Abstract
Listeria monocytogenes is a food pathogen that can attach on most of the surfaces encountered in the food industry. Biofilms are three-dimensional microbial structures that facilitate the persistence of pathogens on surfaces, their resistance toward antimicrobials, and the final contamination of processed goods. So far, little is known about the structural dynamics of L. monocytogenes biofilm formation and its regulation. The aims of this study were, by combining genetics and time-lapse laser-scanning confocal microscopy (LSCM), (i) to characterize the structural dynamics of L. monocytogenes EGD-e sessile growth in two nutritional environments (with or without a nutrient flow), and (ii) to evaluate the possible role of the L. monocytogenes agr system during biofilm formation by tracking the spatiotemporal fluorescence expression of a green fluorescent protein (GFP) reporter system. In the absence of nutrient flow (static conditions), unstructured biofilms composed of a few layers of cells that covered the substratum were observed. In contrast, when grown under dynamic conditions, L. monocytogenes EGD-e biofilms were highly organized. Indeed, ball-shaped microcolonies were surrounded by a network of knitted chains. The spatiotemporal tracking of fluorescence emitted by the GFP reporter system revealed that agr expression was barely detectable under static conditions, but it progressively increased during 40 h under dynamic conditions. Moreover, spatial analysis revealed that agr was expressed preferentially in cells located outside the microcolonies. Finally, the in-frame deletion of agrA, which encodes a transcriptional regulator, resulted in a decrease in initial adherence without affecting the subsequent biofilm development.
Biofilms are communities of microorganisms attached to a surface (44). Several steps can be identified during biofilm development. After an initial step of reversible, then irreversible, adherence, bacteria grow as microcolonies and spread on the surface; also, biofilms develop as complex, three-dimensional (3D) structures during the maturation step (17). The microorganisms undergo profound changes during their transition from planktonic cells (free-floating organisms) to cells that are part of a complex, surface-attached community (sessile organisms) (44), and cells develop an increased resistance to antimicrobial agents (34, 38, 64). For this reason, the removal of established biofilms requires harsh treatments, with most using oxidizing biocides (18, 20, 25, 54). The presence of biofilms raises safety issues in the food industry, especially when biofilms are located on food-processing surfaces and pipelines that are unreachable by washing agents (8). Clearly, biofilms can become a health hazard by harboring pathogenic bacteria such as Listeria monocytogenes (4). Moreover, L. monocytogenes is capable of attaching and developing biofilms on a variety of surfaces, such as stainless steel, polymers, and rubber gaskets (2, 3, 13, 36, 40). L. monocytogenes is a gram-positive human pathogen that is responsible for serious infections among immunocompromised individuals and pregnant women (19).
Several systems to study biofilms have been used to identify and characterize the bacterial elements and genetic determinants involved in biofilm development. For instance, plate counting has been used to quantify sessile cells of L. monocytogenes on abiotic surfaces (1, 5-7, 10, 12, 13, 32, 43, 47, 52, 53). Also, biofilms of L. monocytogenes have been quantified by using the microplate adhesion method (3, 11, 23, 33, 48), while their structures have been investigated by scanning electron microscopy (SEM) (3, 12, 13, 28, 32, 39, 49), epifluorescence microscopy (9, 24, 28, 36, 37, 41, 46), or laser-scanning confocal microscopy (LSCM) (10, 31, 51). LSCM has allowed the visualization of fully hydrated samples and has revealed the elaborate 3D structure of biofilms (14). The generally accepted structure of Pseudomonas aeruginosa biofilm is composed of mushroom-shaped microcolonies, in which cells are embedded in an exopolymeric cement and the microcolonies are separated by channels and voids (29, 58). Reports concerning the structure of L. monocytogenes biofilm are available. Few studies have used LSCM to investigate the structure of L. monocytogenes biofilm (10, 31, 51).
Biofilm development and maturation requires complex cellular mechanisms in which cell-cell communication is involved. Among these, the agr system plays a role during the biofilm development of Staphylococcus aureus (61, 65), Enterococcus faecalis (22), and Lactobacillus plantarum (57) and during the early stages of the biofilm formation of L. monocytogenes (48). Indeed, the deletion of the gene coding for the response regulator AgrA impaired the ability of L. monocytogenes EGD-e to adhere to abiotic surfaces (48).
