Abstract
Avian pathogenic Escherichia coli (APEC) strains are a subset of extraintestinal pathogenic E. coli (ExPEC) strains associated with respiratory infections and septicemia in poultry. The iroBCDEN genes encode the salmochelin siderophore system present in Salmonella enterica and some ExPEC strains. Roles of the iro genes for virulence in chickens and production of salmochelins were assessed by introducing plasmids carrying different combinations of iro genes into an attenuated salmochelin- and aerobactin-negative mutant of O78 strain χ7122. Complementation with the iroBCDEN genes resulted in a regaining of virulence, whereas the absence of iroC, iroDE, or iroN abrogated restoration of virulence. The iroE gene was not required for virulence, since introduction of iroBCDN restored the capacity to cause lesions and colonize extraintestinal tissues. Prevalence studies indicated that iro sequences were associated with virulent APEC strains. Liquid chromatography-mass spectrometry analysis of supernatants of APEC χ7122 and the complemented mutants indicated that (i) for χ7122, salmochelins comprised 14 to 27% of the siderophores present in iron-limited medium or infected tissues; (ii) complementation of the mutant with the iro locus increased levels of glucosylated dimers (S1 and S5) and monomer (SX) compared to APEC strain χ7122; (iii) the iroDE genes were important for generation of S1, S5, and SX; (iv) iroC was required for export of salmochelin trimers and dimers; and (v) iroB was required for generation of salmochelins. Overall, efficient glucosylation (IroB), transport (IroC and IroN), and processing (IroD and IroE) of salmochelins are required for APEC virulence, although IroE appears to serve an ancillary role.
Escherichia coli is a commensal resident of the intestine as well as a pathogen that can cause both enteric and systemic diseases of humans and animals (11, 31, 52). Certain commensal intestinal Escherichia coli isolates can cause disease at extraintestinal sites, and these strains have been collectively termed extraintestinal pathogenic E. coli (ExPEC) (53). Among ExPEC strains, uropathogenic E. coli (UPEC) is the most common cause of human urinary tract infections (UTIs) (37, 52). Avian pathogenic E. coli (APEC) strains share some virulence traits with ExPEC from human infections and are responsible for extraintestinal infections of economic importance to the poultry industry (11, 15, 49, 50). One of the most common forms of disease associated with APEC is avian colibacillosis, which starts as a respiratory infection (airsacculitis) that is frequently followed by generalized infections such as perihepatitis, pericarditis, and septicemia (11, 25).
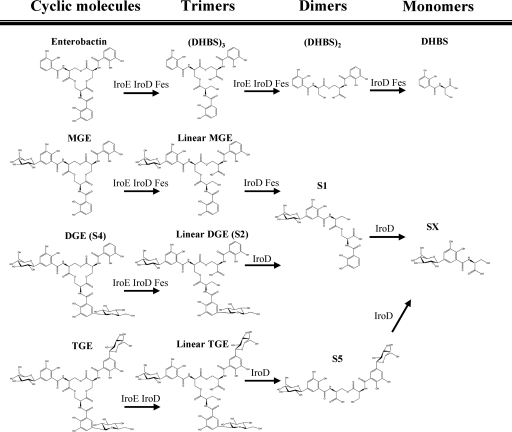
One attribute associated with APEC and other ExPEC strains is the presence of multiple iron uptake systems, including siderophores, which play an important role in extraintestinal virulence (13, 55, 57). Siderophores are high-affinity iron-chelating molecules that can contribute to bacterial survival during infection by sequestering iron, which is an essential trace element for most bacteria (24, 42, 46). Most E. coli strains, including nonpathogenic E. coli K-12, and other enterobacteria such as Salmonella enterica and Klebsiella spp. produce the catecholate siderophore enterobactin, which is a cyclic trimer of 2,3-dihydroxybenzoyl serine (DHBS) (24, 42). Enterobactin is very efficient at sequestering iron in vitro; however, it is less able to compete for iron during infection, as it is inhibited by serum albumin (32) and specifically binds to the host innate defense protein neutrophil gelatinase-associated lipocalin (NGAL; also called lipocalin 2 or siderocalin) (22, 39). In