Abstract
Marine sediments of coastal margins are important sites of carbon sequestration and nitrogen cycling. To determine the metabolic potential and structure of marine sediment microbial communities, two cores were collected each from the two stations (GMT at a depth of 200 m and GMS at 800 m) in the Gulf of Mexico, and six subsamples representing different depths were analyzed from each of these two cores using functional gene arrays containing ∼2,000 probes targeting genes involved in carbon fixation; organic carbon degradation; contaminant degradation; metal resistance; and nitrogen, sulfur, and phosphorous cycling. The geochemistry was highly variable for the sediments based on both site and depth. A total of 930 (47.1%) probes belonging to various functional gene categories showed significant hybridization with at least 1 of the 12 samples. The overall functional gene diversity of the samples from shallow depths was in general lower than those from deep depths at both stations. Also high microbial heterogeneity existed in these marine sediments. In general, the microbial community structure was more similar when the samples were spatially closer. The number of unique genes at GMT increased with depth, from 1.7% at 0.75 cm to 18.9% at 25 cm. The same trend occurred at GMS, from 1.2% at 0.25 cm to 15.2% at 16 cm. In addition, a broad diversity of geochemically important metabolic functional genes related to carbon degradation, nitrification, denitrification, nitrogen fixation, sulfur reduction, phosphorus utilization, contaminant degradation, and metal resistance were observed, implying that marine sediments could play important roles in biogeochemical cycling of carbon, nitrogen, phosphorus, sulfate, and various metals. Finally, the Mantel test revealed significant positive correlations between various specific functional genes and functional processes, and canonical correspondence analysis suggested that sediment depth, PO43−, NH4+, Mn(II), porosity, and Si(OH)4 might play major roles in shaping the microbial community structure in the marine sediments.
The world's global oceans are a major site of carbon cycling and sequestration (31). The sunlit layers exchange gases with the atmosphere such that CO2 removal from the ocean is accompanied by CO2 removal from the atmosphere. The time scale of this sequestration is short, however, unless the fixed carbon is transported to deeper layers not in direct contact with the atmosphere. If the fixed carbon is remineralized back to CO2 in these deeper waters, it is sequestered there on the time scale of ocean mixing—500 to 1,000 years. If the sinking fixed carbon reaches the sediments, however, it can be permanently buried (sequestration time scale, >106 years), but the burial efficiency of marine sediments varies, apparently as a function of the mechanisms of metabolism. The different carbon degradation metabolisms in marine sediments also affect the cycling of the nutrient elements incorporated into organic tissue along with carbon. The prime example here is denitrification, which consumes combined nitrogen (14) and thereby reduces both the ocean's overall productivity and carbon sequestration potential. Another nutrient, phosphorus (P), has often been overshadowed by N, even though P limits or colimits many ecosystems, including some marine systems (32, 48). Therefore, continental margin sediments are important sites of carbon sequestration and nitrogen cycling (2, 30) as well as other biogeochemical processes.
Microorganisms in marine sediments play critical roles in biogeochemical cycling of carbon, nitrogen, phosphorus, sulfur, and various metals as well as contaminants (6, 37, 38, 54). So far, predicting global dynamics of biogeochemical cycles remains difficult due to uncertainty in estimating the rates of various processes. Such difficulty is compounded by the considerable ambiguity surrounding the organisms that control the dynamics of carbon and nitrogen in the marine sediment environment. Thus, understanding the diversity of microbial populations in marine environments is critical for understanding global C and N and nutrient dynamics and predicting their response to global change.
The last 20 years have provided a vast amount of data on the microbial diversity in marine environments, both planktonic (16, 18, 23, 24, 27, 42, 45, 49) and sedimentary (8-10, 37, 38, 40, 57, 58). The lion's share of this data is based on the small-subunit rRNA gene (8, 16-18, 23, 24, 27, 42, 45, 49, 57, 58). However, these analyses are limited to phylogenetic information with little information on potential functional diversity within the community, unless the phylogenetic group is closely linked to the known organisms of narrowly defined metabolic capabilities (50, 56). The information on the functional genes involved in biogeochemical cyclings provides a window into the potential metabolic functioning within a community and the functional guilds present within a community (21, 50, 52, 56). In addition, little is known about the heterogeneity and distributional characteristics of different microbial functional groups in marine sediments.
DNA microarray technologies have emerged as the most promising technology to characterize complex microbial communities (1, 5, 12, 29, 43, 50, 53, 59, 63, 65). In contrast to conventional studies constrained by a limited number of targeted genes, microarray-based analysis allows high-throughput analysis and quantitation of multiple functional genes of interest. However, our previous results showed that roughly 107 cells are needed to achieve reasonably strong hybridization (43). If these values can be directly applicable to natural environmental samples, the level of the 50-mer-based functional gene array (FGA) detection sensitivity is sufficient to detect dominant members of a microbial community but not sensitive enough to detect less-abundant microbial populations. Thus, amplification of the target DNA prior to hybridization is needed. In this study, we incorporated a newly developed amplification approach, termed whole community genome amplification (WCGA) (59), for analyzing microbial community functional structure of marine sediment cores from two stations in the Gulf of Mexico using FGAs. Geochemistry was also examined for potential correlations to provide insight into potential community structure. Our results indicated that the marine sediment microbial communities are diverse and spatially heterogeneous and are capable of performing various biogeochemical functions such as carbon degradation, nitrification, denitrification, nitrogen fixation, sulfur reduction, phosphorus utilization, and contaminant degradation.
MATERIALS AND METHODS
Sample collection and geochemical analysis of sediments.
Marine sediment samples were collected from two stations of the Gulf of Mexico: station 3 (GMT), 200-m water column depth (latitude, 26°41.87′N; longitude, 96°24.913′W); and station 6 (GMS), 800-m water column depth (latitude, 26°40.37′N; longitude, 96°06.84′W). Sediment cores, with overlying water, were collected with a Soutar box core and subcores at different sediment depths were taken using 7.5- and 10-cm cast-acrylic tubes (Table 1) (38). Chemical analysis of the sediment samples was performed as described previously (11, 37, 38). Sediment samples for microbiological analysis were frozen in liquid N2, shipped to Oak Ridge National Laboratory, and stored at −70°C until use.
TABLE 1.
Geochemistry measured in each depth increment by station
| Station and sample code (water column depth) | Sediment depth (cm)a | PO4 concn (μM) | NO3 + NO2 concn (μM) | NH4 concn (μM) | SO4 concn (μM) | SO4 reduction rate (cm−3 day−1) | Mn(II) concn (μM) | Porosity | Si(OH)4 concn (μM) | O2 concn (μM) |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 (200 m) | ||||||||||
| GMT-1 | 0.75 | 1.62 | 20.07 | 5.20 | 28.5 | 82 | NDb | 0.899 | 111.3 | 0 |
| GMT-3 | 2.50 | 6.05 | −0.04 | 8.86 | 28.4 | 819 | 21.0 | 0.869 | 93.1 | 0 |
| GMT-5 | 4.50 | 7.20 | 0.40 | 27.21 | 28.4 | ND | 22.6 | 0.849 | 133.6 | 0 |
| GMT-7 | 9.00 | 6.16 | 0.04 | 39.22 | 28.5 | 547 | 20.5 | 0.827 | 117.2 | 0 |
| GMT-9 | 16.00 | 7.25 | 0.04 | 38.35 | 28.5 | 444 | 19.4 | 0.785 | 154.5 | 0 |
| GMT-11 | 25.50 | 7.03 | 0.11 | 110.42 | 28.6 | 591 | 20.3 | 0.798 | 111.3 | 0 |
| 6 (800 m) | ||||||||||
| GMS-1 | 0.25 | 4.27 | 34.90 | 11.46 | 28.8 | 26 | ND | 0.907 | 85.4 | 105.4 |
| GMS-3 | 1.25 | 6.34 | 25.20 | 3.77 | 28.8 | ND | 2.6 | 0.871 | 127.7 | 55.1 |
| GMS-5 | 2.50 | 9.65 | 2.08 | 2.28 | 28.8 | ND | 64.7 | 0.863 | 149.5 | 15.4 |
| GMS-7 | 4.50 | 5.98 | ND | 17.04 | 28.7 | 21 | 137.1 | 0.847 | 125.9 | 0 |
| GMS-9 | 9.00 | 12.08 | 0.00 | 44.76 | 28.6 | 371 | 214.8 | 0.849 | 185.9 | 0 |
| GMS-11 | 16.00 | 15.80 | 0.00 | 116.03 | 28.8 | 87 | ND | 0.843 | 250.0 | 0 |
Data are shown only for sediment depths for which microarray analyses were performed.