To characterize the biofilm development of L. monocytogenes, several models of growth, media, and bacterial strains from the literature were used, and the influence of several environmental parameters were assessed. For example, the temperature of incubation affects L. monocytogenes biofilm growth (7, 52, 53). In the present study, we investigated the kinetics of L. monocytogenes EGD-e development in biofilms by LSCM under two different environmental conditions; i.e., static and flowing systems. Second, we determined the agr expression in situ during biofilm development by using the green fluorescent protein (GFP) reporter.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study and their characteristics are shown in Table 1. L. monocytogenes was cultivated in tryptic soy broth (TSB; Biokar Diagnostics, Pantin, France) at 25°C for biofilm and planktonic cultures and in brain heart infusion broth (BHI; Biokar Diagnostics) at 40°C for strain construction. Escherichia coli TOP10 (Invitrogen, Cergy Pontoise, France) cells were grown aerobically in Luria-Bertani broth (Biokar Diagnostics) at 37°C. When appropriate, antibiotics (Sigma, St. Quentin Fallavier, France) were added as follows: kanamycin, 50 μg·ml−1 (for E. coli); ampicillin, 200 μg·ml−1 (for E. coli); chloramphenicol, 10 μg·ml−1 or 7.5 μg·ml−1 (for L. monocytogenes) (Table 1).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Bacterial strain, plasmid, or oligonucleotide | Relevant property(ies)a | Created restriction site | Reference or source |
|---|---|---|---|
| Bacteria | |||
| E. coli TOP10 | Cloning host | Invitrogen | |
| L. monocytogenes EGD-e | Wild type of serotype 1/2a, for which the genome sequence is available | 42 | |
| L. monocytogenes DG125A | EGD-e with in-frame deletion of agrA gene | 48 | |
| L. monocytogenes AR009 | Emr, Cmr derivative of EGD-e harboring pGID128 in integrative form | This work | |
| L. monocytogenes AR011 | Emr, Cmr derivative of DG125A harboring pGID128 in integrative form | This work | |
| Plasmids | |||
| pGF-EM | Emr, Cmr, Amr; 9.4-kb derivative of pCON-1 carrying 0.8-kb gfp gene of pKV111 and erm gene of Tn917 | 35 | |
| pGID113 | Kmr Amr, 4.7-kb derivative of pCRII-TOPO carrying 0.7-kb Pagr region promoter | This work | |
| pGID120 | Emr,Cmr, Amr; 10.4-kb derivative of pGF-EM carrying 0.6-kb Pagr region promoter inserted into HindIII/XbaI site of pGID113 | This work | |
| pGID128 | Emr, Cmr, Amr; derivative of pGID120 carrying the Pagr-gfp translational fusion obtained after restriction by XbaI/NheI | This work | |
| Primers (5′→3′)b | |||
| AGRB5 | TGAGCTATGAAGACGCGATT | ||
| AGRB6 | ATCTAGACAACTAATTCACCTCCACTA | XbaI |
Emr, erythromycin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; and Amr, ampicillin resistant.
Specific restriction sites are underlined, and extra nucleotides added to include restriction sites in the PCR product are shown in boldface.
Strain and plasmid construction.
The reporter plasmid pGID128 contains a fusion of the agr promoter region (located 690 bp upstream of the start codon) with gfp, the gene coding for GFP. First, the agr promoter region was amplified by PCR with primers AGRB5 and AGRB6 (Invitrogen) (Table 1) and Taq DNA polymerase (Q-Biogene, MP Biomedicals, Illkirch, France) using L. monocytogenes EGD-e genomic DNA as a template. This PCR product was cloned into the pCRII-TOPO vector (Invitrogen) to obtain pGID113, and this vector was transferred into chemically competent E. coli TOP10 cells as recommended by the manufacturer (Invitrogen). Second, plasmid pGID113 was digested with HindIII/XbaI (Invitrogen), and the resulting 617-bp fragment containing the promoter region of the agr operon (Pagr) was ligated into pGF-EM (35) that had been restricted with HindIII/XbaI (Invitrogen) to obtain pGID120. Third, this pGID120 plasmid was digested with XbaI/NheI (Invitrogen) to obtain pGID128. The strains EGD-e and DG125A (ΔagrA) (48) were transformed by pGID128, and the transformants were selected on BHI agar plates (Biokar diagnostics) containing 10 μg·ml−1 chloramphenicol. These strains were named AR009 and AR011 (ΔagrA), respectively, and harbored plasmid pGID128 in the integrative form.
Biofilm growth conditions. (i) Static biofilm experiments.
