Abstract
The abundance, vertical distribution, and diversity of aerobic anoxygenic phototrophic bacteria (AAP) were studied at four basins of the Baltic Sea. AAP were enumerated by infrared epifluorescence microscopy, and their diversity was analyzed by using pufM gene clone libraries. In addition, numbers of CFU containing the pufM gene were determined, and representative strains were isolated. Both approaches indicated that AAP reached maximal abundance in the euphotic zone. Maximal AAP abundance was 2.5 × 105 cells ml−1 (11% of total prokaryotes) or 1.0 × 103 CFU ml−1 (9 to 10% of total CFU). Environmental pufM clone sequences were grouped into 11 operational taxonomic units phylogenetically related to cultivated members of the Alpha-, Beta-, and Gammaproteobacteria. In spite of varying pufM compositions, five clones were present in all libraries. Of these, Jannaschia-related clones were always found in relative abundances representing 25 to 30% of the total AAP clones. The abundances of the other clones varied. Clones potentially affiliated with typical freshwater Betaproteobacteria sequences were present at three Baltic Sea stations, whereas clones grouping with Loktanella represented 40% of the total cell numbers in the Gotland Basin. For three alphaproteobacterial clones, probable pufM phylogenetic relationships were supported by 16S rRNA gene analyses of Baltic AAP isolates, which showed nearly identical pufM sequences. Our data indicate that the studied AAP assemblages represented a mixture of marine and freshwater taxa, thus characterizing the Baltic Sea as a “melting pot” of abundant, polyphyletic aerobic photoheterotrophic bacteria.
Aerobic bacteriochlorophyll a-producing bacteria, or so-called aerobic anoxygenic phototrophic bacteria (AAP), are strict aerobes, carrying out a photoheterotrophic metabolism. They require organic substrates for growth, but they can supplement a significant portion of their metabolic requirements by light-derived energy (8, 16, 26, 31, 39). AAP were discovered in the Bay of Tokyo by Shiba et al. (34) in 1970s. Later, their wide distribution in marine environments was documented by infrared (IR) kinetic fluorometry (16, 18), IR epifluorescence microscopy (IREM) (9, 14, 19, 23, 33) and pigment analyses (12), as well as cultivation-based (1, 15, 38) and genetic (3, 5, 13, 28, 41) studies. The planktonic bacterial assemblages are known to contain various AAP groups belonging to Alpha-, Beta-, and Gammaproteobacteria (41). Microscopic analyses have shown that AAP abundances vary in different environments from 0 to 24% of total prokaryotes (9, 21, 33).
The ecological role of aerobic anoxygenic phototrophy is still unclear. A recent study suggested that AAP significantly contribute to bacterial production in oligotrophic oceans (17). Their ability to use light energy in addition to respiration could provide them an advantage over their heterotrophic bacterial relatives (2). In contrast, Schwalbach et al. (32) found no effect of light on microbial community structure. Clearly, more data on AAP abundance and activity are required.
The Baltic Sea is a brackish, rather heterogeneous, enclosed-shelf sea system (27) harboring periodic anoxic conditions in its depths. During early summer three strata are formed with a stable thermocline around 20 m and a permanent halocline at 50 to 60 m. The presence and concentration of AAP in these surface waters of the central Baltic has been documented in two studies. Using IR kinetic fluorometry, bacteriochlorophyll a concentrations of between 8 and 50 ng liter−1 were detected in samples in the late summer of 2003 (18). IREM analyses demonstrated seasonal changes in AAP abundance ranging from 0 to 12% of total prokaryotes (23). The highest cell counts were detected between April and September. Using IREM coupled with fluorescence in situ hybridization, it was found that the Baltic Sea AAP community consisted of Alpha- and Gammaproteobacteria. However, representative studies of AAP depth distribution and a reliable phylogenetic identification of such bacteria are still lacking. In the present study AAP abundances and depth distributions in four different basins of the central Baltic Sea were investigated by using IREM coupled with analysis of AAP diversity using environmental pufM gene clone libraries and cultivation-dependent methods.
MATERIALS AND METHODS
Sampling.
