allowing germ tubes to emerge. Each point with the error bars represents the mean ± standard deviation of 15 observations from three repeated experiments.
No significant germ tube emergence (direct evidence for sporangial germination) occurred in the cucumber spermosphere until 2 to 2.5 h of exposure to the seed (Fig. 1B). By 5 to 6 h after exposure to cucumber seeds, the percentage of sporangia with emerged germ tubes reached a maximum level of 60 to 70%. In contrast, a significant percentage of sporangia with emerged germ tubes were observed in the corn spermosphere as early as 1 to 1.5 h after exposure to seeds. Maximum germ tube emergence (>90% germination) was observed after 4 h of exposure to corn seeds.
Sporangial activation and germ tube emergence in response to corn and cucumber seeds increased linearly and significantly (P < 0.0001) over the first few hours of seed germination until the response was maximized (Table 1). The rate of increase in the cucumber spermosphere was significantly lower than that in the corn spermosphere.
TABLE 1.
Regression analysis of P. ultimum sporangial activation and germ tube emergence in response to cucumber and corn seeds during the first 4 h of seed germination
| Assay and seed type | r2a | LS meanc |
|---|---|---|
| Activation | ||
| Corn | 0.803**b | 97.84 a |
| Cucumber | 0.822** | 23.65 b |
| Germ tube emergence | ||
| Corn | 0.910** | 72.16 a |
| Cucumber | 0.922** | 15.59 b |
Regression coefficient for response (activation and germ tube emergence) versus time.
**, significant (P < 0.001) by F test for the association between the response variable and time.
Least-squares (LS) mean estimates from ANCOVA comparing the slopes for the activation and germination assays. Estimates followed by different letters are significantly different.
Effects of E. cloacae cell density and fad mutants on P. ultimum sporangial activation in the spermosphere.
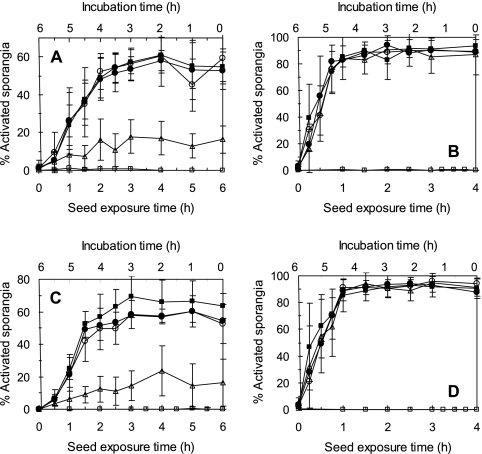
These experiments focused on sporangial activation and not germ tube emergence with the reasoning that the exudates responsible for inducing initial sporangial activation are those that E. cloacae must inactivate to effectively prevent germ tube emergence. Significant reductions in sporangial activation were observed upon introducing E. cloacae cells to cucumber seeds but only when cells were applied at the highest rate of 108 cells/cm3 substrate (Fig. 2A and C). Significant reductions in percentages of activated sporangia were observed at all sampling points up to 6 h (Table 2). The wild-type strain of E. cloacae (strain EcCT-501R3) reduced the percentage of activated sporangia by approximately 45% in the cucumber spermosphere over that for seeds not treated with E. cloacae. However, fad mutants of E. cloacae (strains EcL1 and Ec31) applied at 108 cells/cm3 substrate were ineffective in reducing sporangial activation in the cucumber spermosphere at any time point tested (Fig. 2C; Table 2). No reductions in sporangial activation at any cell density by any of the E. cloacae strains were observed in the corn spermosphere at any sampling time (Fig. 2B and D and Table 2).
FIG. 2.
Activation of Pythium ultimum sporangia in the spermospheres of cucumber and corn in the presence of E. cloacae EcCT-501R3 and fatty acid degradation mutant derivative strains EcL1 and Ec31 during the first 4 (corn) to 6 (cucumber) h of seed germination. (A and B) Sporangial activation in the cucumber (A) and corn (B) spermospheres in response to seeds treated with 0 (▪), 104 (○), 106 (•), or 108 (▵) cells of E. cloacae EcCT-501R3 per cm3 of sand. (C and D) Sporangial activation in the cucumber (C) and corn (D) spermospheres in response to nontested seeds (▪) or seeds treated with 108 cells of E. cloacae EcL1 (○), Ec31 (•), or EcCT-501R3 (▵) per cm3 substrate. Each point represents the mean of nine observations from three repeated experiments. No-seed treatments (□) served as negative controls.