AISI 304 stainless steel chips (25 by 25 by 1 mm; Goodfellow SARL, Lille, France) were inserted in separate 55-mm-diameter petri dishes (Dominique Dutscher S.A., Brumath, France) that contained 8 ml of TSB. Chloramphenicol (7.5 μg·ml−1) was added to the growth medium to ensure the stable carriage of plasmid pGID128. An overnight culture of strains AR009 and AR011 (ΔagrA) in TSB at 25°C was used to inoculate (1/100, vol/vol) TSB in petri dishes, which were incubated at 25°C. The medium was removed after 2 h, and then every 24 h fresh TSB (8 ml) was added. Cell adhesion and biofilm development were evaluated by microscopic observations after 2, 24, and 48 h at 25°C. Following incubation, the medium was removed and 8 ml of saline solution (150 mM NaCl) was gently poured onto the chips to remove loosely adhering bacteria. Sessile cells were stained with a 0.01% SYTO61 (Molecular Probes, Invitrogen) solution. This stain penetrates and stains nucleic acids in both live and dead cells. Three independent experiments were performed.
(ii) Flow-cell biofilm experiments.
Biofilms were cultivated in flow cell BST FC 81 (Biosurface Technologies Corporation, Bozeman, MT), with channel dimensions of 1.6 by 12.7 by 47.5 mm. The flow cells are small continuous-flow systems with a glass viewing port that allows the direct observation of the biofilms without disrupting the community. Flow chambers were inoculated with overnight cultures of the AR009 and AR011 (ΔagrA) strains in TSB medium (1/100, vol/vol). After inoculation, the medium flow was stopped for 1 h to allow bacterial adhesion, and thereafter the medium was pumped through the flow cells at 10 ml/h by using a peristaltic pump (model 205S; Watson Marlow, Falmouth, England, United Kingdom). Flow-cell biofilms were stained with the nucleic acid stain SYTO61 (http://probes.invitrogen.com/lit/catalog/3/sections/2426.html) after 16, 24, and 40 h of incubation at 25°C before microscopic observations. Two independent experiments with two replicates each were made.
LSCM and image processing.
Image acquisition was performed on a Leica TCS SP2 AOBS (Leica Microsystems, France) laser-scanning confocal laser microscope on the MIMA2 microscopic platform (http://voxel.jouy.inra.fr/mima2). LSCM allowed the simultaneous 3D monitoring of GFP and SYTO61 dyes. The excitation wavelength used for GFP was 488 nm, and the emitted fluorescence was collected in the range of 500 to 600 nm. The red fluorescent nucleic acid stain SYTO61 was excited at 633 nm, and the emitted fluorescence was collected in the range of 650 to 700 nm. Images were collected through a ×63 Leica oil immersion objective (numeric aperture, 1.4). Simulated 3D fluorescence projections and vertical cross-sections through the biofilms were generated using the LSCM 3D package of the Leica SP2 software. The number of sessile cells and the percentage of cells expressing GFP were quantified with light emission intensity using PHLIP and the Matlab toolbox developed by Joao Xavier (www.phlip.org). This software was used to evaluate biofilm structural parameters (biovolume and thickness). For each experiment, five microscopic fields, chosen haphazardly, were analyzed.
The fluorescence loss in photobleaching (FLIP) of intracytoplasmic GFP was used to evaluate if the chains were single elongated cells or separate short rods (59). A fluorescent L. monocytogenes chain was repeatedly photobleached for ∼60 s within a small region while the whole chain was continuously imaged. Any connected cell in the area of the chain being bleached gradually loses fluorescence due to the lateral movement of mobile GFP into this area. By contrast, the fluorescence in unconnected cells is not affected.
Statistical analysis.
A one-way analysis of variance was performed using the SigmaStat, version 3.0.1, software (SPSS Inc.) in order to test the significance of the differences in biovolume or the thickness of biofilms. When the result of the one-way analysis of variance was significant, the Holm-Sidak test (P < 0.05) was used to locate significant differences.
RESULTS
A network of knitted chains of L. monocytogenes EGD-e were observed only under flow conditions.