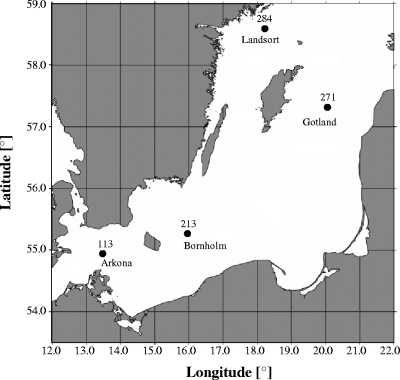
Samples were obtained from onboard the R/V Professor Albrecht Penck during cruise 40/06/16 in 2006 to the Arkona Basin (station 113, 7 July; 54°55.42′N; 13°30.17′E), Bornholm Basin (station 213, 8 July; 55°14.76′N; 15°59.22′E), Gotland Basin (station 271, 10 July; 57°19.82N; 20°02.82′E), and Landsort Deep (station 284, 11 July; 58°34.95′N; 18°14.62′E) (Fig. 1). Water samples from different depths were collected in 5-liter Free-Flow bottles (Hydrobios) attached to a polyvinyl chloride-coated stainless steel rosette equipped with conductivity, temperature, and depth sondes (SBE 911plus; Seabird), which included an electrode for measuring oxygen. For microscopy, water samples were fixed with 2% formaldehyde (0.2-μm pore size, prefiltered), gently mixed, and incubated at 4°C.
FIG. 1.
Map of the central Baltic Sea and positions of the sampling stations.
Preparation of cultivation-based samples occurred during cruise 44/03/08 (9 August 2003) to the eastern Gotland Basin (station 271) onboard the R/V Alexander von Humboldt.
Microscopic quantification of AAP.
Determination of the total cell numbers (TCN) and determination of AAP was done by IREM analyses as described by Mašín et al. (23).
Environmental pufM gene clone library construction and RFLP.
Samples obtained at depths of maximal AAP concentration in the Arkona Basin (0.5 m), Bornholm Basin (4.0 m), Gotland Basin (9.0 m), and Landsort Deep (3.0 m) were filtered immediately onto Nuclepore filters (pore size, 0.2 μm) and stored frozen at −80°C. In the laboratory, nucleic acids were extracted according to the procedure described by Weinbauer et al. (37). The pufM gene was amplified by using the pufL (forward, 5′-CTKTTCGACTTCTGGGTSGG-3′) and pufM_uniR (reverse, 5′-YCCATNGTCCANCKCCARAA-3′) primer system (40), which included parts of the pufL and nearly the entire pufM gene. PCR mixtures contained 1× JumpStart REDTaq ReadyMix reaction mix (Sigma), 100 nM concentrations of the forward and reverse primers, and usually 10 ng of template. The reactions were incubated in a MyCycler (Bio-Rad) with initial denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 60 s, followed in turn by one 72°C step for 10 min. PCR products were purified (MiniElute PCR purification kit; Qiagen) and cloned in competent E. coli JM109 cells according to the manufacturer's instructions for the pGEM-T Easy vector system (Promega). Vector-specific primers T7 and SP6 were used to amplify PCR products for restriction fragment length polymorphism (RFLP) analyses and sequencing. The restriction enzymes MspI and Hin6I were used to digest and visualize the unpurified PCR products as described previously (4). Operational taxonomic units (OTUs) were grouped by identical RFLP patterns, and representative fragments were sequenced by using the reverse primer pufM_uniR (40) by MWG Biotech AG (Martinsried, Germany).
Isolation and cultivation of AAP.
Water samples from 13 different depths (6 to 180 m) at station 271 were taken in August 2003 (see Fig. S1 in the supplemental material). Dilutions (1 to 1:10,000) were prepared in 100-μm-pore-size-filtered and autoclaved Baltic seawater. Volumes of 100 μl were spread on Baltic seawater medium agar plates (three replicates) which had been prepared in Baltic seawater of 12‰ salinity amended with (per liter) 0.1 g NH4-acetate, 0.05 g of K2HPO4, 0.05 g of KH2PO4, and 15 g of Bacto agar. CFU were determined after an 8-month incubation period at 10°C in the dark.
Nucleic acids of all colonies originating from one representative replicate plate out of three were extracted according to the method of Weinbauer et al. (37). pufM gene analyses, RFLP, and sequencing of representative OTUs were carried out as described for the environmental pufM gene. Dilution series were used to isolate all pufM-positive colonies on the Baltic seawater agar medium. These were then stored deep-frozen in glycerol (15% [vol/vol]) at −80°C.
Additional water samples were taken from the surface layer of different areas of the Baltic Sea, resulting in the isolation of strains B04, B09, and B11 on agar plates (15), and strains WM1 and WM2 using a two-step dilution technique with subsequent fluorescence screening (M. Koblížek et al., unpublished data). Additional information regarding the water samples for AAP cultures is summarized in Table 1.
TABLE 1.