TABLE 2.
Regression analysis of sporangial activation, seed colonization incidence, and seed colonization biomass development by P. ultimum on cucumber and corn seeds treated with E. cloacae strain EcCT-501R3, EcL1, or Ec31
| Seed type and treatment | Activation
|
Colonization
|
||||
|---|---|---|---|---|---|---|
| Incidence
|
Biomass
|
|||||
| r2a | LS meanb | r2a | LS meanb | r2a | LS meanb | |
| Cucumber | ||||||
| Noninoculation | 0.047 (NS) | 0.106 c | 0.000 (NS) | 0.000 a | 0.000 (NS) | 0.000 b |
| Inoculation with strain | 0.920** | 34.05 a | 0.829** | 52.78 a | 0.547** | 406.83 a |
| EcCT-501R3 | 0.538** | 7.14 b | 0.532** | 30.56 b | 0.111 (NS) | 44.02 b |
| EcL1 | 0.853** | 28.21 a | 0.654** | 47.22 a | NDc | ND |
| Ec31 | 0.873** | 28.21 a | 0.709** | 42.59 ab | 0.150 (NS) | 131.22 a |
| Corn | ||||||
| Noninoculation | 0.035 (NS) | 0.079 b | 0.004 (NS) | 1.85 b | 0.000 (NS) | 0.000 b |
| Inoculation with strain | 0.783** | 53.54 a | 0.814** | 48.15 a | 0.797** | 1420.92 a |
| EcCT-501R3 | 0.860** | 48.66 a | 0.797** | 51.85 a | 0.417* | 971.65 a |
| EcL1 | 0.906** | 47.35 a | 0.788** | 53.71 a | ND | ND |
| Ec31 | 0.954** | 47.96 a | 0.555** | 51.85 a | 0.593** | 972.26 a |
Regression coefficients for response (activation, colonization incidence, and colonization biomass) versus time, followed by the significance level of the F test for the association between the response variable and time. NS, nonsignificant; *, P < 0.01; **, P < 0.001.
Least-squares (LS) mean estimates from ANCOVA comparing treatment slopes. Estimates followed by different letters are significantly different.
ND, not determined.
P. ultimum colonization of seeds treated with E. cloacae.
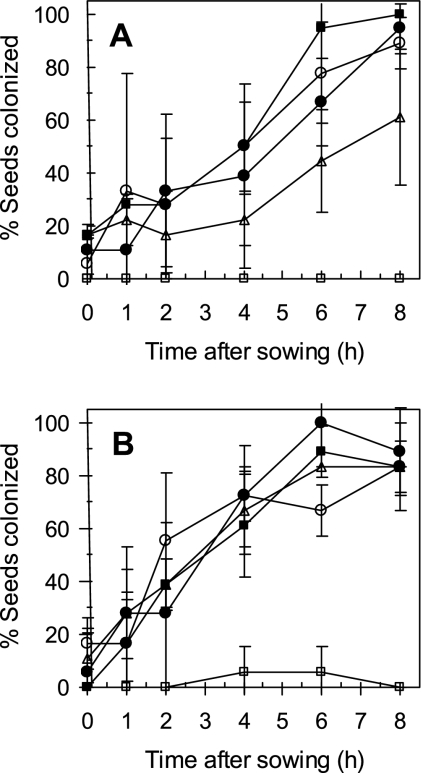
The incidence of corn and cucumber seeds colonized by P. ultimum increased at a constant rate during seed germination, reaching 100% colonization incidence by 6 h after sowing (Fig. 3A and B). No significant differences in colonization incidence between corn and cucumber seeds were observed at any sampling point. Neither strain EcL1 nor Ec31 reduced the incidence of P. ultimum seed colonization in the cucumber spermosphere. E. cloacae strain EcCT-501R3 reduced Pythium colonization incidence in the cucumber spermosphere only at the 8-h sampling point. The rate of increase in seed colonization incidence in the cucumber spermosphere was significantly lower for seeds treated with E. cloacae strain EcCT-501R3 than for nontreated seeds (Table 2). None of the E. cloacae strains reduced seed colonization incidence in the corn spermosphere at any sampling time (Fig. 3B and Table 2).
FIG. 3.
Frequency of cucumber (A) and corn (B) seeds colonized by Pythium ultimum in the presence of E. cloacae strain EcL1 (○), Ec31 (•), or EcCT-501R3 (▵). Controls consisted of noninoculated seeds (□) as well as inoculated seeds not treated with bacteria (▪). Each point represents the mean of three observations (six replicates each) from three repeated experiments.