contrast, aerobactin and salmochelins, which are siderophores associated with APEC and UPEC strains (15, 42, 49, 57), can effectively acquire iron in the presence of serum albumin or NGAL (3, 19, 32). Aerobactin is a hydroxamate siderophore produced by most APEC strains (11) and certain pathogenic E. coli, Klebsiella pneumoniae, and Shigella strains (9, 23, 42). Aerobactin is synthesized by the iucABCD-encoded gene products, and aerobactin uptake occurs via the iutA-encoded receptor protein (23, 42). Salmochelins are C-glucosylated derivatives of enterobactin and DHBS molecules (glucosylated linear trimers, dimers, and monomers of DHBS) that were initially described by Hantke et al. (27). Salmochelins are produced by Salmonella enterica and certain ExPEC strains (3, 18, 27, 36, 62). In E. coli and Salmonella enterica, the salmochelin-encoding system comprises two divergently transcribed sets of genes, iroBCDE and iroN, which constitute the iro gene cluster (13, 17, 42, 56) (Fig. 1). The iroB gene encodes a glucosyltransferase that glucosylates enterobactin, iroC encodes an ABC transporter required for transport of salmochelins, and iroD, iroE, and iroN code, respectively, for a cytoplasmic esterase, a periplasmic hydrolase, and an outer membrane siderophore receptor (10, 18, 27, 36, 62).
FIG. 1.
Schematic diagram of the region comprising the iro locus and flanking sequences carried on plasmid pAPEC-1 of APEC strain χ7122. Numbers within arrows correspond to designated ORFs within the sequence (accession no. AF449498). Black arrows indicate the iro genes. Nucleotide identities between segments of the χ7122 iro coding region and those of UPEC strain CFT073 (accession no. AE014075) (60) and Klebsiella pneumoniae plasmid pLVKP (accession no. AY378100) (9) are illustrated within boxed segments, with numbers representing the percent nucleotide identity between segments. Additional sequences of CFT073 and pLVKP that are absent from the pAPEC-1 region are illustrated with diagonal lines and described in text boxes. Plasmid constructs described in the text are illustrated above pAPEC-1 containing the iro locus region. Primers used to generate clones by PCR are indicated with the symbol > for sense primers or < for antisense primers. Corresponding restriction endonuclease sites used to generate the plasmids are indicated. See Materials and Methods for details.
APEC strain χ7122 (O78:K80:H9) has been used as a model strain to study molecular mechanisms of APEC pathogenicity (4, 13). Strain χ7122 possesses the chromosome-encoded enterobactin siderophore system and also produces the aerobactin and salmochelin siderophores, which are encoded by genes present on a large virulence plasmid, pAPEC-1 (13). The salmochelin and aerobactin systems are required for full virulence of the strain in a chicken infection model (13). The loss of either of these systems reduced the capacity of the APEC strain to colonize extraintestinal tissues. Moreover, loss of both the aerobactin- and salmochelin-encoding gene clusters resulted in an avirulent strain that was unable to colonize extraintestinal sites, such as the lungs and liver, compared to the wild-type parent strain. Complementation of the salmochelin- and aerobactin-negative APEC strain with a plasmid containing the iro gene cluster resulted in a regaining of virulence comparable to that of the wild-type parent strain, despite the lack of the aerobactin gene cluster (13). However, the specific roles of iro genes in complementation of the attenuated strain have thus far not been established. Furthermore, to our knowledge, the quantities of enterobactin, DHBS molecules, and the salmochelins produced by APEC or other pathogenic E. coli strains have not been assessed.