16S rRNA gene sequence length, nucleotide accession number and closest phylogenetic neighbor, as well as the location, temperature, and sampling date of isolated Baltic AAP
| Isolate | Locationa | Latitude | Longitude | Date | Temp (°C) | 16S rRNA sequence length (bp) | Nucleotide accession no. | Closest phylogenetic neighbor | Similarity (%)b |
|---|---|---|---|---|---|---|---|---|---|
| B04 | Eastern Gotland Basin, station F1* | 58°39′N | 21°24′E | 27 August 2003 | 16 | 666 | EU564435 | Erythromicrobium ramosum DSM 8510 | 99 |
| B09 | Gulf of Finland, station F4* | 59°39′N | 24°09′E | 28 August 2003 | 17 | 1430 | DQ648518 | Roseobacter sp. strain BS90 | 98 |
| B11 | Northern Baltic Proper, station F5* | 59°20′N | 22°33′E | 30 August 2003 | 16 | 1434 | DQ659411 | Marine alphaproteobacterium AS-11 | 98 |
| WM1 | Warnemünde beach | 54°11′N | 12°06′E | 7 July 2006 | 19 | 1416 | EF421433 | Loktanella vestfoldensis LMG 22003T | 99 |
| WM2 | Warnemünde beach | 54°11′N | 12°06′E | 7 July 2006 | 19 | 1421 | EF044234 | Uncultured Rhodobacter group bacterium clone LA4-B3 | 97 |
| IOW 11b | Gotland Basin, station 271† | 58°00′N | 19°54′E | 9 August 2003 | 21 | 1344 | EU168097 | Marine bacterium BS110 | 96 |
| IOW 23b | Gotland Basin, station 271† | 58°00′N | 19°54′E | 9 August 2003 | 21 | 1334 | EU168099 | Loktanella vestfoldensis LMG 22003T | 99 |
*, samples were obtained during the “Biooptics cruise” aboard R/V Oceania in August 2003, as described previously (19); †, samples were obtained onboard R/V Alexander von Humboldt during cruise 44/03/08.
As determined by the program BLAST.
Identification of AAP isolates.
The full-length 16S rRNA gene was amplified from pufM-positive isolates by using the bacterial primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′). PCR mixtures contained 1× PCR buffer, 250 μmol of each deoxynucleoside triphosphate, 150 nM concentrations of the forward and reverse primers, and 0.025 U of Taq polymerase (Fermentas) μl−1. PCR was done in a MyCycler (Bio-Rad) with initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 45°C for 45 s, and 72°C for 90 s, followed by one 72°C step for 2 min. PCR products were sequenced by MWG Biotech AG (Martinsried, Germany).
Phylogenetic analysis.
Using ARB (http://www.arb-home.de), pufM sequences from the present study, together with sequences from public databases, were translated into amino acids and aligned. The protein alignment was used to realign nucleic acids, and phylogenetic trees were reconstructed based on sequences of approximately 400 nucleotides. These were reduced to unambiguously alignable positions by using group-specific filters. An evolutionary-distance dendrogram was constructed by using the Jukes-Cantor correction and neighbor-joining. The reliability of the branching patterns was evaluated by using 500 random bootstrap resamplings.
Statistical analyses.
To investigate relationships of temperature and salinity to AAP, as well as of picocyanobacteria and heterotrophic prokaryotic cell numbers, Spearman rank correlations were used. Levels of significance were defined as P ≤ 0.05 and P ≤ 0.001.
Nucleotide sequence accession numbers.
Partial pufM gene sequences were deposited in the GenBank database under accession numbers EU564436 (isolate B04), EU009365 (isolate B09), EU009366 (isolate B11), EU009373 (isolate WM1), EU026123 (isolate WM2), EU1680083 (isolate IOW23b), EU1680098 (isolate IOW11b), and EU1680084 to EU1680096 (pufM clones).
RESULTS
AAP cell distribution.