Biomass of P. ultimum on corn and cucumber seeds treated with E. cloacae.
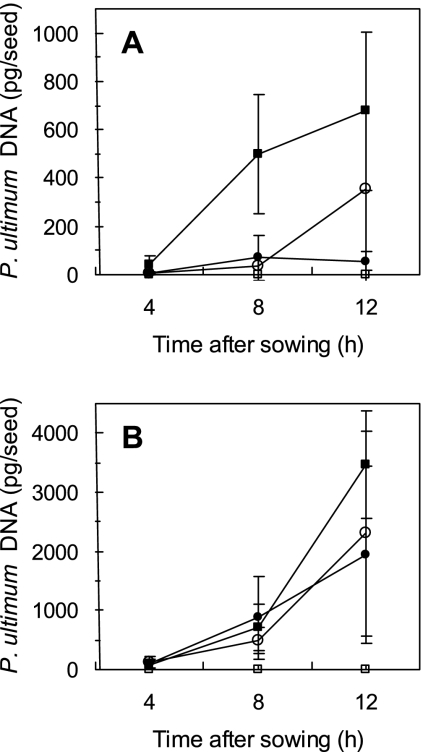
The incidence of seeds colonized by P. ultimum provided little information on the absolute amount of P. ultimum seed colonization. On the other hand, quantitative estimates of P. ultimum biomass provided a better indication of the overall level of seed colonization. P. ultimum was detected on both cucumber (43 pg P. ultimum DNA/seed) and corn (75 pg P. ultimum DNA/seed) seeds as early as 4 h after sowing (Fig. 4A and B). By 8 h after sowing, an approximately 10-fold increase of P. ultimum biomass was detected (501 and 721 pg DNA per cucumber and corn seed, respectively). However, these levels were not significantly different from those at 4 h after sowing. Levels of P. ultimum biomass on cucumber seeds at 12 h after sowing (677 pg P. ultimum DNA/seed at 12 h) did not differ significantly from levels detected at 8 h after sowing. In contrast, levels of P. ultimum biomass on corn seeds were significantly greater by 12 h after sowing (3,470 pg P. ultimum DNA/seed) than those observed at 8 h after sowing. No P. ultimum DNA was detected on noninoculated seeds at any of the sampling times.
FIG. 4.
Pythium ultimum biomass development on cucumber (A) and corn (B) seeds in the presence of E. cloacae strain Ec31 (○) or EcCT-501R3 (•). Seeds were exposed to Pythium for 4, 8, and 12 h, after which amounts of P. ultimum DNA were determined using qPCR. Seeds that were inoculated but not treated with E. cloacae (▪) and noninoculated seeds (□) served as positive and negative controls, respectively. Each point with the error bars represents the mean ± standard deviation of six seeds from two repeated experiments. Note the different y axes.
Introducing E. cloacae strain EcCT-501R3 to cucumber seeds significantly reduced P. ultimum biomass development compared to that observed on nontreated seeds at all time points tested (Fig. 4A). The rate of biomass increase over time was significantly lower on seeds treated with strain EcCT-501R3 than on nontreated seeds (Table 2). P. ultimum biomass on corn was not significantly reduced by either of the two E. cloacae strains (Fig. 4B and Table 2).
Reduction of P. ultimum biomass development on cucumber seeds by E. cloacae strain Ec31 was variable (Fig. 4A). By 4 and 8 h after sowing, significantly lower levels of P. ultimum biomass developed on seeds treated with strain Ec31. However, by 12 h after sowing, levels of P. ultimum biomass developing on treated and nontreated seeds did not differ. Furthermore, the linear rate of biomass increase was not significantly lower in the presence of strain Ec31 than in its absence (Table 2).
Relationship between P. ultimum sporangial activation and cucumber seed colonization.
Adding wild-type cells of E. cloacae to seeds after sporangia were fully activated (2 h after sowing) resulted in no significant decreases in Pythium biomass on seed surfaces compared to that on nontreated seeds at any sampling time after inoculation (Table 3). The rate at which Pythium biomass increased on seeds over time was significantly higher (P < 0.0001) when E. cloacae cells were introduced 2 h after sowing than when cells were added at the time of sowing. Strain Ec31 of E. cloacae behaved similarly to the wild-type strain.
TABLE 3.