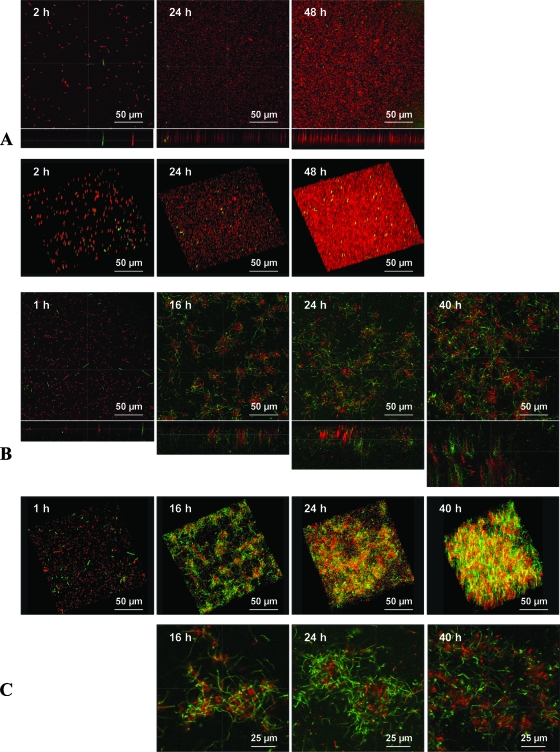
The sessile growth of L. monocytogenes AR009 was monitored under static and flowing conditions using LSCM. Under static conditions, after 2 h of contact with the surface, a few cells were scattered on the surface (Fig. 1A). After 24 h of growth, most of the surface was covered by a monolayer of cells (Fig. 1A). After 48 h of incubation, several layers of cells covered the surface as a uniform biofilm. The 3D reconstruction of the LSCM image stacks confirmed these results of a uniform biofilm that can be described as unorganized multicellular layers of cells covering the surface (Fig. 1A). The structure of the biofilm grown under flowing conditions was different. In fact, after the initial adhesion of L. monocytogenes AR009 as single cells, the biofilm developed as a complex structure in which dense, ball-shaped microcolonies separated by poorly colonized zones were observed (16 h) (Fig. 1B). These structures were thicker after 24 and 40 h of incubation (Fig. 1B). Observations under a stronger magnification and 3D analysis evidenced that these microcolonies were surrounded by a network of knitted chains (Fig. 1 and C).
FIG. 1.
Scanning confocal micrographs (upper lines) and 3D reconstruction of a z stack (lower lines) of biofilms formed by L. monocytogenes AR009 in TSB medium at 25°C under static conditions after 2, 24, and 48 h of incubation (A) and under flowing conditions after 1, 16, 24, and 40 h of incubation (B). (C) Biofilms grown under flowing conditions and observed at a higher magnification. Green indicates those cells expressing GFP from the reporter plasmid pGID128, which contains a fusion of the agr promoter region with gfp. The red cells are stained with the nucleic acid SYTO61 dye (cells lacking agr activity).
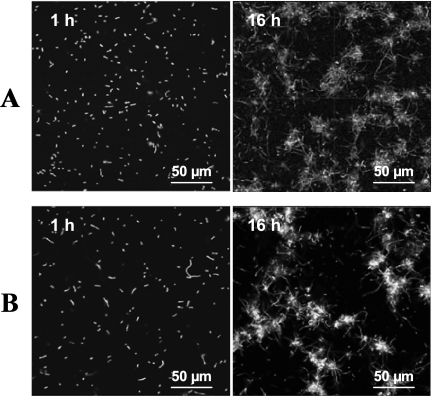
Surprisingly, the morphology of L. monocytogenes was affected by the environmental growth conditions; while short rods typical of L. monocytogenes morphology were seen under static conditions (Fig. 1A), long chains formed the network of knitted chains under dynamic conditions (Fig. 1B and C). FLIP results confirmed by photobleaching that these structures were chains of short rods and not elongated filamentous cells (Fig. 2). These results pointed to a distinct structure of biofilm that was composed of a network of knitted chains when L. monocytogenes was grown under flow conditions.
FIG. 2.
FLIP of intracytoplasmic GFP within chains. A fluorescent L. monocytogenes chain was repeatedly photobleached within a small region (indicated by a gray circle) for ∼60 s while the whole chain was continuously imaged. Single elongated cells being photobleached would gradually lose fluorescence due to the lateral movement of mobile GFP into this area. By contrast, the fluorescence in separate short rods near the area being bleached would not be affected.
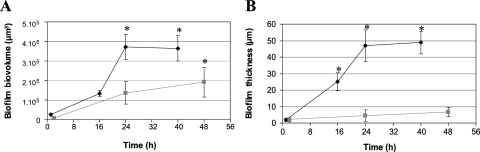
The influence of the hydrodynamic conditions on the structure of the biofilm also was observed using two structural parameters: biofilm volume (Fig. 3A) and thickness (Fig. 3B). Indeed, the biofilm's volume increased during incubation irrespective of the hydrodynamic conditions. Under dynamic conditions, a significant increase (P < 0.05) was observed 24 h after the beginning of the experiment, but the differences were no longer significant after that time. Under static conditions, the increase was significant (P < 0.05) at 48 h (Fig. 3A). Interestingly, under dynamic conditions, the biofilm's volume was significantly higher than the volume that was calculated under static conditions from 24 to 48 h (P < 0.05) (Fig. 3A). On the other hand, biofilm thickness increased during growth under dynamic conditions (P < 0.05), but the differences were not significant under static conditions (Fig. 3B). Furthermore, under dynamic conditions, the biofilm's thickness was significantly increased (P < 0.05) after 16, 24, and 40 h of sessile growth (Fig. 3B).