ND, not determined.
DNA extraction and purification.
The bulk community DNA was directly extracted from 2 g of sediment by combining grinding, freeze-thawing, and sodium dodecyl sulfate (SDS) for cell lysis (64). The crude DNA was purified by the minicolumn purification method (64), except that the DNA was eluted twice from the resin column with 50 μl of hot water (80°C) both times. DNA quantity was determined with an ND-1000 spectrophotometer (Nanodrop, Inc.). All nucleic acids were stored at −70°C until used.
Oligonucleotide probe (50-mer) design and array construction.
The FGA (FGA-I) was constructed using a diverse set of functional genes involved in various geochemical processes, such as carbon and nitrogen cycling, phosphorus utilization, organic contaminant degradation, and metal resistance (Table 2). The sequences used for oligonucleotide probe design were downloaded from the GenBank database (September 2002) through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) by key word search followed by manual sequence check of their functional description.
TABLE 2.
Numbers of probes used in the FGA-I functional groups
| Functional group | No. of gene categories | No. of probes |
|---|---|---|
| Carbon degradation | 13 | 422 |
| Carbon fixation | 4 | 131 |
| Contaminant degradation | 90 | 733 |
| Phosphorous metabolism | 4 | 81 |
| Nitrogen cycle | 3 | 319 |
| Sulfur cycle | 2 | 204 |
| Metal resistance | 1 | 83 |
| 16S rRNA (positive control) | 2 | |
| Human (negative control) | 20 | |
| Arabidopsis thaliana (negative control) | 11 | |
| Total | 127 | 2,006 |
The 50-mer oligonucleotide probes were designed using the software PRIMEGENS (62) with modified parameters. To design 50-mer oligonucleotide probes, each gene sequence was compared against the entire downloaded sequence database using pairwise BLAST and aligned with the other sequences using dynamic programming. Based on these global optimal alignments, segments of 50-mer with <85% nucleotide identity to the corresponding aligned regions of any of BLAST hit sequences were selected as potential probes. We also considered stretches of matching sequences between aligned regions. Probes with more than 15-bp matching regions were removed from the potential probes (43). Among the potential probes identified, one probe was finally selected by considering melting temperature and self-complementarity. All probes selected were searched against the GenBank database and proved to be unique in the database. In summary, a total of 2,006 oligonucleotide probes were designed.
The designed oligonucleotide probes (50-mers) were synthesized without modification (MWG Biotech, Inc.) in a 96-well plate format. The oligonucleotides were diluted to a final concentration of 50 pmol μl−1 in 50% dimethyl sulfoxide (Sigma Chemical, Co.). Ten microliters of each probe was transferred to a 384-well microtiter plate for printing. The probes were arrayed with 16 pins (Stealth SMP2.5; TeleChem International, Inc.) at a spacing of 210 μm onto 25- by 75-mm UltraGAPS, gamma amino propyl silane-coated glass slides (Corning, Inc.) using a MicroGridII microarrayer (Genomic Solutions, Ann Arbor, MI) at 60% relative humidity. Each probe set was printed in duplicate on a different section of the slide. The slides were cross-linked by exposure to 600 mJ of UV irradiation in a UV Stratalinker 1800 (Stratagene, La Jolla, CA) and washed at room temperature with 0.1% SDS for 4 min followed by water for 2 min. The slides were dried by centrifugation at 500 × g for 5 min and stored in a clean slide box at room temperature.
WCGA and fluorescent labeling of target DNA.
The amplification of the community DNAs was carried out using the WCGA approach as described previously (59). Briefly, 100 ng DNA from each sample was amplified in triplicate using the Templiphi 500 amplification kit (Amersham Biosciences, Piscataway, NJ) in a modified buffer containing Escherichia coli single-strand binding protein (200 ng/μl) and spermidine (0.04 mM) for 2- to 6-h incubations at 30°C (59). The community DNA (100 ng, 1 to 2 μl) was mixed thoroughly with 10 μl of sample buffer containing random hexamers and set at room temperature for 10 min prior to adding the solution to 10 μl of reaction buffer containing deoxynucleotides, 1 μl single-strand binding protein (5 μg/μl), 1 μl spermidine (1 mM), and 1 μl enzyme mix. Reactions were stopped by heating at 65°C for 10 min, and the amplified products were quantified as above and visualized on 1% agarose gels.
All of the amplified DNA from each sample was denatured by boiling for 2 min and immediately chilled on ice. The denatured genomic DNA solution was then used for a 40-μl labeling reaction solution containing 50 μM dATP, dCTP, and dGTP; 20 μM dTTP (USB Corporation, Cleveland, OH); 25 μM Cy5-dUTP (Amersham Pharmacia Biotech, Piscataway, NJ); 40 U of Klenow fragment (Invitrogen, Carlsbad, CA); and 200 ng/μl of RecA. The reaction mixture was incubated at 37°C for 3 h. The labeled target DNA was purified using a QIAquick PCR purification column (Qiagen, Valencia, CA). Absorption (A260, A280, and A650) of 1 μl of the labeled samples was measured with an ND-1000 spectrophotometer (Nanodrop, Inc.) for evaluation of fluorescent dye incorporation efficiencies. All subsequent manipulations with Cy5-dUTP-labeled community DNAs were performed in the dark.
Microarray hybridization.
After purification, the labeled sample DNA was concentrated in a Speedvac at 40°C for 1 h and resuspended in an appropriate volume of distilled water and then suspended in a 40-μl hybridization solution. The hybridization solution contained 50% formamide, 3× SSC (1× SSC is 150 mM NaCl and 15 mM trisodium citrate), 10 μg of unlabeled herring sperm DNA (Promega, Madison, WI), and 0.31% SDS in a total volume of 17.5 μl. The hybridization solution was denatured at 95°C for 5 min. After heat denaturation, the hybridization solution was kept at >50°C until washing to prevent cross-hybridization. The hybridization mixture was deposited directly onto slides which were prewarmed to 50°C and covered with a coverslip. The microarray was placed into a self-contained flow cell (Telechem International) and plunged into the 50°C water bath immediately for overnight hybridization. Each microarray slide was taken out, and the coverslip was immediately removed in wash solution A (1× SSC and 0.2% SDS). Slides were washed using wash solutions A, B (0.1× SSC and 0.2% SDS), and C (0.1× SSC) for 5 min each at ambient temperature prior to drying. The slide was dried using centrifugation as described above.
Microarray scanning and data processing.
Microarray scanning and data processing were carried out as previously described (59). Briefly, a ScanArray 5000 microarray analysis system (PerkinElmer, Wellesley, MA) was used for scanning microarrays at a resolution of 10 μm. Scanned image displays were saved as 16-bit TIFF files and analyzed by quantifying the pixel density (intensity) of each spot using ImaGene version 5.0 (Biodiscovery, Inc., Los Angeles, CA). Mean signal intensity was determined for each spot, and the local background signals were subtracted automatically from the hybridization signal of each spot. Fluorescence intensity values for all replicates of the negative control genes (human genes) were averaged and then subtracted from the background-corrected intensity values for each hybridization signal. The signal/noise ratio was also calculated (59). Outliers were defined as those spots whose absolute signal value of the spot − the mean value of all replicates is larger than 2.90 σ. All outlying spots were removed from subsequent analysis. Any gene with more than 1/3 probe spots hybridized was considered positive.
The signal intensities were normalized based on the mean signal intensity across all genes on the arrays. Since the same amounts of DNA from all samples were used for amplification, labeling, and hybridization, it is expected the average signal intensities across all of the genes should be approximately equal. Thus, the across-arrays mean was calculated based on all intensities on the arrays after correction for empty and poor spots and outliers was made. A ratio was calculated for each positive spot by dividing the signal intensity of the spot by the mean signal intensity to obtain the normalized ratio. The normalized microarray data were then used for further analysis.
Data analysis.