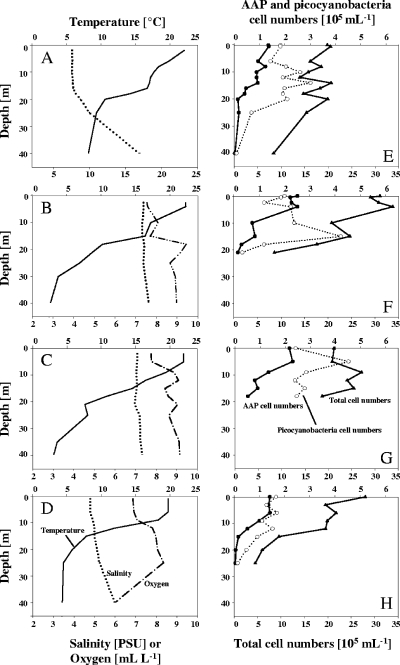
AAP abundances were assessed in the major basins of the Arkona, Bornholm, Landsort, and Gotland basins. Depth profiles showed the typical summer stratification of the upper water column for each of the investigated sites (Fig. 2A to D). Salinity usually increased slightly (5 to 8‰) with depth, except for the Arkona Basin where salinity reached 17‰ at a depth of 40 m (Fig. 2A). The highest prokaryotic cell numbers were measured in the upper water column ranging from 2.1 × 106 (Fig. 2E) to 3.4 × 106 cells ml−1 (Fig. 2F) but declined below the thermocline (Fig. 2E to H). Picocyanobacteria showed maximum abundance of 21.7% of the TCN in concentrations of up to 4.5 × 105 cells ml−1 (Fig. 2G). The AAP cell numbers ranged from 4 × 104 cells ml−1 up to a maximal level of 25 × 104 cells ml−1 observed in the Bornholm Basin (Fig. 2F). In relative numbers AAP formed 2 to 11% of the total prokaryotes present in the euphotic zone. The highest numbers (5 to 11%) were detected at the upper 10 m at the Gotland Basin station (Fig. 2G). Zones for maximal cell numbers of AAP, picocyanobacteria, and TCN varied between the surface and depths of 5 to 17 m (Fig. 2E to G). TCN, picocyanobacteria, and AAP abundances at different depths were correlated in terms of salinity and temperature. Salinity did not significantly correlate with AAP abundance, but Spearman rank correlations between water temperature and heterotrophic bacteria (TCN minus cyanobacteria and AAP), picocyanobacteria, and AAP cell counts confirmed that, at a significance of P ≤ 0.05, temperature significantly correlated with all three parameters. At a higher significance level (P ≤ 0.001) only AAP cell numbers were influenced by temperature (see Table S1 in the supplemental material).
FIG. 2.
Depth profiles throughout the Arkona Basin (A and E), Bornholm Basin (B and F), Gotland Basin (C and G), and Landsort Deep (D and H) in July 2006: temperature, oxygen, and salinity (A to D), as well as total, picocyanobacteria, and AAP cell numbers (E to H).
Environmental pufM gene diversity.
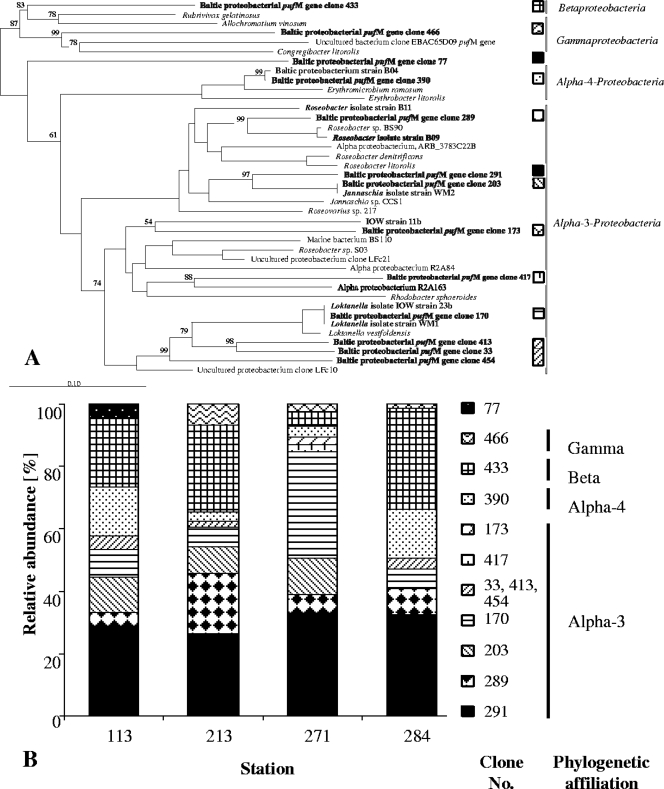
Based on restriction patterns and partial pufM sequencing 274 pufM clones could be grouped into 11 different OTUs using RFLP analysis. These grouped together with cultivated members of the Alpha-, Beta-, and Gammaproteobacteria (Fig. 3). The pufM phylogenetic tree exhibited three principal clusters: OTUs similar to Alpha-3-proteobacteria, namely, Loktanella vestfoldensis (represented by clone 170), Rhodobacter sphaeroides (clones 173 and 417), Jannaschia sp. (clones 203 and 291), and Roseobacter denitrificans (clone 289), as well as OTUs from two other clusters which were related to Alpha-4-proteobacteria (clone 390) or Gammaproteobacteria (clone 466). Clone 433 was most similar to the pufM sequence of Rubrivivax gelatinosus (80% sequence similarity) according to BLAST. However, this betaproteobacterial relationship was not reflected in the phylogenetic tree where Beta- and Gammaproteobacteria clustered together (Fig. 3A) The phylogenetic affiliation of pufM clone 77 could not be determined.