FIG. 3.
Biovolume (A) and thickness (B) of L. monocytogenes AR009 biofilms in TSB medium at 25°C under static conditions after 2, 24, and 48 h of incubation (square on the gray line) and under flowing conditions after 1, 16, 24, and 48 h of incubation (diamond on the black line). For these quantitative analyses, images obtained by LSCM were analyzed using the PHLIP Matlab routine. The data represent the means and standard deviations from three independent experiments with five measurements for each point. Asterisks indicate statistically significant differences (P < 0.05).
agr expression in L. monocytogenes biofilms.
We have recently demonstrated that the agr system is involved in the onset of L. monocytogenes biofilm (48). In order to investigate the expression of the agr genes, we used GFP as a reporter. Strain AR009 harbors the reporter plasmid pGID128, in which the promoter region required for agr-dependent expression is fused to the gfp gene. Under static conditions, irrespective of the length of incubation (2, 24, or 48 h), only a few cells expressing GFP were detected (no more than 1% of the total number of cells expressing agr after 48 h) (Fig. 1A). In contrast, during the 40 h of incubation in flowing conditions, agr expression increased progressively over time. Indeed, the percentages of cells expressing agr were, respectively, 15, 50, 76, and 80% of the total number of cells after 1, 16, 24, and 40 h of growth (Fig. 1B). Most of the GFP fluorescence was observed within the network of knitted chains surrounding the ball-shaped microcolonies, whereas bacteria inside the microcolonies showed very little GFP fluorescence (Fig. 1C).
The agr system in biofilm development of L. monocytogenes.
The role of the agr system during biofilm development in flow cells was examined by comparing the behavior of strain AR011, an agrA in-frame deletion mutant, to the behavior of AR009. The deletion of agrA, the gene coding for the response regulator AgrA, affected the adhesion of L. monocytogenes to the surface, as previously described (48) (Fig. 4). The number of adhered cells of strain AR011 (ΔagrA) was 3.5-fold lower. However, after 16 h of incubation, the differences between the sessile growth of AR009 and AR011 were not significant (Fig. 4). Moreover, a similar network of knitted chains was observed irrespective of the strain examined. These results confirmed the involvement of the agr system during the adhesion of L. monocytogenes but probably not during the later stages of biofilm development.
FIG. 4.
Scanning confocal micrographs of biofilms formed by L. monocytogenes AR009 (A) and L. monocytogenes AR011 (ΔagrA) (B) in TSB medium at 25°C under flowing conditions after 1 and 16 h of incubation. The images were acquired using LSCM settings with a ×63 objective.
DISCUSSION
The dynamics of L. monocytogenes biofilm formation and its regulation still is mostly unknown. The literature related to the biofilm structures of L. monocytogenes deals with, in most cases, static experiments using SEM (3, 12, 13, 28, 32, 39, 49). LSCM is a powerful in situ visualization tool for the temporal study of the 3D structures of biofilms (27, 29, 30, 45, 55, 58) and has been used to study the biofilm structure of a variety of microorganisms, such as Pseudomonas aeruginosa and S. aureus (26, 29). Other studies often mention that the structure of microbial biofilms can vary in response to environmental conditions such as nutrient limitation, flow rate, shear, and pressure (56, 63). In this study, we used LSCM to characterize and compare L. monocytogenes biofilms grown under two environmental conditions (growth medium flow or no flow) and to investigate agr-dependent gene expression in L. monocytogenes biofilms.
Significantly greater biofilm volume and thickness were observed under flowing conditions than under static conditions. Our observations of the biofilm under static conditions showed unorganized 3D structures that were similar to those observed previously (3, 12, 13). In contrast, for the biofilm under flow conditions we described another organization of L. monocytogenes EGD-e biofilms that consisted of a network of knitted chains that could structure the microcolonies. The mature biofilms of many species, such as P. aeruginosa, that are grown in flow cells display structures that have been described as mushroom-like, with a cohesive polymeric matrix interconnected between water channels (29). These studies indicated that surface-associated motility and biosurfactant production both affect the structure of P. aeruginosa biofilms (30, 45). In this study, the newly identified structure that we observed in L. monocytogenes EGD-e biofilms could contribute to the 3D structure by interconnecting the ball-shaped microcolonies.