Regression analysis of P. ultimum biomass associated with cucumber seeds inoculated 2 h after sowing with E. cloacae strain EcCT-501R3 or Ec31
| Treatment | r2a | LS meanb |
|---|---|---|
| Noninoculation | 0 (NS) | −0.000 b |
| Inoculation with strain | 0.575** | 813.96 a |
| EcCT-501R3 | 0.600** | 485.34 a |
| Ec31 | 0.605** | 610.34 a |
Regression coefficient of the regression line for biomass versus time, followed by the significance level of the F test testing the association between the response variable and time. NS, nonsignificant; **, P < 0.001.
Least-squares (LS) mean estimates from ANCOVA comparing the slopes of treatments. Estimates followed by different letters are significantly different.
Adjusting the potential number of activated sporangia in the spermosphere by exposing cucumber seeds to increasing dosages of sporangia ranging from 0 to 600 sporangia per seed resulted in increasing levels of P. ultimum biomass on cucumber seeds for each sampling time point (Table 4). Biomass levels at each of the sampling times were approximately 10% of those from experiments where sporangial inoculum was not manipulated (i.e., in sporangial disks) (data not shown). Within a given dosage, P. ultimum biomass levels on seeds did not increase significantly over time between 4 and 12 h.
TABLE 4.
Regression analysis of P. ultimum biomass in and on cucumber seeds inoculated with different numbers of sporangia per seed
Regression coefficient of the regression line for biomass versus number of sporangia, followed by the significance level of the F test testing the association between the response variable and time. *, P < 0.01; **, P < 0.001.
Least-squares (LS) mean estimates from ANCOVA comparing the slopes of treatments. Estimates followed by the same letter are significantly similar.
DISCUSSION
The results of this study revealed that P. ultimum sporangia respond to exudates released into the spermospheres of corn and cucumber within 30 min after exposure to seeds, nearly 45 to 75 min before germ tubes emerge. Such an early response indicates that sufficient concentrations of LCUFA must be present in the spermosphere within 30 min after sowing since LCUFA are the only known elicitors of P. ultimum sporangial germination (10). Recent studies of fatty acid exudation from seeds have confirmed that LCUFA, especially oleic and linoleic acids, are released from both corn and cucumber seeds as early as 15 min after sowing (23). Although it is difficult to know the precise concentration and distribution of LCUFA in the spermosphere at any point in time, the calculated concentrations of oleic and linoleic acids are well within the concentrations previously reported to induce high levels of sporangial germination (18, 20). Assuming a spermosphere radius of 5 mm (10) surrounding a 5-mm-diameter seed, the volume of the spermosphere would be approximately 0.5 cm3. In this scenario the combined levels of oleic and linoleic acids released over the first 6 h of germination would yield around 226 μg/cm3, approximately the level known to induce maximum sporangial germination (18, 20).
Given that LCUFA are present in the corn and cucumber spermospheres at stimulatory concentrations as early as 15 to 30 min after sowing, we would predict that the percentage of activated sporangia should be reduced in the cucumber spermosphere but not in the corn spermosphere in the presence of E. cloacae. Although previous studies demonstrated that E. cloacae can reduce the stimulatory activity of cucumber but not corn seed exudates collected for as little as 2 h after initiating seed imbibition in vitro (4), it was not previously known whether exudate inactivation can occur within 30 min in the spermosphere. Our results confirm that exudate inactivation occurs in the cucumber spermosphere but not the corn spermosphere in the presence of E. cloacae within 30 min of sowing, suggesting that E. cloacae actively degrades fatty acid elicitors present in the spermosphere, which can explain the observed reduction in sporangial activation.
Sporangial activation experiments with cucumber and corn in the presence of fatty acid degradation mutants of E. cloacae were carried out to provide further evidence that the failure of E. cloacae to suppress sporangial activation in the corn spermosphere is due to the failure to degrade fatty acid elicitors in the spermosphere. Previous in vitro studies with cottonseed exudates demonstrated that fatty acid degradation by E. cloacae was required to prevent P. ultimum sporangial germination and eliminate subsequent disease development (20, 21). If the degradation of fatty acid elicitors by E. cloacae can explain the reduced sporangial activation in the cucumber spermosphere, such reduced activation should be eliminated in the cucumber spermosphere in the presence of the E. cloacae mutants. Our results support these predictions since both the fatty acid mutants failed to reduce sporangial activation in the cucumber or corn spermospheres at any time after sowing.