Functional gene diversity was calculated using Simpson's reciprocal index (1/D) and Shannon-Weaver index (H′) using freely available software (http://www2.biology.ualberta.ca/jbrzusto/krebswin.html). Cluster analysis was performed using the pairwise average-linkage hierarchical clustering algorithm (22) in the CLUSTER software (http://rana.stanford.edu), and the results of hierarchical clustering were visualized using TREEVIEW software (http://rana.stanford.edu/). Several multivariate statistical methods were employed to analyze the microarray data. The Mantel test (39) was used for inferring the association between sediment depth or site geochemistry and functional gene diversity. All of the matrices required for the Mantel test were constructed based on Euclidean distance measurements—i.e., dissimilarity matrices of all microbiological (functional gene diversity, richness, and composition)—and geochemical measurements. For the Mantel test, both R package version 4.0 (13) and vegan package in R were used for comparison. The majority of analyses were done by functions in vegan packages with some additional code utilizing vegan package functions. We also used hierarchical cluster analysis to determine various groupings based on hybridization data.
Canonical correspondence analysis (CCA) was performed using the program package Canoco for Windows 4.5 (Biometris, The Netherlands). Environmental factors (explanatory variables) included sediment depth, phosphate, nitrate/nitrite, ammonium, sulfate, sulfate reduction rate, manganese(II), porosity, silica, and oxygen (Table 1). The analysis was performed without transformation of data, with focus scaling on interspecies distances and automatic selection of environmental variables, applying a partial Monte Carlo permutation test (499 permutations) including unrestricted permutation.
RESULTS
Sediment geochemistry.
The geochemistry varied both with the location of the site as well as the depth of sediment, with striking differences for some of the elements, such as ammonium, nitrate, and manganese. The depth of penetration for oxygen (O2) was only to 0.6 cm at GMT, while it was slightly more than 3 cm into the sediment at GMS (Table 1). As a result, no O2 was present at any of the sediment depths sampled for microarray hybridization analysis at GMT. Conversely, the first three sediment depths sampled for hybridization analysis at GMS contained oxygen, although the 2.5-cm sediment depth contained less than 15% of the concentration at the surface. The deepest three sediments from GMS (4.5, 9, and 16 cm) were similar to those of GMT, which was devoid of O2 (Table 1).
The amounts of nitrate (NO3−) and nitrite (NO2−), ammonium (NH4+), and sulfate (SO42−) are shown in Table 1. Much like O2, NO3−/NO2− disappeared rapidly in the sediment core from GMT; however, NO3−/NO2− was found up to 2.5 cm in the GMS core. Ammonium generally increased with sediment depth at both stations, potentially as a consequence of both anaerobiosis and ammonification. Although the measured sulfate concentration was constant at all depths at both stations, the sulfate reduction rates, as measured with radioactive isotopes, varied along the depth of the sediment core. In addition, Mn2+ differed markedly between the stations. Mn2+ at GMT stayed relatively stable through the sediment core; however, Mn2+ at GMS increased substantially almost 100-fold from 2.5 to 16 cm.
FGA.
All together, FGA-I (59) consisted of 2,006 probes (Table 2), which included 1,973 functional gene probes based on 7,418 sequences downloaded from multiple databases, 2 16S rRNA gene probes as positive controls, 20 human gene probes, and 11 probes of Arabidopsis thaliana genes as negative controls. A sequence-specific probe was designed for any sequence with <85% nucleic acid sequence identity. When a group of sequences had >85% identity, one probe was designed to represent that group. The target genes of the probes are involved in numerous biogeochemical processes, including carbon degradation, carbon fixation, nitrogen cycling (nitrogen fixation, nitrification, and denitrification), sulfate reduction, and phosphorus utilization. Other probes on this array are involved in degradation and transformation processes of a variety of chemical compounds such as monoaromatic compound oxidation and polycyclic aromatic hydrocarbon oxidation, biphenyl oxidation, ring cleavage reaction, aliphatic compound transformation, anaerobic degradation, and heavy metal reduction and export (Table 2). Almost all genes were from bacteria and archaea, although some were from fungi.
Functional gene diversity in the sediments.
A total of 930 or 47.1% of the probes showed significant hybridization with at least one of the 12 samples. At GMT, 723 genes were detected. This number was somewhat higher at GMS (846 genes). The number of functional genes detected varied considerably among the samples. For instance, twice as many genes on the array hybridized in the deep sediments (GMT-11 and GMS-11) as those in the top sediment (GMT-1 and GMS-1; Tables 3 and 4). Simpson's reciprocal diversity index (1/D) indicated higher levels of genetic diversity in the deep-sediment samples (e.g., 204.9 for GMT-11 and 201.6 for GMS-11) than the top sediment samples (e.g., 118.4 for GMT-1 and 121.9 for GMS-1; Tables 3 and 4). Similar results were obtained when Shannon-Weaver index was used as the diversity index. In addition, the evenness was comparable among all sediment samples examined (Table 3 and 4), suggesting that the evenness does not vary too much with sediment depth.
TABLE 3.
Among-depth gene results for station 3 (water column depth of 200 m), including gene overlap, gene uniqueness, and diversity indices for each depth
| Variable | Result at depth (cm)a:
|
|||||
|---|---|---|---|---|---|---|
| 0.75 | 2.50 | 4.50 | 9.00 | 16.00 | 25.50 | |
| % of genes detected at depth (cm): | ||||||
| 0.75 | 1.70 | 66.40 | 70.30 | 63 | 50 | 44.70 |
| 2.50 | 4.60 | 65.90 | 65.30 | 58.90 | 54.40 | |
| 4.50 | 1.90 | 67.70 | 57.50 | 50.90 | ||
| 9.00 | 1.80 | 62.10 | 59.80 | |||
| 16.00 | 12.10 | 63.80 | ||||
| 25.50 | 18.90 | |||||
| Total no. of genes detected | 291 | 373 | 324 | 394 | 522 | 593 |
| Diversity indices | ||||||
| Simpson's (1/D) | 118.4 | 133.1 | 134.6 | 136.4 | 168.2 | 201.6 |
| Shannon-Weaver H′ | 5.185 | 5.266 | 5.175 | 5.203 | 5.311 | 5.282 |
| Shannon-Weaver evenness | 0.797 | 0.809 | 0.795 | 0.8 | 0.816 | 0.812 |
Values in boldface represent gene uniqueness. Italic values represent gene overlap.
TABLE 4.
Among-depth gene results for station 6 (water column depth of 800 m), including gene overlap, gene uniqueness, and diversity indices for each depth
| Variable | Result at depth (cm)a:
|
|||||
|---|---|---|---|---|---|---|
| 0.25 | 1.25 | 2.50 | 4.50 | 9.00 | 16.00 | |
| % of genes detected at depth (cm): | ||||||
| 0.25 | 1.20 | 58.40 | 60.50 | 58 | 61 | 44.40 |
| 1.25 | 5.50 | 65.80 | 67.10 | 68.80 | 63.90 | |
| 2.50 | 4.00 | 70.10 | 69.70 | 60.60 | ||
| 4.50 | 3.20 | 68.50 | 66.60 | |||
| 9.00 | 2.80 | 61.20 | ||||
| 16.00 | 15.20 | |||||
| Total no. of genes detected | 342 | 531 | 504 | 539 | 494 | 746 |
| Diversity indices | ||||||
| Simpson's (1/D) | 121.9 | 185.4 | 199 | 168.2 | 179.7 | 204.9 |
| Shannon-Weaver H′ | 5.216 | 5.294 | 5.365 | 5.355 | 5.336 | 5.392 |
| Shannon-Weaver evenness | 0.801 | 0.813 | 0.824 | 0.823 | 0.82 | 0.829 |
Values in boldface represent gene uniqueness. Italic values represent gene overlap.
A significant percentage of the genes detected were shared across the sites, ranging from 38 to 72% (Table 5). A total of 223 genes were shared across all the samples at these two stations. Similarly, pairwise comparisons of the genes detected indicated that the proportions of the overlapped genes correlated (r = 0.60) with the sediment sample's depth. As the spatial distances increased, the proportion of the gene overlap decreased (Table 5). These results indicated that site factors may have greater impact on the community structure once buried within the sediment than the horizontal distance between stations.
TABLE 5.