FIG. 3.
(A) Unrooted tree showing the phylogenetic relationships of pufM gene sequences detected in the central Baltic Sea and their closest relatives. The tree was reconstructed by using the neighbor-joining method and is based on a comparison of approximately 400 nucleotides. Bootstrap values, expressed as a percentage of 500 replications, are given at branching points. Only values greater than 50% are shown. Chloroflexus aurantiacus was used as the outgroup. The pufM sequence of L. vestfoldensis DSM16212T was obtained in the present study. Names in boldface indicate clones or isolates obtained from the present study. Patterns indicate representative clones that were retrieved throughout the stations. Bar, 10 substitutions per 100 nucleotides. (B) Relative abundance of different pufM genes in clone libraries, reflected by patterns identical to those from panel A. Clones derived from the depths of the maximal AAP concentration of the Arkona Basin (station 113), Bornholm Basin (station 213), Gotland Basin (station 271), and Landsort Deep (station 284).
Associations between seven to nine different OTUs could be formed for each of the basin-specific pufM clone libraries (Fig. 3B). Five OTUs were present at all stations (clones 170, 289, 291, 340, and 433). Of these, Jannaschia-related clone 291 accounted for 25 to 30% of the analyzed sequences, whereas the relative abundances of the other groups varied among the stations. High abundances of 20 to 30% were found for betaproteobacterial clone 433 in the Arkona, Bornholm, and Landsort libraries, as well as in the Gotland library for Loktanella-related clone 170 (40%). The pufM sequences in Bornholm, Gotland, and Landsort libraries related to Gammaproteobacteria (clone 466) contributed less than 10%.
AAP CFU.
The depth profiles for temperature and oxygen recorded in August 2003 confirmed the typical summer stratification of the upper water column for the Gotland Basin (see Fig. S1 in the supplemental material). Highest TCN (8.9 × 106 cells ml−1) and CFU numbers (1.2 × 104 colonies ml−1) were found at a depth of 6 m. A total of 224 colonies from 13 different depths between 6 and 180 m were screened for the pufM gene. Only colonies of 1.0 × 103 and 5.0 × 102 CFU ml−1 (9 to 10% of total CFU), originating from depths of 6 and 14 m, respectively, were pufM positive. Based on RFLP analyses, the pufM-positive colonies were grouped into two OTUs, represented by strains IOW11b and IOW23b (Table 1), which were phylogenetically located within the Roseobacter cluster (Table 1 and Fig. 3A).
Five more AAP strains were isolated from other areas of the Baltic Sea (Table 1). In accordance with their position in the phylogenetic pufM tree (Fig. 3A), their partial 16S rRNA gene strain sequences were most closely related to members of the Alpha-3- and Alpha-4-proteobacteria (Table 1). Interestingly, the pufM gene sequences of obtained isolates showed high similarities to environmental sequences. The pufM gene sequences of strain B04 and clone 390 and of strain WM2 and clone 203, as well as strains WM1, IOW 23b, and clone 170, were identical or nearly identical (Fig. 3A). No gamma- or betaproteobacterial AAP strains could be isolated.
DISCUSSION
The aim of this study was to identify the diversity and distribution of AAP in the deepest stations of four major Baltic basins. The Baltic Sea was chosen for AAP analyses because its distinct vertical and horizontal salinity gradients allow the coexistence of freshwater and marine species (29). An increasing number of reports concerning the AAP presence in estuarine, coastal (33, 35), or fresh (24) waters formed by nonmarine species (41) suggested that these environments are inhabited by distinct AAP assemblages. In accordance with this, the diversity of pufM genes detected and high concentrations of AAP cells measured in the present study reflected specific brackish shelf conditions in the Baltic Sea.
AAP diversity.
The Roseobacter group plays an important role in diverse marine surface systems (7), forming the major part of AAP communities in the Mediterranean and the Red Sea (28). The Roseobacter group comprised a substantial portion of the bacterial community in the Skagerrak-Kattegat front as well (30), which is characterized by its surface brackish outflow from the Baltic Sea and deep saline inflow from the Skagerrak and North Sea. However, its occurrence in the central Baltic Sea was supported only by a few isolates, and confirmation by molecular methods is still lacking (6, 20). In the present study, potential members of the Roseobacter clade represented 50 to 90% of all clones investigated (Fig. 3). Three clones of this clade (Table 1) contained pufM sequences nearly identical to those of Baltic AAP isolates, enabling us to support the pufM phylogenetic relationships by 16S rRNA gene analyses (Fig. 3A).