In this report, we introduced plasmids containing different combinations of iro genes into an attenuated APEC salmochelin- and aerobactin-deficient strain in order to determine which iro genes were required to complement virulence of this strain in a chicken experimental infection model. By using this approach, we were able to specifically investigate the contribution of iro-encoded gene products for APEC virulence without the influence of the other virulence-associated siderophore, aerobactin. Furthermore, we used liquid chromatography-tandem mass spectrometry (LC-MS/MS) to determine the relative levels of siderophores produced by strain χ7122 from the tissues of infected chickens and to quantitate siderophores produced directly from culture supernatants of strain χ7122 and complemented mutants.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. APEC strain χ7122 is an O78:K80:H9 strain and produces enterobactin, salmochelins, and aerobactin. In addition, APEC and E. coli fecal isolates from healthy poultry were used to screen for the presence of iro genes. The 298 APEC isolates were previously described elsewhere (14). Thirty-two E. coli fecal isolates from healthy poultry were kindly provided by J. M. Fairbrother (University of Montreal, Montreal, Quebec, Canada). APEC strains and commensal fecal poultry isolates were previously classified for virulence based on lethality for 1-day-old chicks following subcutaneous inoculation, where LC1 corresponds to the high-lethality class, LC2 to the low-lethality class, and LC3 to the nonlethal class (14).
TABLE 1.
Bacterial strains and plasmids used for this study
| Bacterial strain or plasmid | Genotype or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| χ7122 | Avian pathogenic; O78:K80:H9; gyrA Nalr | 44 |
| χ7304 | ΔiroBCDEN::nptII ΔiucABCD iutA::xylE Kmr Nalr | 13 |
| DH5α | F− (Δ80d lacZΔM15) Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 gyrA96 relA1 | Bethesda Research Laboratories |
| Plasmids | ||
| pACYC184 | p15A replicon; Cmr Tcr | 6 |
| pBC SK (−) | ColE1 origin; Cmr | Stratagene |
| pYA3661 | pACYC184::iroBCDEN; Cmr | 13 |
| pIJ33 | pACYC184::iroN; Cmr | This study |
| pIJ34 | pACYC184::iroBCDE; Cmr | This study |
| pIJ37 | pACYC184::iroBDEN; Cmr | This study |
| pIJ53 | pACYC184::iroB; Cmr | This study |
| pIJ121 | pACYC184::iroBC; Cmr | This study |
| pIJ135 | pACYC184::iroBCN; Cmr | This study |
| pIJ136 | pACYC184::iroBCEN; Cmr | This study |
| pIJ137 | pACYC184::iroBCDN; Cmr | This study |
Cm, chloramphenicol; Km, kanamycin; Nal, nalidixic acid.
Luria-Bertani (LB) broth and tryptic soy agar (Difco Laboratories, Detroit, MI) were routinely used for growing E. coli strains and clones. E. coli strain DH5α was used for plasmid cloning and recovery. For infection studies, strain χ7122 and derivatives were grown in brain heart infusion broth (Difco). For production and detection of catecholate siderophores, bacteria were grown at 37°C for 17 h in iron-poor M63-glycerol minimal medium containing the following per liter: 5.3 g KH2PO4, 13.9 g K2HPO4·3H2O, and 2.0 g (NH4)2SO4. The pH was adjusted to 7.5 with KOH, and medium was supplemented with 1 mM MgSO4, 1 mM CaCl2, 1 mM thiamine, and 0.6% (wt/vol) glycerol before inoculation. The minimal medium was inoculated with 0.6% of a 5-h culture grown in TY medium containing (per liter) 5 g yeast extract, 5 g sodium chloride, and 8 g tryptone according to previous reports (27, 58, 59, 62). Iron-poor medium was prepared in plastic bottles to reduce trace contamination of iron. Antibiotics were added as required at the following concentrations: kanamycin, 30 μg ml−1; chloramphenicol, 30 μg ml−1; and nalidixic acid, 30 μg ml−1.
Construction of plasmids.