Percentage of genes overlapping between similar depth increments between stations and overall
| Station (depth increment in cm) | % of genes overlapping (depth increment in cm)
|
||||||
|---|---|---|---|---|---|---|---|
| GMS (overall) | GMS-1 (0.25) | GMS-3 (1.25) | GMS-5 (2.50) | GMS-7 (4.50) | GMS-9 (9.00) | GMS-11 (16.00) | |
| GMT (overall) | 68.7 | ||||||
| GMT-1 (0.75) | 72.0 | 52.5 | 51.7 | 49.6 | 53.3 | 37.7 | |
| GMT-3 (0.25) | 68.6 | 61.1 | 62.7 | 59.7 | 63.9 | 47.8 | |
| GMT-5 (4.5) | 68.2 | 56.1 | 57.7 | 54.1 | 59.1 | 42.1 | |
| GMT-7 (9.0) | 66.9 | 63.7 | 61.5 | 63.7 | 63.5 | 51.6 | |
| GMT-9 (16.0) | 56.8 | 65.3 | 69.3 | 71.7 | 68.2 | 62.8 | |
| GMT-11 (25.5) | 51.3 | 66.3 | 62.3 | 65.5 | 64.9 | 64.09 | |
A total of 233 genes were shared among the sediment depths at GMT and 300 genes were shared at GMS. The proportions of overlapped genes ranged from 44% to 70% in both stations (Table 3 and 4). As expected, the proportions of overlapping genes between samples were consistent with the spatial distances of these samples. In general, the proportions of the overlapped genes decreased as the distances increased (r = −0.9 and P < 0.01 for GMT; r = −0.4 and P < 0.05 for GMS). For instance, at GMT, about 66% of the genes were shared between GMT-1 (0.75 cm) and GMT-3 (2.5 cm), whereas ∼45% of genes were common between GMT-1 and GMT-11 (25.5 cm) (Table 3). A similar trend was observed among different samples from GMS (Table 4).
To understand the heterogeneity of microbial populations in these sediment samples, the proportions of unique genes detected were calculated. An average of 6.8% genes (within the range of 1.7% to 18.9%) were unique to the samples analyzed at GMT (Table 3). Similarly, at GMS, the average proportion of the genes unique to these samples is 5.3%, ranging from 1.2% to 15.2% (Table 4). The proportions of the unique genes in the deepest layer of the sediments examined (18.9% for GST-11 and 15.2% for GMS-11) are more than 10 times higher than those in the top sediments (1.7% for GMT-1 and 1.2% for GMS-1).
Cluster analysis of functional genes.
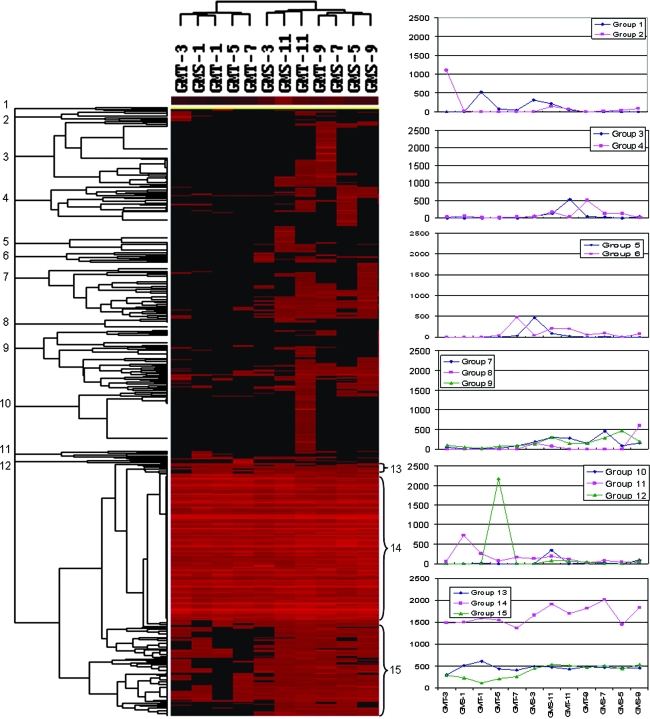
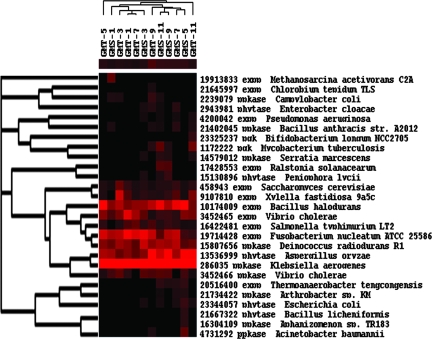
To visualize how the functional genes detected change across different sediment samples, a cluster analysis of all of the genes detected was performed (Fig. 1). Cluster analysis revealed that, in general, the samples from the deep layers of sediments clustered together and separated from the samples from top layers of sediments, except for samples GMT-7 and GMS-3. These results suggested the overall community structure appears to be more similar for the samples close together.
FIG. 1.
Hierarchical cluster analysis of genes based on hybridization signals for both sites and all depths. The figure was generated using CLUSTER and visualized with TREEVIEW. Black represents no hybridization above background level and red represents positive hybridization. The color intensity indicates differences in hybridization patters. The numbers equal groupings found among the hybridization patterns. A total of 15 different patterns of genes were observed, of which the most obvious were groups 14 and 15, which were abundant across all of the samples. Group 8 and 10 were most abundant in the deep layers of marine sediments. Groups 3 and 10 existed in low abundance and appeared to be unique to the deep-sediment samples (GMT-11 and GMS-11, respectively). The remaining groups were present in low abundance in different samples.
A total of 15 different patterns of genes were observed. The most obvious pattern was group 14 (223 [23.9%]), whose members were abundant across all of the samples (Fig. 1 and see Table S1 in the supplemental material). The genes in this group were from functional categories involved in nitrification; nitrogen fixation; denitrification; sulfate reduction; degradation of cellulose, lignin, xylan, chitin, and catechol, as well as many other organic contaminants; and metal resistance (see Table S1 in the supplemental material). The next obvious group is group 15 (15.7%), whose members are present in most of the sediment samples. Similarly this group also consists of most of the genes belonging to the functional categories mentioned above (see Table S1 in the supplemental material). The third distinct patterns are groups 8 (1.8%) and 10 (9.8%), which appear to exist in the deep layers of marine sediments. The genes in this group are found more toward those related to degradation of cellulose, liginin, xylan, and catechol. Groups 3 and 10 existed in low abundance and appear to be unique to the deep-sediment samples, GMT-11 and GMS-11, respectively, primarily including genes involved in denitrification, sulfate reduction, and xylan and catechol degradation (Fig. 1 and see Table S1 in the supplemental material). Other groups (groups 1, 2, 4, 5, 6, 7, 8, and 9) were present in low abundance in different samples (Fig. 1 and see Table S1 in the supplemental material).
Since nitrogen and carbon dynamics are of major concern in recent studies in global changes and biogeochemistry, the genes involved in nitrogen fixation, nitrification, denitrification, sulfate reduction, carbon degradation, and phosphorus utilization were examined in more detail.
(i) Nitrogen cycling genes.
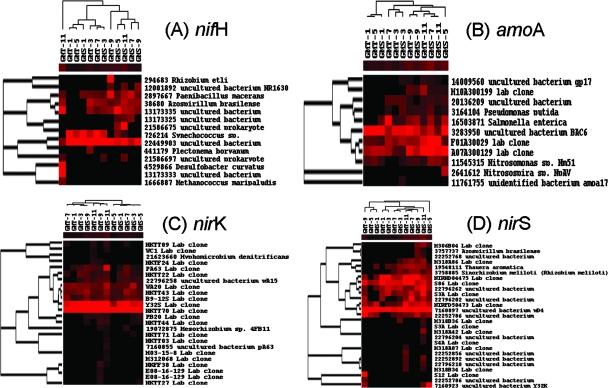
As nitrogen may be limiting in marine systems, we examined the relationship among nitrogen cycling genes and nitrogen sediment concentrations. The majority of the ammonia monooxygenase genes observed were environmental clones. The amoA clones—3283950 from biofilters originating from nitrifying activated sludge (44) and A07A300129 and F01A30029 from the groundwater of Environmental Remediation Science Program Field Research Center—were observed across all samples (Fig. 2). The Mantel test showed that the changes of many of the amoA genes (e.g., F0130029 and A07A300129) were positively correlated with NH4+ concentrations and/or NO3−/NO2− concentrations (P < 0.10). These results suggest that the bacteria containing these amoA genes could play important roles in nitrogen cycling of the marine sediments. However, the relationship between amoA and N concentration varied between stations. For instance, differences found in amoA clones (F01A30029 and A07A300129) were positively associated with differences found in NH4+ concentration at GMT, but no association was observed at GMS.