To examine the role of the L. monocytogenes agr system during biofilm development and its possible role in the biofilm structure observed in flowing conditions, an agr fusion to a GFP reporter was generated. Using LSCM, this construction allowed us to determine both temporal and spatial gene expression patterns throughout biofilm formation. Our findings revealed that agr gene expression increased progressively over the incubation period. In terms of spatial expression throughout the stratified biofilm, the agr gene activity of L. monocytogenes was maximal in cells outside ball-shaped microcolonies. It is intriguing that although the in-frame deletion of agrA, which encodes a transcriptional regulator, affected adhesion, no differences were observed after the first step of biofilm development or in biofilm structure (48). agr-dependent regulation may be transitory; it may suggest that following adhesion, L. monocytogenes undergoes profound gene expression alterations during sessile growth, as has been proposed for other microorganisms (21, 24, 60, 62). In the orthologous agr system of S. aureus, the expression of agr was patched within cell clusters and oscillated with time (65). Moreover, the role of the agr system during S. aureus development depends on the hydrodynamic conditions of the experimental setup; under static conditions, the disruption of agr expression enhanced biofilm formation (61, 65), while under dynamic conditions it had no influence on biofilm formation (65). In P. aeruginosa, las and rhl, two systems of cell-to-cell signaling, control biofilm formation; the las system is involved in early biofilm development, whereas the rhl system is implicated in the maturation of the biofilm (15, 50). De Kievit et al. (16), demonstrated by spatial analysis that lasI and rhlI were maximally expressed in cells located at the substratum and that expression decreased with increasing biofilm height. These authors suggested that the increased accumulation of autoinducers at the substratum resulted in the expression of both of these systems. For L. monocytogenes, it is proposed that cells were not enclosed in a matrix and the autoinducers could diffuse away from the cells.
In conclusion, our work has yielded a better understanding of L. monocytogenes EGD-e biofilm formation. Our data described, for the first time, the existence of a new structure that consists of a network of knitted chains during the growth of L. monocytogenes EGD-e in a flowthrough system. We demonstrate that the model used to grow L. monocytogenes biofilms (dynamic flow-cell and static models) deeply affects their structure and the spatiotemporal patterns of gene expression. The agr-dependent expression of L. monocytogenes in biofilm differed from that of an S. aureus orthologous system previously described. Deciphering the mechanisms involved in the development of the network of knitted chains that could structure the sessile growth of L. monocytogenes EGD-e is an exciting prospect and will contribute to our better understanding of the ecology of this pathogen.
Acknowledgments
This work was supported by the Ministère de l'Education Nationale de la Recherche et de la Technologie, the Institut National de la Recherche Agronomique, the Université de Bourgogne, and the PRA Listeria program. We thank the local government of the Département de Essonnes for the financial support for a laser-scanning confocal microscope (ASTRE no. A02137).
We thank S. Kathariou for providing the pGF-EM vector.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Beresford, M. R., P. W. Andrew, and G. Shama. 2001. Listeria monocytogenes adheres to many materials found in food-processing environments. J. Appl. Microbiol. 90:1000-1005. [DOI] [PubMed] [Google Scholar]
- 2.Blackman, I. C., and J. F. Frank. 1996. Growth of Listeria monocytogenes as a biofilm an various food-processing surfaces. J. Food Prot. 59:827-831. [DOI] [PubMed] [Google Scholar]
- 3.Borucki, M. K., J. D. Peppin, D. White, F. Loge, and D. R. Call. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 69:7336-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bower, C. K., J. McGuire, and M. A. Daeschel. 1996. The adhesion and detachment of bacteria and spores on food-contact surfaces. Trends Food Sci. Technol. 71:152-157. [Google Scholar]
- 5.Brackett, R. E. 1992. Shelf stability and safety of fresh produce as influenced by sanitation and disinfection. J. Food Prot. 55:808-814. [DOI] [PubMed] [Google Scholar]
- 6.Briandet, R., V. Leriche, B. Carpentier, and M.-N. Bellon-Fontaine. 1999. Effect of the growth procedure on the surface hydrophobicity of Listeria monocytogenes cells and their adhesion to stainless steel. J. Food Prot. 62:994-998. [DOI] [PubMed] [Google Scholar]