The presence of genes grouping with L. vestfoldensis in all four basins (Fig. 3B) was remarkable and supported by independent isolation of two Loktanella strains (Table 1). Metagenomic data from the global ocean sampling expedition detected members of the Loktanella cluster in a sample of a suboxic-to-anoxic hypersaline lagoon, but not in oxic samples (41). In the central Baltic, pufM genes probably affiliated with Loktanella were especially abundant in the clone library from the Gotland Basin (Fig. 3B). The Gotland Basin is the largest basin area with anoxic bottom water; during the sampling the water column at station 271 was anoxic below 140 m (unpublished data). However, how local physicochemical conditions could influence the presence of the Loktanella cluster remains to be investigated.
Analogously, high abundances of potentially betaproteobacterial clones from the Arkona, Bornholm, and Landsort basins were found in the pufM clone libraries. Interesting is the presence of potential Erythromicrobium related genes (clone 390) in all of the studied libraries. Erythromicrobium- and Erythrobacter-related species are frequently found in cultivated material (15, 34, 39); however, they have not been found in genetic studies (3, 28, 41). Based on our analyses sequences clustering with Erythromicrobium/Erythrobacter accounted for 3 to 18% of Baltic pufM sequences, which is in fair agreement with previously published fluorescence in situ hybridization analyses (23). In the phylogenetic pufM tree clone 433 is positioned within the Gamma- and Betaproteobacteria (Fig. 3A). In general, the distinction between Gamma- and Betaproteobacteria is not simple. Indeed, recent principal-component analyses have shown that Betaproteobacteria could be a subgroup of much larger Gammaproteobacteria (11). Since BLAST analyses showed the closest similarity to the betaproteobacterium Rubrivivax gelatinosus, clone 433 was assumed to be of betaproteobacterial origin. R. gelatinosus is a freshwater bacterium that was first described in 1907 as “Rhodocystis gelatinosa” (25). Since the drainage area of the Baltic Sea is more than four times as large as the sea itself (10), the high abundances of freshwater-related betaproteobacterial pufM clones was expected.
Structure of bacterial communities based on phylogenetic relationships does not necessarily provide insight into their physiological properties and should be interpreted with caution. In addition, nucleic acid extraction, PCR, and cloning techniques are biased, which may have affected the determinations of pufM abundances. This might explain why more gammaproteobacterial AAP cells may have been detected by using fluorescence in situ hybridization (23) than in our study. Nevertheless, considering the whole pufM-based data set, the clones derived from the four basins were indicative of AAP assemblages formed by a mixture of marine and freshwater influences characteristic for brackish water environments such as the Baltic Sea. In summary, our study demonstrated a broad spectrum of AAP diversity, which is in contrast to the very low diversity of AAP in eutrophic environments suggested by Jiao et al. (14).
AAP abundance.
AAP cell concentrations of up to 11% of TCN were determined in 2006 by IREM (Fig. 2E to H), a level similar to those for the surface water in the summer of 2005 (23) and to the CFU of pufM-containing colonies compared to the total CFU (9 to 10%). This relatively high percentage is in agreement with other reports of high AAP abundances in coastal and shelf waters (35, 42).
IREM analyses showed the highest AAP cell numbers in the euphotic zone (upper 20 to 30 m of the water column) (Fig. 2E to H). Colonies containing pufM were only detected at comparable depths (Fig. S1 in the supplemental material), consistent with the phototrophic character of AAP. Light influence was already documented in different environments including the mid-Atlantic bight and the North Pacific gyre, where AAP cells were most abundant above a 1% light depth (9). In the Delaware and Chesapeake estuaries, AAP abundance was significantly correlated with light attenuation, as well as ammonium and nitrate concentrations (36), indicating that light is not the only factor influencing AAP assemblages. Thus, additional important ecological factors should also be considered. Analyses of AAP depth distributions in the Baltic showed a strong correlation between temperature and AAP abundance in the water column (Fig. 2). The highest AAP cell numbers were usually found at sites where temperatures were ≥20°C. Sieracki et al. (35) proposed that low temperatures (in this case, <6°C) limited AAP abundances in the Northwest Atlantic, which would support our studies. On the other hand, temperature is linked to absorbed light energy, and it directly influences many other parameters. Thus, the extent of direct influence by temperature on AAP abundance in the central Baltic Sea during our sampling is unknown but should be taken into account in further analyses.
Supplementary Material
Acknowledgments
We are grateful to the captains and crews of R/V Alexander von Humboldt and R/V Professor Albrecht Penck. The excellent technical assistance of Heike Brockmöller, Bärbel Buuk, and Annett Grüttmüller is greatly appreciated.