FIG. 2.
Hierarchical cluster analysis of genes involved in the nitrogen cycle, including nitrogen fixation, nitrification, and denitrification. See the Fig. 1 legend for explanation.
Most of the nifH genes observed were from environmental clones rather than from pure culture studies (Fig. 2). The clone 22449903 was abundant and present across all samples. Our analysis showed that the change of this clone was positively associated with the differences of NH4+ concentration at GMT, but no correlation existed at GMS. In addition, several other nifH genes were significantly correlated with the changes of NH4+ and NO3−/NO2− concentrations (Table 6), but their abundance varied across different samples (Fig. 2).
TABLE 6.
Correlations among variables
| Variable | Correlation with variable:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Depth | PO4 | NO3 + NO2 | NH4 | SO4 | SO4 reduction rate (cm−3 day−1) | Mn(II) | Porosity | Si(OH)4 | |
| PO4 | 0.420 | ||||||||
| NO3 + NO2 | −0.533 | −0.488 | |||||||
| NH4 | 0.888 | 0.630 | −0.440 | ||||||
| SO4 | −0.076 | 0.371 | 0.384 | 0.120 | |||||
| SO4 reduction rare | 0.367 | −0.086 | −0.507 | 0.194 | −0.832 | ||||
| Mn(II) | 0.220 | 0.826 | −0.415 | 0.481 | 0.366 | −0.297 | |||
| Porosity | −0.836 | −0.268 | 0.689 | −0.561 | 0.279 | −0.468 | −0.048 | ||
| Si(OH)4 | 0.368 | 0.922 | −0.405 | 0.580 | 0.346 | −0.250 | 0.844 | −0.238 | |
| O2 | −0.437 | −0.284 | 0.880 | −0.352 | 0.520 | −0.436 | −0.347 | 0.557 | −0.346 |
The majority of the nirS and nirK genes observed were also environmental clones. Although nirS environmental clones of S86 and 7160897 were abundant across all different samples, their changes are not significantly associated with the changes of NO3−/NO2− and NH4+ in these samples (Fig. 2). Several other nirS clones (2279262, 19548111, and 22796202) showed a strong significant correlation with the changes of NH4+ and/or NO3−/NO2− concentrations. Similarly, the nirK clones NKTT70 and Y32S were found to be dominant across all of the samples (Fig. 2) but were not correlated with NH4+ or NO3−/NO2− concentrations. Several other clones (e.g., PA63 and 22796258) were less abundant but were significantly correlated with N concentrations.
(ii) Sulfur reduction genes.
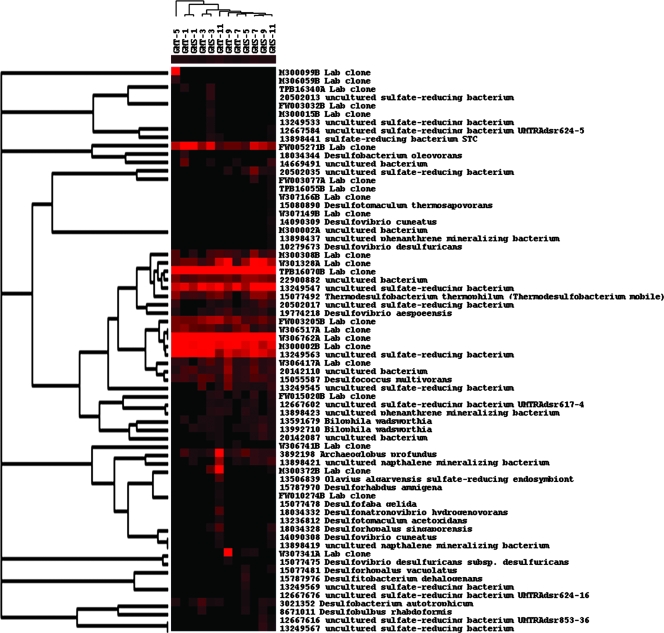
Of the genes detected involved in sulfur reduction, many were from either environmental clones or uncultured organisms. For example, the clones W306517A and W306762A are from the sediment of the continental margin off the Pacific coast of Mexico, and the clones FW005271B and TPB16070B are from groundwater samples. Also, several clones were observed across both sites as well as within the depths, such as TPB16070B, W306762A, and M300002B (Fig. 3). The results from the Mantel test showed that 7 (FW005271B, M300308B, M300002B, W306762A, GI13591679, GI13898441, and GI13249563) of the 56 genes in GMT and 3 (GI18034157, GI6561489, and GI20142087) of the 69 genes detected in GMS were found to correlate with the sulfur reduction rates. These results indicate that these particular genes, such as FW005271B or M300002B from laboratory clone libraries, may be the ones responsible for the observed rate changes. While a few of the genes were shared across sites and depths, one originating from Bilophila wadsworthia correlated with sulfur reduction for GMT but, though it was found at GMS, was not correlated with reduction rates.
FIG. 3.
Hierarchical cluster analysis of sulfate reduction genes (dsr). See the Fig. 1 legend for explanation.
Many of these genes were also correlated with porosity and/or depth. In the GMT site, eight of these gene probes were from lab clones, while a number of them were from isolates such as Desulfovibrio aespoeensis. For GMS, fewer were correlated with porosity and/or depth. A few were shared between GMS and GMT (FW005271B and W306762A), but most were different for GMS, such as 22900882, a dissimilatory sulfur reductase from an uncultured organism.
Additionally, some dissimilatory sulfur reduction genes were correlated with nitrogen concentrations. In GMT, the correlated genes were associated with either NO3− (six genes) or NH4+ (five genes). The genes associated with nitrogen concentrations for GMS were almost entirely correlated with NO3− levels. The probe derived from Bilophila wadsworthia was found to correlate with NO3− concentrations in both GMT (P = 0.088) and GMS (P = 0.061). Because the correlation was positive, with increasing presence of the gene with increasing nitrogen, there may be a possible connection between sulfur reduction and nitrogen limitation (Fig. 3).
(iii) Carbon degradation genes.
Carbon degradation is a major biogeochemical process in these sediments. The rate of degradation depends on a number of factors, including availability and types of carbon substrates as well as the microbial consortium present. Carbon degradation has often been considered potentially nitrogen limited (33). When we estimated correlations among the gene probes and nitrogen concentration, we found a number of the genes were strongly correlated with nitrogen concentrations, although some of these were negative associations. Negative correlations with nitrogen concentrations may have less to do with nitrogen per se. Rather, these correlations could indicate a change in carbon substrate. Terrestrial and marine ecosystem studies of carbon have shown that as carbon complexity increases, nitrogen concentration often decreases (41).
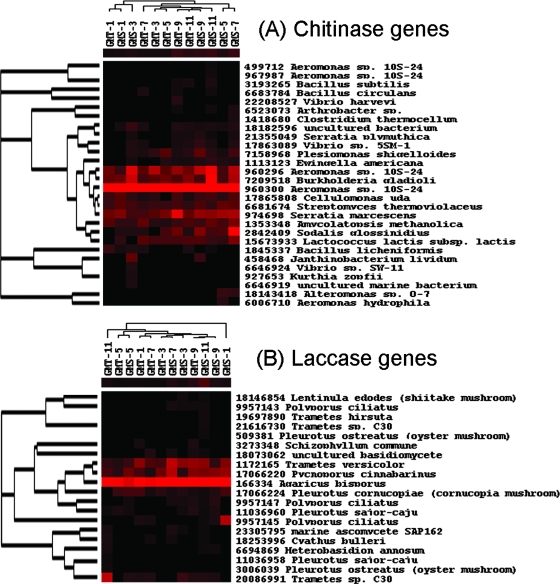
Chitin as a carbon source is abundant in marine ecosystems, with billions of tons generated annually from diverse sources (7). Turnover of chitin occurs rapidly in marine waters, but a significant portion may reach sediments (7). As expected a number of chitinase genes were found in these sediments; 19 and 28 genes were detected in GMT and GMS, respectively. Of these, at least three were common across sites and depths, all from isolations (960300 from Aeromonas sp., 960296 from Aeromonas sp., and 7209578 from Burkolderia gladioli; Fig. 4A). Six of the 19 genes found at GMT were correlated with nitrogen concentrations. Among them, two genes (7158968 from Plesiomonas shigelloides and 15673933 from Lactococcus lactis) were negatively associated with NO3− and the other four were positively associated with NH4+. Also, 4 of the 28 genes found at GMS were correlated with nitrogen concentrations. Among them, one (6681674 from Streptomyces thermoviolaceus) was positively associated with NH4+ and the other three, all from isolates, were weakly negatively associated with NO3−.