- 7.Briandet, R., T. Meylheuc, C. Maher, and M.-N. Bellon-Fontaine. 1999. Listeria monocytogenes ScottA: cell surface charge, hydrophobicity, and electron donor and acceptor characteristics under different environmental growth conditions. Appl. Environ. Microbiol. 65:5328-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpentier, B., and O. Cerf. 1993. Biofilms and their consequences, with particular reference to hygiene in the food industry. J. Appl. Microbiol. 75:499-511. [DOI] [PubMed] [Google Scholar]
- 9.Carpentier, B., and D. Chassaing. 2004. Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int. J. Food Microbiol. 97:111-122. [DOI] [PubMed] [Google Scholar]
- 10.Chae, M. S., and H. Schraft. 2000. Comparative evaluation of adhesion and biofilm formation of different Listeria monocytogenes strains. Int. J. Food Microbiol. 62:103-111. [DOI] [PubMed] [Google Scholar]
- 11.Challan Belval, S., L. Gal, S. Margiewes, D. Garmyn, P. Piveteau, and J. Guzzo. 2006. Assessment of the roles of LuxS, S-ribosyl homocysteine, and autoinducer 2 in cell attachment during biofilm formation by Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 72:2644-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chavant, P., B. Gaillard-Martinie, and M. Hébraud. 2004. Antimicrobial effects of sanitizers against planktonic and sessile Listeria monocytogenes cells according to the growth phase. FEMS Microbiol. Lett. 236:241-248. [DOI] [PubMed] [Google Scholar]
- 13.Chavant, P., B. Martinie, T. Meylheuc, M.-N. Bellon-Fontaine, and M. Hebraud. 2002. Listeria monocytogenes LO28: surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl. Environ. Microbiol. 68:728-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 16.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dornseiffen, J. W. 1998. Residue aspects of disinfectants used in the food industry. Int. Biodeterior. Biodegrad. 41:309-312. [Google Scholar]
- 19.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank, J. F., and R. A. Chmielewski. 1997. Effectiveness of sanitation with quaternary ammonium compound or chlorine on stainless steel and other domestic food-preparation surfaces. J. Food Prot. 60:43-47. [DOI] [PubMed] [Google Scholar]
- 21.Gilmore, K. S., P. Srinivas, D. R. Akins, K. L. Hatter, and M. S. Gilmore. 2003. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect. Immun. 71:4759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harvey, J., K. P. Keenan, and A. Gilmour. 2007. Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol. 24:380-392. [DOI] [PubMed] [Google Scholar]
- 24.Hefford, M. A., S. D'Aoust, T. D. Cyr, J. W. Austin, G. Sanders, E. Kheradpir, and M. L. Kalmokoff. 2005. Proteomic and microscopic analysis of biofilms formed by Listeria monocytogenes 568. Can. J. Microbiol. 51:197-208. [DOI] [PubMed] [Google Scholar]
- 25.Holah, J. T., J. H. Taylor, D. J. Dawson, and K. E. Hall. 2002. Biocide use in the food industry and the disinfectant resistance of persistent strains of Listeria monocytogenes and Escherichia coli. J. Appl. Microbiol. 92(Symp. Suppl.):111S-120S. [PubMed] [Google Scholar]
- 26.Jefferson, K. K., D. A. Goldmann, and G. B. Pier. 2005. Use of confocal microscopy to analyse the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 49:2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, S. M., J. Yerly, Y. Hu, H. Ceri, and R. Martinuzzi. 2007. Structure of Proteus mirabilis biofilms grown in artificial urine and standard laboratory media. FEMS Microbiol. Lett. 268:16-21. [DOI] [PubMed] [Google Scholar]
- 28.Kalmokoff, M. L., J. W. Austin, X. D. Wan, G. Sanders, S. Banerjee, and J. M. Farber. 2001. Adsorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J. Appl. Microbiol. 91:725-734. [DOI] [PubMed] [Google Scholar]
- 29.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 30.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 31.Korber, D. R., G. G. Greer, G. M. Wolfaardt, and S. Kohlman. 2002. Efficacy enhancement of trisodium phosphate against spoilage and pathogenic bacteria in model biofilms and on adipose tissue. J. Food Prot. 65:627-635. [DOI] [PubMed] [Google Scholar]
- 32.Krysinski, E. P., L. J. Brown, and T. J. Marchisello. 1992. Effect of cleaners and sanitizers on Listeria monocytogenes attached to product contact surfaces. J. Food Prot. 55:246-251. [DOI] [PubMed] [Google Scholar]
- 33.Lemon, K. P., D. E. Higgins, and R. Koler. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189:4418-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, G., and S. Kathariou. 2003. An improved cloning vector for construction of gene replacements in Listeria monocytogenes. Appl. Environ. Microbiol. 69:3020-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundén, J. M., T. J. Autio, and H. J. Korkeala. 2002. Transfer of persistent Listeria monocytogenes contamination between food-processing plants associated with a dicing machine. J. Food Prot. 65:1129-1133. [DOI] [PubMed] [Google Scholar]