Collectively, our observations of sporangial activation in the presence of E. cloacae indicate that the failure of E. cloacae to protect corn seedlings from Pythium damping-off is due not to a lack of synchronization between E. cloacae fatty acid degradation and seed exudation of fatty acids but to other factors. It is more likely that the inability of E. cloacae to protect corn seeds from infection is due to the failure of E. cloacae to degrade fatty acid elicitors released into the spermosphere soon after sowing. It has been shown in other studies (16, 17) as well as in preliminary studies in our lab (S. Windstam and E. B. Nelson, unpublished data) that corn seeds support a high level of growth and activity of E. cloacae in the spermosphere. This suggests that the growth and development of E. cloacae are not inhibited in the corn spermosphere, making it unlikely that the lack of seed protection is due to the general inhibition of E. cloacae metabolism by corn seed exudates. We hypothesize, therefore, that the inability of E. cloacae to reduce P. ultimum infections in the corn spermosphere is due to the suppression of β-oxidation by other components of corn seed exudates. This hypothesis is explored more fully in a companion paper (23).
Successful disease development by P. ultimum is dependent not only on the activation and germination of sporangia in the spermosphere but also on the rapid colonization of seeds and seedling surfaces, ultimately resulting in embryo or seedling infection (11). Our results have shown that P. ultimum colonization levels on cucumber seeds are reduced when E. cloacae is active. Yet, either such reductions in colonization could be due to reduced sporangial activation and germ tube emergence or E. cloacae may continue to interact with P. ultimum mycelia after germ tube emergence to reduce seed colonization and subsequent disease development.
Two approaches were used to determine whether the reduction in P. ultimum biomass by E. cloacae on cucumber seeds was due solely to reductions of sporangial activation. In the first approach, E. cloacae cells were added to cucumber spermospheres only after sporangia were fully activated (2 h). If disruption of sporangial activation is the principal cause of reduced P. ultimum biomass, adding E. cloacae cells after full activation should result in seed colonization levels greater than or equal to those observed on nontreated seeds or seeds coated with E. cloacae at the time of sowing. In the second approach, seeds were exposed to increasing levels of activated sporangia at the time of sowing. If biomass development on the seed was dependent solely on the numbers of activated and germinated sporangia in the spermosphere, there should be a direct correlation between sporangial dosage and biomass development. Such dose-dependent responses have been frequently described for Pythium species (6). Using both approaches, our results confirmed the experimental predictions—P. ultimum seed colonization was not reduced if E. cloacae cells were added after full sporangial activation. Furthermore, increasing the numbers of sporangia to which seeds were exposed resulted in increasing amounts of seed colonization biomass. These findings are significant because they provide further evidence that the behavior of sporangia alone can establish the ultimate levels of seed colonization and subsequent disease incidence and severity. Therefore, reductions in sporangial activation should translate directly into reductions in disease development.
Acknowledgments
This work was supported by grant no. 2002-02343 from the U.S. Department of Agriculture National Research Initiative competitive grants program.
We gratefully acknowledge the technical input from Monica Vencato and Kevin Myers. We are grateful for the technical assistance of Scott Nelson, Allison Jack, and Mary Ann Karp.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Fukui, R., G. S. Campbell, and R. J. Cook. 1994. Factors influencing the incidence of embryo infection by Pythium spp. during germination of wheat seeds in soils. Phytopathology 84:695-702. [Google Scholar]
- 2.Fukui, R., M. N. Schroth, M. Hendson, and J. G. Hancock. 1994. Interaction between strains of pseudomonads in sugar beet spermospheres and their relationship to pericarp colonization by Pythium ultimum in soil. Phytopathology 84:1322-1330. [Google Scholar]
- 3.Hadar, Y., G. E. Harman, A. G. Taylor, and J. M. Norton. 1983. Effects of pregermination of pea and cucumber seeds and of seed treatment with Enterobacter cloacae on rots caused by Pythium spp. Phytopathology 73:1322-1325. [Google Scholar]
- 4.Kageyama, K., and E. B. Nelson. 2003. Differential inactivation of seed exudate stimulation of Pythium ultimum sporangium germination by Enterobacter cloacae influences biological control efficacy on different plant species. Appl. Environ. Microbiol. 69:1114-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lievens, B., M. Brouwer, A. C. R. C. Vanachter, C. A. Levesque, B. P. A. Cammue, and B. P. H. J. Thomma. 2005. Quantitative assessment of phytopathogenic fungi in various substrates using a DNA macroarray. Environ. Microbiol. 7:1698-1710. [DOI] [PubMed] [Google Scholar]
- 6.Martin, F. N., and J. E. Loper. 1999. Soilborne plant diseases caused by Pythium spp.: ecology, epidemiology, and prospects for biological control. Crit. Rev. Plant Sci. 18:111-181. [Google Scholar]
- 7.McKellar, M. E., and E. B. Nelson. 2003. Compost-induced suppression of Pythium damping-off is mediated by fatty-acid-metabolizing seed-colonizing microbial communities. Appl. Environ. Microbiol. 69:452-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson, E. B. 1988. Biological control of Pythium seed rot and preemergence damping-off of cotton with Enterobacter cloacae and Erwinia herbicola applied as seed treatments. Plant Dis. 72:140-142. [Google Scholar]
- 9.Nelson, E. B. 1990. Exudate molecules initiating fungal responses to seeds and roots. Plant Soil 129:61-73. [Google Scholar]
- 10.Nelson, E. B. 2004. Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 42:271-309. [DOI] [PubMed] [Google Scholar]
- 11.Nelson, E. B. 2006. Rhizosphere regulation of preinfection behavior of oomycete plant pathogens, p. 311-341. In K. G. Mukerji, C. Manoharachary, and J. Singh (ed.), Microbial activity in the rhizosphere. Springer-Verlag, Berlin, Germany.