FIG. 4.
Hierarchical cluster analysis of carbon degradation genes. See the Fig. 1 legend for explanation.
In anaerobic as well as aerobic cellulose degradation, endoglucanases are responsible for the initial steps; therefore, endoglucanase genes would be expected to be highly represented in these marine sediments. We detected 38 and 33 endoglucanase genes for GMT and GMS, respectively, with 9 and 6 genes significantly correlated with nitrogen levels. At the GMT site, five of the nine genes were correlated with increasing NO3−, while the other four were correlated with increasing NH4+. The endoglucanase-positive gene probes at GMS were also primarily derived from isolated organisms, such as 11595860 from Xanthomonas campestris. At GMS, two of the six genes showing significant correlations with nitrogen were negatively correlated with NO3−, three were correlated with increasing NH4+, and one (666885 from Fibrobacter intestinalis) was negatively correlated with NO3− and positively correlated with NH4+.
Lignin is the most abundant and widely found aromatic polymer, second only to cellulose. This compound requires a suite of enzymes for degradation, one of which is laccase, a lignolytic enzyme found in a number of efficient lignin degraders. Totals of 16 and 17 laccase genes were detected at GMS and GMT. The vast majority of the probes were derived from isolated organisms such as Pleurotus. The laccase gene, 166334, from Agaricus bisporus, appeared to be dominant across all sediment samples (Fig. 4B). Three laccase genes at GMT were significantly correlated with NH4+. At GMS, the changes of the genes (17066224 from Pleurotus cornucopiae and 166334 from Agaricus bisporus) were significantly correlated with NO3− or NH4+, suggesting that nitrogen availability could be tightly linked with lignin mineralization.
Xylanase, an important hemicellulase, separates lignin from the hemicellulose fibers in numerous types of plant materials. Because of the importance in carbon degradation as well as xylanase function in complex substrate utilization, xylanase was expected to be highly represented in these sediments. Totals of 39 and 43 xylanase genes were found in GMT and GMS, respectively. Of these genes, the vast majority were derived from isolated organisms, many from Clostridium sp., with only two from other than isolations. A small number (five for GMT and six for GMS) were correlated with nitrogen. Of the five genes in GMT, one (18086520) was negatively correlated with NO3− while the other four were positively correlated with increasing NH4+ (7594905, 6176558, 974180, and 2980618). For GMS, four genes (10802606, 2624008, 144932, and 499714) were negatively correlated with NO3− and two (2980618 and 2645418) were positively correlated with NH4+.
(iv) Phosphorus utilization genes.
Phosphorus limits primary productivity in many ecosystems, including marine sediments, either singularly or as a colimiting nutrient with nitrogen (32, 48). The phosphorus cycle in any ecosystem includes both a major abiotic component as well as a biotic component; however, remineralization remains a major pathway for phosphorus availability (4). Mineralization of organic phosphorus is accomplished through phosphatase enzymes, such as exopolyphosphatase. Additionally, many microorganisms are capable of dissolving adsorbed or chemically bound phosphorus through use of phosphatase enzymes (3).
Phosphorus metabolism genes were well represented at both sites, with 19 found at GMT and 25 at GMS (Fig. 5). A total of seven genes were found across both sites and all depths. Four genes for exopolyphosphatase were observed, including 3452465 from Vibrio cholerae, 19714428 from Fusobacterium nucleatum, 10174009 from Bacillus halodurans, and 458943 from Saccharomyces cerevisiae. One phytase gene in common across the sites and depths was 13536999 from Aspergillus oryzae, and two polyphosphate kinase genes in common were 15807656 from Deinococcus radiodurans and 286035 from Klebsiella aerogenes.
FIG. 5.
Hierarchical cluster analysis of phosphorus metabolism genes. See the Fig. 1 legend for explanation.
Of the total number of genes found, one at GMT and two at GMS were associated with depth (higher representation at deeper depths). This may be due to alterations in phosphorus substrates, either changes in the abiotic (i.e., more chemically bound phosphorus) or biotic (i.e., alterations in the organic substrates) component. At GMT, of the three genes correlated with nitrogen concentrations, all three were correlated with NH4+. However, the correlational directions differed. The one exopolyphosphatase enzyme (19714428 from Fusobacterium nucleatum) was negatively correlated with amounts of NH4+. The other two genes (1172222 from an uncultured organism and 21402045 from Bacillus anthracis) were positively correlated with NH4+. Unlike the relationship found between phosphatase enzymes and phosphorus availability (3), there was no relationship among the phosphorus metabolism genes and phosphorus concentrations, which may be due to the ubiquitous nature of these genes.
Relationships between community structure and environmental variables.
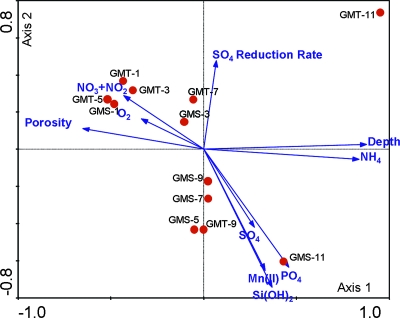
To examine the relationships between microbial community structure and geochemistry, CCA was used, which is a multivariate ordination method that combines correspondence analysis and multiple regression using environmental variables to “constrain” the ordination, leading to a more realistic, direct gradient analysis associated with biological variables (25, 36). CCA results revealed significant correlation between microbial communities in the sediments and environmental factors, as indicated by the species-environment correlations. The total canonical eigenvalue was 0.510, which is significant (P < 0.02). The first four canonical axes explained 52.6% of the variance of the species data (functional genes) and explained 55.8% variance of the species-environment relation. The CCA biplot (Fig. 6) of the first two canonical axes reflects the effect of geochemical factors on the microbial communities. The projection of environmental variables revealed that the first canonical axis is positively correlated with sample depth and NH4+ concentration and negatively correlated with porosity and O2 and NO3−/NO2− concentrations (Fig. 6). The second canonical axis is positively correlated with sulfate reduction rate and negatively correlated with the concentrations of SO42−, PO43−, Mn(II), and Si(OH)4, which are also positively related to NH4+ and sediment depth. Since important variables are represented by longer arrows in the biplot (51), the following six environmental factors appeared to play major roles in shaping the microbial community structure in the marine sediments: sediment depth, PO43−, NH4+, Mn(II), porosity, and Si(OH)4 (Fig. 6). Second, porosity and sediment depth were negatively related (r = −0.836; Table 6), but positively related to NO3− plus NO2− and O2 (r = 0.689, 0.557; Table 6). The microbial community structures in GMT-1, -3, and -5 and GMS-1 and -3 were positively correlated with porosity and negatively correlated with sediment depth and ammonia. Third, microbial community structures in GMS-5, 7, 9, and 11 as well as in GMT-9 were positively affected by manganese(II), silica, and phosphate but negatively affected by sulfate reduction rate. In contrast, microbial community structures in GMT-7 as well as GMS-3 were positively correlated with sulfate reduction rate and negatively correlated with manganese(II), silica, and phosphate. In addition, in some cases, the effects of environmental variables were obvious on certain specific functional gene group: for example, in GMT-9, in which the microbial community structure was governed by PO43−, more phosphorous cycling genes were detected (data not shown) than in other GMT samples. However, in most of cases, environmental variables might affect the entire communities instead of specific species.
FIG. 6.
CCA of geochemistry and community structure. The first canonical axis represents a gradient due to porosity, sediment depth, NH4+, NO3−/NO2−, and O2; the second canonical axis represents a gradient due to SO42− and SO42− reduction rate, Mn(II), Si(OH)4, and PO4+.
DISCUSSION
Functional gene arrays.