- 37.Lundén, J. M., M. K. Miettinen, T. J. Autio, and H. J. Korkeala. 2000. Persistent Listeria monocytogenes strains show enhanced adherence to food contact surface after short contact times. J. Food Prot. 63:1204-1207. [DOI] [PubMed] [Google Scholar]
- 38.Mah, T.-F. C., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-38. [DOI] [PubMed] [Google Scholar]
- 39.Marsh, E. J., H. Luo, and H. Wang. 2003. A three-tiered approach to differentiate Listeria monocytogenes biofilm-forming abilities. FEMS Microbiol. Lett. 228:203-210. [DOI] [PubMed] [Google Scholar]
- 40.Meyer, B. 2003. Approaches to prevention, removal and killing of biofilms. Int. Biodeterior. Biodegrad. 51:249-253. [Google Scholar]
- 41.Monk, I. R., G. M. Cook, B. C. Monk, and P. J. Bremer. 2004. Morphotypic conversion in Listeria monocytogenes biofilm formation: biological significance of rough colony isolates. Appl. Environ. Microbiol. 70:6686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray, E. G. D., R. A. Webb, and M. B. R. Swann. 1926. A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes. J. Pathol. Bacteriol. 29:407-439. [Google Scholar]
- 43.Norwood, D. E., and A. Gilmour. 1999. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 86:576-582. [DOI] [PubMed] [Google Scholar]
- 44.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 45.Pamp, S. J., and T. Tolker-Nielsen. 2007. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 189:2531-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan, Y., F. Breidt, Jr., and S. Kathariou. 2006. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 72:7711-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren, T. J., and J. F. Frank. 1993. Susceptibility of starved planktonic and biofilm Listeria monocytogenes to quaternary ammonium sanitizer as determined by direct viable and agar plate counts. J. Food Prot. 56:573-576. [DOI] [PubMed] [Google Scholar]
- 48.Rieu, A., S. Weidmann, D. Garmyn, P. Piveteau, and J. Guzzo. 2007. agr system of Listeria monocytogenes EGD-e: role in adherence and differential expression pattern. Appl. Environ. Microbiol. 73:6125-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasahara, K. C., and E. A. Zottola. 1993. Biofilm formation by Listeria monocytogenes utilizes a primary colonizing microorganism in flowing systems. J. Food Prot. 56:1022-1028. [DOI] [PubMed] [Google Scholar]
- 50.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sela, S., S. Frank, E. Belausov, and R. Pinto. 2006. A mutation in the luxS gene influences Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 72:5653-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smoot, L. M., and M. D. Pierson. 1998. Effect of environmental stress on the ability of Listeria monocytogenes ScottA to attach to food contact surfaces. J. Food Prot. 61:1293-1298. [DOI] [PubMed] [Google Scholar]
- 53.Smoot, L. M., and M. D. Pierson. 1998. Influence of environmental stress on the kinetics and strength of attachment of Listeria monocytogenes ScottA to Buna-N rubber and stainless steel. J. Food Prot. 61:1286-1292. [DOI] [PubMed] [Google Scholar]
- 54.Somers, E. B., and A. C. Wong. 2004. Efficacy of two cleaning and sanitizing combinations on Listeria monocytogenes biofilms formed at low temperature on a variety of materials in the presence of ready-to-eat meat residue. J. Food Prot. 67:2218-2229. [DOI] [PubMed] [Google Scholar]
- 55.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stickler, D. 1999. Biofilms. Curr. Opin. Microbiol. 2:270-275. [DOI] [PubMed] [Google Scholar]
- 57.Sturme, M. H. J., J. Nakayama, D. Molenaar, Y. Murakami, R. Kunugi, T. Fujii, E. E. Vaughan, M. Kleerebezem, and W. M. de Vos. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187:5224-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Drogen, F., and M. Peter. 2004. Revealing protein dynamics by photobleaching techniques. Methods Mol. Biol. 284:287-306. [DOI] [PubMed] [Google Scholar]
- 60.Vilain, S., P. Cosette, M. Hubert, C. Lange, G.-A. Junter, and T. Jouenne. 2004. Comparative proteomic analysis of planktonic and immobilized Pseudomonas aeruginosa cells: a multivariate statistical approach. Anal. Biochem. 329:120-130. [DOI] [PubMed] [Google Scholar]
- 61.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 62.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 63.Wimpenny, J., W. Manz, and U. Szewzyk. 2000. Heterogeneity in biofilms. FEMS Microbiol. Rev. 24:661-671. [DOI] [PubMed] [Google Scholar]
- 64.Xu, K. D., G. A. McFeters, and P. S. Stewart. 2000. Biofilm resistance to antimicrobial agents. Microbiology 146:547-549. [DOI] [PubMed] [Google Scholar]
- 65.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]