This study was funded by the Leibniz Institute for Baltic Sea Research (IOW), by Czech projects GAČR 204/05/0307 and 206/07/0241 and GAAV project 1QS500200570, and by the Czech-German bilateral AV ČR-DFG project 436 TSE to M.K. and M.L.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allgaier, M., H. Uphoff, A. Felske, and I. Wagner-Döbler. 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 69:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beatty, J. T. 2002. On the natural selection and evolution of the aerobic phototrophic bacteria. Photosynth. Res. 73:109-114. [DOI] [PubMed] [Google Scholar]
- 3.Béjà, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, T. Hamada, J. A. Eisen, C. M. Fraser, and E. F. DeLong. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 4.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britschgi, T. B., and S. J. Giovannoni. 1991. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 57:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchan, A., J. M. Gonzalez, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooney, M. J., W. A. Johnston, S. Pohl, and R. R. Bidigare. 2006. Influence of photoperiod on pigmentation and metabolic efficiency of the marine aerobic anoxygenic photosynthetic bacterium Erythrobacter longus strain NJ3Y. Aquat. Microb. Ecol. 43:303-309. [Google Scholar]
- 9.Cottrell, M. T., A. Mannino, and D. L. Kirchman. 2006. Aerobic anoxygenic phototrophic bacteria in the mid-Atlantic bight and the North Pacific gyre. Appl. Environ. Microbiol. 72:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmgren, R., and U. Larsson. 2001. Nitrogen and the Baltic Sea: managing nitrogen in relation to phosphorus. Sci. World J. 1:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrity, G. M., and J. G. Holt. 2001. The road map to the manual, p. 119-154. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 12.Goericke, R. 2002. Bacteriochlorophyll a in the ocean: is anoxygenic bacterial photosynthesis important? Limnol. Oceanogr. 47:290-295. [Google Scholar]
- 13.Hu, Y., H. Du, N. Jiao, and Y. Zeng. 2006. Abundant presence of the γ-like proteobacterial pufM gene in oxic seawater. FEMS Microbiol. Lett. 263:200-206. [DOI] [PubMed] [Google Scholar]
- 14.Jiao, N., Y. Zhang, Y. Zeng, N. Hong, R. Liu, F. Chen, and P. Wang. 2007. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ. Microbiol. 9:3091-3099. [DOI] [PubMed] [Google Scholar]
- 15.Koblížek, M., O. Béjà, R. R. Bidigare, S. Christensen, B. Benitez-Nelson, C. Vetriani, M. K. Kolber, P. G. Falkowski, and Z. S. Kolber. 2003. Isolation and characterization of Erythrobacter sp. strains from the upper ocean. Arch. Microbiol. 180:327-338. [DOI] [PubMed] [Google Scholar]
- 16.Koblížek, M., P. G. Falkowski, and Z. S. Kolber. 2006. Diversity and distribution of photosynthetic bacteria in the Black Sea. Deep-Sea Res. II 53:1934-1944. [Google Scholar]
- 17.Koblížek, M., M. Mašín, J. Ras, A. J. Poulton, and O. Prášil. 2007. Rapid growth rates of aerobic anoxygenic phototrophs in the ocean. Environ. Microbiol. 9:2401-2406. [DOI] [PubMed] [Google Scholar]
- 18.Koblížek, M., J. Stoń-Egiert, S. Sagan, and Z. S. Kolber. 2005. Diel changes in bacteriochlorophyll a concentration suggest rapid bacterioplankton cycling in the Baltic Sea. FEMS Microbiol. Ecol. 51:353-361. [DOI] [PubMed] [Google Scholar]
- 19.Kolber, Z. S., F. G. Plumley, A. S. Lang, J. T. Beatty, R. E. Blankenship, C. L. VanDover, C. Vetriani, M. Koblížek, C. Rathgeber, and P. G. Falkowski. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492-2495. [DOI] [PubMed] [Google Scholar]
- 20.Labrenz, M., G. Jost, and K. Jürgens. 2007. Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic-anoxic interface. Aquat. Microb. Ecol. 46:177-190. [Google Scholar]
- 21.Lami, R., M. T. Cottrell, J. Ras, O. Ulloa, I. Obernosterer, H. Claustre, D. L. Kirchman, and P. Lebaron. 2007. High abundances of aerobic anoxygenic photosynthetic bacteria in the South Pacific Ocean. Appl. Environ. Microbiol. 73:4198-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane, D. J. 1991. 16S/23S rRNA sequencing. John Wiley & Sons, Chichester, United Kingdom.