The development and application of microarray-based genomic technology for bacterial detection and microbial community analysis have received a great deal of attention in microbial ecology (43, 50, 52, 56, 59-61). Because of their high-density and -throughput capacities, it is expected that microarray-based genomic technologies will revolutionize the analysis of microbial community structure, function, and dynamics. Various microarray formats have been developed and evaluated for bacterial detection and microbial community analysis of environmental samples (43, 50, 52, 56, 63). Due to their high specificity and ease of construction, oligonucleotide-based microarrays are an important array-based approach for microbial ecology (26, 63). In this study, we developed a more comprehensive 50-mer array containing 2,006 probes from the genes involved in various geochemical processes on the basis of our previous studies (43, 52). We have recently expanded our FGA-I further to the second-generation FGA-II, or GeoChip 2.0, containing more probes covering more functional gene categories and functional genes (28). This type of array should be useful for monitoring microbial community structure in natural environments and linking genomic information with the associated biogeochemical processes. This study, along with our previous study, has clearly demonstrated that microarray-based genomic technologies have great potential as a high-throughput tool for microbial detection, identification, and characterization in natural environments.
When using FGAs for detecting bacterial populations in a mixed microbial community of unknown composition, specificity is always a concern. Thus, it is important to use the 50-mer FGAs for relative rather than absolute comparison so that the effects of any potential cross-hybridization can be cancelled out. In this study, we achieved this through hybridization signal normalization by comparing the change of a population relative to other populations within a community. The general consistency of microbial community structure relative to sediment depths and geochemistry suggested that this approach appears to be a useful way to compare microarray data across different samples.
Microbial distributions along sediment depth and stations.
Marine sediments play important roles in many of biogeochemical processes, including carbon sequestration and nitrogen conversions. These processes are largely controlled by the metabolic activities of the microorganisms present in the sediment. Therefore, the distribution of metabolic genes may reflect the metabolic potential of the microbial community. Our FGA results from these marine samples provide valuable insight to the composition, structure, distribution, and potential function of the microbial communities.
The relationship of microbial community structure to sediment depth appears to be a complex interaction. Bowman and McCuaig (6) examined the microbial community structure changes within sediment depths (0 to 0.4, 1.5 to 2.5, and 20 to 21 cm) from an Antarctic continental shelf using 16S rRNA-based cloning approach. While the highest diversity was observed within the 2-cm layer, the diversity was significantly reduced at the 21-cm-deep layer. The reduced diversity at the deep layer could be explained by much lower nutrient availability due to poorer quality of substrate available. Our results showed that richness and diversity of functional genes were generally higher at the deep layers than at the shallow layers at the two stations examined. This is contradictory to conventional wisdom. Based on the energy limitation hypothesis (the amount of energy available to an ecosystem limits the species richness by limiting the density of its individual taxa [34]), higher diversity is expected at shallow layers where organic carbon and nutrients should be higher. Bowman and McCuaig (6) also observed that oxic surface layer (0.4 cm) has lower diversity than the 2-cm-deep layer. This is consistent with our observations that the oxic layers at GMS (GMS-1, -3, and -5) have lower functional gene diversity than the anaerobic deep layers. However, this could not completely explain the observations in the shallow layers at GMT because no oxygen is observed in the shallow layers, such as GMT-1 (NO3−/NO2− rich) and GMT-3 (higher sulfate reduction rate). Some other factors should be also considered to explain the observations. For instance, in the deep layers of the sediments, the limited energy may limit the population of dominant species, and the relative lower porosity in the deep layers may limit the competition of dominant species against rare species. These two factors may lead to the increase of the species evenness in the deep layers, which contribute to their higher microbial diversity. More dedicated experiments with sophisticated designs are needed to test various hypotheses.
The overall community structure significantly varies with sediment depth as indicated by the Mantel test. Based on pairwise comparisons, a range from 30 to 60% of genes in both stations were unique and about 1 to 19% of the functional genes overlapped when the comparison was made across all of the samples. These results suggest that the spatial heterogeneity of microbial community structure in these samples appears to be high. In contrast to our results, marine sediment samples from Antarctica exhibited lower variability in the microbial community structure than expected based on denaturing gradient gel electrophoresis analysis (6). The apparent difference could be due to the low resolution of denaturing gradient gel electrophoresis analysis because generally less than 20 bands were obtained in gel electrophoresis.
Functional potentials in marine sediments.
Although marine denitrification occurs in both the water column when oxygen is absent and sediments, sedimentary denitrification is the largest sink term in the marine N budget, and it is also the most poorly quantified (13, 19). Benthic flux studies in many continental margins have shown that both O2 and NO3− diffuse into the sediments, while NH4+, N2, and sometimes NO3− diffuse out (20, 35) In the coastal sediments, denitrification is tightly coupled to nitrification, and in some areas, nitrification is the dominant supply pathway of NO3− for denitrification (20, 46). Thus, we hypothesized that the genes involved in nitrogen cycling would be tightly linked NH4 and NO3− plus NO2− concentrations. The Mantel tests of microbial community and geochemical data supported this expectation: many genes that are involved in nitrification, dentrification, and nitrogen fixation were strongly correlated with NH4 and NO3− plus NO2−.
The microbial communities in these sediment samples appear to be capable of decomposing complex organic matter. Chitin, cellulose, and lignin could be the major carbon and energy sources for various microbial populations (47). Thus, it is expected that microbial communities would be dominated by various related functional genes encoding chitinase, xylanase, cellulases, and laccases. Our microarray hybridization data supported this with evidence of a wide variety of these types of genes across sites and depths. Many of the genes observed (laccase and chitinase genes) were originally found in fungi. However, little is known about deep-sea fungal species, although fungal organisms, including filamentous fungi, have been isolated from deep-sea sediments (15). Of the carbon-associated genes detected, a small number showed a positive relationship with nitrogen, potentially indicating that nitrogen may limit some of these processes.
Another important biogeochemical process in marine sediments is sulfate reduction. Our results revealed a wide variety of functional genes encoding enzymes involved in sulfate reduction with different degrees of abundance in these marine sediment samples. The dsrAB genes from lab clones, uncultured organisms, and isolates such as Bilophila wadsworthia are prominent members of these communities. Interestingly, although sulfate concentrations were similar for all sediment layers, the sulfate reduction rates differed greatly, based on both depth and site. The results from the Mantel tests suggested that the marine sediments could have an active sulfate-sulfide oxidation-reduction cycle controlled by a few functional genes, even though sulfate remained similar through the depths. This is consistent with Antarctica marine sediments (6) and even terrestrial ecosystems (55).
Microarray hybridization data indicated some nirS, nirK, and amoA genes are abundant across all of the sediment samples, but their changes do not necessarily correlate with the differences of NH4+ and/or NO3−. Further analysis of the functional activity with different approaches such as mRNA-based microarray hybridization or isotope determination of rates is needed. The designed FGA is able to hybridize with cDNA transcribed from mRNA. However, isolating enough high-quality RNAs from a small amount of marine sediments for microarray hybridization is a great challenge.
Supplementary Material
Acknowledgments
This research was supported by the U.S. DOE Office of Science as part of its Biological and Environmental Research Programs in Biotechnology Investigations-Ocean Margins program and by the University of Oklahoma Foundation. Oak Ridge National Laboratory is managed by the University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Published ahead of print on 30 May 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adamczyk, J., M. Hesselsoe, N. Iversen, M. Horn, A. Lehner, P. H. Nielsen, M. Schloter, P. Roslev, and M. Wagner. 2003. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 69:6875-6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnosti, C., and M. Holmer. 2003. Carbon cycling in a continental margin sediment: contrasts between organic matter characteristics and remineralization rates and pathways. Estuarine Coastal Shelf Sci. 58:197-208. [Google Scholar]
- 3.Ayyakkan, K., and D. Chandram. 1971. Occurrence and distribution of phosphate solubilizing bacteria and phosphatase in marine sediments at Porto Novo. Mar. Biol. 11:201-205. [Google Scholar]
- 4.Benitez-Nelson, C. R. 2000. The biogeochemical cycling of phosphorus in marine systems. Earth-Sci. Rev. 51:109-135. [Google Scholar]
- 5.Bodrossy, L., N. Stralis-Pavese, J. C. Murrell, S. Radajewski, A. Weilharter, and A. Sessitsch. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566-582. [DOI] [PubMed] [Google Scholar]
- 6.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, J. N. 1994. Aerobic and anaerobic degradation and mineralization of 14C chitin by water column and sediment inocula of the York River estuary, Virginia. Appl. Environ. Microbiol. 60:174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braker, G., and J. M. Tiedje. 2003. Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. Microbiol. 69:3476-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandes, J. A., and A. H. Devol. 1997. Isotopic fractionation of oxygen and nitrogen in coastal marine sediments. Geochim. Cosmochim. Acta 61:1793-1801. [Google Scholar]
- 12.Brodie, E. L., T. Z. DeSantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casgrain, P., and P. Legendre. 2001. The R package for multivariate and spatial analysis, version 4.0 (development release 6). User's manual. Universite de Montreal, Montreal, Quebec, Canada.