- 12.Nelson, E. B., W. L. Chao, J. M. Norton, G. T. Nash, and G. E. Harman. 1986. Attachment of Enterobacter cloacae to hyphae of Pythium ultimum: possible role in biological control of Pythium pre-emergence damping-off. Phytopathology 76:327-335. [Google Scholar]
- 13.Nelson, E. B., and C. M. Craft. 1989. Comparative germination of culture-produced and plant-produced sporangia of Pythium ultimum in response to soluble seed exudates and exudate components. Phytopathology 79:1009-1013. [Google Scholar]
- 14.Nelson, E. B., and J. S. T. Hsu. 1994. Nutritional factors affecting responses of sporangia of Pythium ultimum to germination stimulants. Phytopathology 84:677-683. [Google Scholar]
- 15.Osburn, R. M., and M. N. Schroth. 1988. Effect of osmopriming sugar beet seed on exudation and subsequent damping-off caused by Pythium ultimum. Phytopathology 78:1246-1250. [Google Scholar]
- 16.Roberts, D. P., P. D. Dery, I. Yucel, J. Buyer, M. A. Holtman, and D. Y. Kobayashi. 1999. Role of pfkA and general carbohydrate catabolism in seed colonization by Enterobacter cloacae. Appl. Environ. Microbiol. 65:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts, D. P., P. D. Dery, I. Yucel, and J. S. Buyer. 2000. Importance of pfkA for rapid growth of Enterobacter cloacae during colonization of crop seeds. Appl. Environ. Microbiol. 66:87-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruttledge, T. R., and E. B. Nelson. 1997. Extracted fatty acids from Gossypium hirsutum stimulatory to the seed-rotting fungus, Pythium ultimum. Phytochemistry 46:77-82. [Google Scholar]
- 19.Stanghellini, M. E., and J. G. Hancock. 1971. Radial extent of the bean spermosphere and its relation to the behavior of Pythium ultimum. Phytopathology 61:165-168. [Google Scholar]
- 20.van Dijk, K., and E. B. Nelson. 2000. Fatty acid competition as a mechanism by which Enterobacter cloacae suppresses Pythium ultimum sporangium germination and damping-off. Appl. Environ. Microbiol. 66:5340-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Dijk, K., and E. B. Nelson. 1998. Inactivation of seed exudate stimulants of Pythium ultimum sporangium germination by biocontrol strains of Enterobacter cloacae and other seed-associated bacteria. Soil Biol. Biochem. 30:183-192. [Google Scholar]
- 22.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis et al. (ed.), PCR protocols: a guide to methods and applications, vol. 18. Academic Press, Inc., San Diego, CA. [Google Scholar]
- 23.Windstam, S., and E. B. Nelson. 2008. Temporal release of fatty acids and sugars in the spermosphere: impacts on Enterobacter cloacae-induced biological control. Appl. Environ. Microbiol. 74:4292-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xi, K., J. H. G. Stephens, and S. F. Hwang. 1995. Dynamics of pea seed infection by Pythium ultimum and Rhizoctonia solani—effects of inoculum density and temperature on seed rot and preemergence damping-off. Can. J. Plant Pathol. 17:19-24. [Google Scholar]