- 23.Mašín, M., A. Zdun, J. Stoń-Egiert, M. Nausch, M. Labrenz, V. Moulisová, and M. Koblížek. 2006. Seasonal changes and diversity of aerobic anoxygenic phototrophs in the Baltic Sea. Aquat. Microb. Ecol. 45:247-254. [Google Scholar]
- 24.Mašín, M., J. Nedoma, L. Pechar, and M. Koblížek. Distribution of aerobic anoxygenic phototrophs in temperate freshwater systems. Environ. Microbiol., in press. [DOI] [PubMed]
- 25.Molisch, H. 1907. Die Purpurbakterien nach neuen Untersuchungen. G. Fischer, Jena, Germany.
- 26.Moran, M. A., and W. L. Miller. 2007. Resourceful heterotrophs make the most of light in the coastal ocean. Nat. Rev. Microbiol. 5:792-800. [DOI] [PubMed] [Google Scholar]
- 27.Omstedt, A., J. Elken, A. Lehmann, and J. Piechura. 2004. Knowledge of the Baltic Sea physics gained during the BALTEX and related programmes. Prog. Oceanogr. 63:1-28. [Google Scholar]
- 28.Oz, A., G. Sabehi, M. Koblížek, R. Massana, and O. Béjà. 2005. Roseobacter-like bacteria in Red and Mediterranean Sea aerobic anoxygenic photosynthetic populations. Appl. Environ. Microbiol. 71:344-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paavola, M., S. Olenin, and E. Leppäkoski. 2005. Are invasive species most successful in habitats of low native species richness across European brackish water seas? Est. Coast. Shelf Sci. 64:738-750. [Google Scholar]
- 30.Pinhassi, J., A. Winding, S. J. Binnerup, U. L. Zweifel, B. Riemann, and Å. Hagström. 2003. Spatial variability in bacterioplankton community composition at the Skagerrak-Kattegat Front. Mar. Ecol. Prog. Ser. 255:1-13. [Google Scholar]
- 31.Rathgeber, C., J. T. Beatty, and V. Yurkov. 2004. Aerobic phototrophic bacteria: new evidence for the diversity, ecological importance and applied potential of this previously overlooked group. Photosynth. Res. 81:113-128. [Google Scholar]
- 32.Schwalbach, M. S., M. Brown, and J. A. Fuhrman. 2005. Impact of light on marine bacterioplankton community structure. Aquat. Microb. Ecol. 39:235-245. [Google Scholar]
- 33.Schwalbach, M. S., and J. A. Fuhrman. 2005. Wide-ranging abundances of aerobic anoxygenic phototrophic bacteria in the world ocean revealed by epifluorescence microscopy and quantitative PCR. Limnol. Oceanogr. 50:620-628. [Google Scholar]
- 34.Shiba, T., U. Simidu, and N. Taga. 1979. Distribution of aerobic bacteria which contain bacteriochlorophyll a. Appl. Environ. Microbiol. 38:43-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sieracki, M. E., I. C. Gilg, E. C. Thier, N. J. Poulton, and R. Goericke. 2006. Distribution of planktonic aerobic anoxygenic photoheterotrophic bacteria in the northwest Atlantic. Limnol. Oceanogr. 51:38-46. [Google Scholar]
- 36.Waidner, L. A., and D. L. Kirchman. 2007. Aerobic anoxygenic phototrophic bacteria attached to particles in turbid waters of the Delaware and Chesapeake estuaries. Appl. Environ. Microbiol. 73:3936-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinbauer, M. G., I. Fritz, D. F. Wenderoth, and M. G. Höfle. 2002. Simultaneous extraction from bacterioplankton of total RNA and DNA suitable for quantitative structure and function analyses. Appl. Environ. Microbiol. 68:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yurkov, V., and J. T. Beatty. 1998. Isolation of aerobic anoxygenic photosynthetic bacteria from black smoker plume waters of the Juan-de-Fuca Ridge in the pacific-ocean. Appl. Environ. Microbiol. 64:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yurkov, V. V., and J. T. Beatty. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 62:695-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yutin, N., M. T. Suzuki, and O. Béjà. 2005. Novel primers reveal wider diversity among marine aerobic anoxygenic phototrophs. Appl. Environ. Microbiol. 71:8958-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yutin, N., M. T. Suzuki, H. Teeling, M. Weber, J. C. Venter, D. B. Rusch, and O. Béjà. 2007. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the global ocean sampling expedition metagenomes. Environ. Microbiol. 9:1464-1475. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Y., and N. Jiao. 2007. Dynamics of aerobic anoxygenic phototrophic bacteria in the East China Sea. FEMS Microbiol. Ecol. 61:459-469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.