- 14.Codispoti, L. A. 1995. Biogeochemical cycles—is the ocean losing nitrate. Nature 376:724. [Google Scholar]
- 15.Das, S., P. S. Lyla, and S. A. Khan. 2006. Marine microbial diversity and ecology: importance and future perspectives. Curr. Sci. 90:1325-1335. [Google Scholar]
- 16.Delong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delong, E. F., K. Y. Wu, B. B. Prezelin, and R. V. M. Jovine. 1994. High abundance of Archaea in Antarctic marine picoplankton. Nature 371:695-697. [DOI] [PubMed] [Google Scholar]
- 19.Devol, A. H. 1991. Direct measurement of nitrogen gas fluxes from continental-shelf sediments. Nature 349:319-321. [Google Scholar]
- 20.Devol, A. H., and J. P. Christensen. 1993. Benthic fluxes and nitrogen cycling in sediments of the continental-margin of the Eastern North Pacific. J. Mar. Res. 51:345-372. [Google Scholar]
- 21.Doney, S. C., M. R. Abbott, J. J. Cullen, D. M. Karl, and L. Rothstein. 2004. From genes to ecosystems: the ocean's new frontier. Front. Ecol. Environ. 2:457-466. [Google Scholar]
- 22.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1992. Novel major Archaebacterial group from marine plankton. Nature 356:148-149. [DOI] [PubMed] [Google Scholar]
- 24.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 59:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gauch, H. G. J. 1982. Multivariate analysis in community ecology. Cambridge University Press, Cambridge, United Kingdom.
- 26.Gentry, T. J., G. S. Wickham, C. W. Schadt, Z. He, and J. Zhou. 2006. Microarray applications in microbial ecology research. Microb. Ecol. 52:159-175. [DOI] [PubMed] [Google Scholar]
- 27.Giovannoni, S. J., E. F. Delong, T. M. Schmidt, and N. R. Pace. 1990. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl. Environ. Microbiol. 56:2572-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, Z., T. J. Gentry, C. W. Schadt, L. Wu, J. Liebich, S. C. Chong, Z. Huang, W. Wu, B. Gu, P. Jardine, C. Criddle, and J. Zhou. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67-77. [DOI] [PubMed] [Google Scholar]
- 29.He, Z., L. Wu, X. Li, M. W. Fields, and J. Zhou. 2005. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol. 71:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hedges, J. I., and R. G. Keil. 1995. Sedimentary organic-matter preservation—an assessment and speculative synthesis. Mar. Chem. 49:81-115. [Google Scholar]
- 31.Houghton, J. T., Y. Ding, D. J. Griggs, M. Noguer, P. J. van der Linden, and D. Xiaosu (ed.). 2001. Climate change 2001: the scientific basis. Cambridge University Press, Cambridge, United Kingdom.
- 32.Howarth, R. W. 1988. Nutrient limitation of net primary production in marine ecosystems. Annu. Rev. Ecol. Syst. 19:89-110. [Google Scholar]
- 33.Hu, S., F. S. Chapin, M. K. Firestone, C. B. Field, and N. R. Chiariello. 2001. Nitrogen limitation of microbial decomposition in a grassland under elevated CO2. Nature 409:188-191. [DOI] [PubMed] [Google Scholar]
- 34.Kaspari, M., S. O'Donnell, and J. R. Kercher. 2000. Energy, density, and constraints to species richness: ant assemblages along a productivity gradient. Am. Nat. 155:280-293. [DOI] [PubMed] [Google Scholar]
- 35.Kristensen, E., and T. H. Blackburn. 1987. The fate of organic-carbon and nitrogen in experimental marine sediment systems—influence of bioturbation and anoxia. J. Mar. Res. 45:231-257. [Google Scholar]
- 36.Legendre, P., and E. D. Gallagher. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129:271-280. [DOI] [PubMed] [Google Scholar]
- 37.Liu, X., C. E. Bagwell, L. Wu, A. H. Devol, and J. Zhou. 2003. Molecular diversity of sulfate-reducing bacteria from two different continental margin habitats. Appl. Environ. Microbiol. 69:6073-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, X., S. M. Tiquia, G. Holguin, L. Wu, S. C. Nold, A. H. Devol, K. Luo, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2003. Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the Pacific coast of Mexico. Appl. Environ. Microbiol. 69:3549-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209-220. [PubMed] [Google Scholar]
- 40.Nold, S. C., J. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the β subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orwin, K. H., D. A. Wardle, and L. G. Greenfield. 2006. Ecological consequences of carbon substrate identity and diversity in a laboratory study. Ecology 87:580-593. [DOI] [PubMed] [Google Scholar]
- 42.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhee, S.-K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakano, Y., and L. Kerkhof. 1998. Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl. Environ. Microbiol. 64:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, T. M., E. F. Delong, and N. R. Pace. 1991. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seitzinger, S. P., and A. E. Giblin. 1996. Estimating denitrification in North Atlantic continental shelf sediments. Biogeochemistry 35:235-260. [Google Scholar]
- 47.Sieburth, J. M. 1976. Bacterial substrates and productivity in marine ecosystems. Annu. Rev. Ecol. Syst. 7:259-285. [Google Scholar]
- 48.Smith, S. V. 1984. Phosphorus versus nitrogen limitation in the marine-environment. Limnol. Oceanogr. 29:1149-1160. [Google Scholar]
- 49.Suzuki, M. T., O. Beja, L. T. Taylor, and E. F. DeLong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 50.Taroncher-Oldenburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terbraak, C. J. F. 1986. Canonical correspondence-analysis—a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167-1179. [Google Scholar]
- 52.Tiquia, S. M., L. Wu, S. C. Chong, S. Passovets, D. Xu, Y. Xu, and J. Zhou. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. BioTechniques 36:664-670. [DOI] [PubMed] [Google Scholar]
- 53.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 54.Urakawa, H., T. Yoshida, M. Nishimura, and K. Ohwada. 2000. Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16S rDNA-based approaches and quinone profiling. Environ. Microbiol. 2:542-554. [DOI] [PubMed] [Google Scholar]
- 55.Vile, M. A., S. D. Bridgham, and R. K. Wieder. 2003. Response of anaerobic carbon mineralization rates to sulfate amendments in a boreal peatland. Ecol. Appl. 13:720-734. [Google Scholar]
- 56.Ward, B. B. 2005. Molecular approaches to marine microbial ecology and the marine nitrogen cycle. Annu. Rev. Earth Planet. Sci. 33:301-333. [Google Scholar]
- 57.Webster, G., C. J. Newberry, J. C. Fry, and A. J. Weightman. 2003. Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J. Microbiol. Methods 55:155-164. [DOI] [PubMed] [Google Scholar]
- 58.Webster, G., R. J. Parkes, J. C. Fry, and A. J. Weightman. 2004. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl. Environ. Microbiol. 70:5708-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, L., X. Liu, C. W. Schadt, and J. Zhou. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, L., D. K. Thompson, G. S. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, L., D. K. Thompson, X. Liu, M. W. Fields, C. E. Bagwell, J. M. Tiedje, and J. Zhou. 2004. Development and evaluation of microarray-based whole-genome hybridization for detection of microorganisms within the context of environmental applications. Environ. Sci. Technol. 38:6775-6782. [DOI] [PubMed] [Google Scholar]
- 62.Xu, D., G. Li, L. Wu, J. Zhou, and Y. Xu. 2002. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics 18:1432-1437. [DOI] [PubMed] [Google Scholar]
- 63.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou, J., and D. K. Thompson. 2002. Challenges in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 13:204-207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.