Enzymes used for generation of constructs were purchased from New England Biolabs. For PCR-generated fragments, Elongase DNA polymerase (Invitrogen) was used at an annealing temperature of 52°C. Plasmid constructs were derived from pYA3661 and are all illustrated in Fig. 1. pYA3661 (13) carries the full iro gene cluster (iroBCDEN) of APEC strain χ7122 on an 11.5-kb HindIII fragment cloned into vector pACYC184 (6) (Fig. 1). Plasmid pIJ34 (iroBCDE) was produced by digesting pYA3661 with HindIII and SspI and cloning the appropriate fragment into the HindIII and EcoRV sites of pACYC184. Plasmid pIJ37 (iroBDEN) was produced by digesting pYA3661 with KpnI, which results in a 2,861-bp in-frame deletion of the 3′ end of iroC. Plasmid pIJ53 was generated by digesting pYA3661 with HindIII and PvuII and cloning the fragment containing iroB into the HindIII and EcoRV sites of pACYC184. Plasmid pIJ121 (iroBC) was generated by digesting pYA3661 with HindIII and AclI and cloning the iroBC-containing fragment into the HindIII and NarI sites of pACYC184. Plasmids pIJ135 (iroBCN) and pIJ136 (iroBCEN) were produced by amplifying two PCR fragments from pYA3661, ligating them with XhoI linkers within the primers, and cloning them into the HindIII site of pACYC184. For pIJ135, the iroBC genes were amplified using primers CMD81 (5′-TCAGAGCAAGAGATTACGCGCAGAC-3′) and CMD91 (5′-TTCTCGAGACCGCCGCTTCTGACTGTGT-3′) and the iroN gene was amplified using primers CMD277 (5′-CGCCCTCGAGACTACGATCAGAATGATGCGGT-3′) and CMD87 (5′-CAGTACCGGCATAACCAAGCCTAT-3′). Primers CMD91 and CMD277 each contain an XhoI site (underlined). Primers CMD81 and CMD87 correspond to pACYC184 vector sequences flanking either side of the cloned iro locus in pYA3661 and generate fragments containing HindIII sites. The pIJ136 construct containing iroBCEN was similarly obtained using primers CMD81 and CMD91 for amplification and generation of the XhoI-HindIII fragment containing iroBC and primers CMD90 (5′-ACCTCGAGGAAACGGTACAGACTTCCTG-3′) (XhoI site underlined) and CMD87 for amplification and generation of the XhoI-HindIII fragment carrying iroEN. Plasmid pIJ137 (iroBCDN), was generated by three cloning steps. First the iroN gene was obtained from pYA3661 following digestion with SphI and cloned into the SphI site of pACYC184, resulting in plasmid pIJ33. The fragment carrying iroBCD was amplified by PCR with primers CMD81 and CMD278 (5′-AAAGTCTCGAGGGGTCAACTCAACCC-3′) (XhoI site underlined) and cloned in vector pBC SK(−) at the HindIII and XhoI sites. This intermediate vector was then digested with SspI and EagI, and the appropriate fragment was then cloned at the NruI and EagI sites of pIJ33, generating plasmid pIJ137. The specific deletions in the iro cluster for each of the plasmids generated were confirmed by PCR in the cloned strains and in χ7304 complemented derivatives.
Prevalence of the iro genes in APEC and fecal commensal strains.
Two hundred ninety-eight APEC and 32 commensal fecal isolates were screened by PCR for the presence of the iroB and iroN genes. Crude bacterial lysates were obtained as described elsewhere (47). The primers used to generate a 663-bp fragment corresponding to iroB were IROBKO1 (5′-AGGCGCGCCTCTCTATGGGC-3′) and IROBKO2 (5′-CTCTAGATCAAGGCCGTCAACC-3′). The primers used to specifically amplify a 549-bp fragment of the iroN gene were IRON1 (5′-TATTCGTGGTATGGGGCCGGA-3′) and IRON2 (5′-GCCCGCATAGATATTCCCCTG-3′). The PCRs were achieved using Taq DNA polymerase (New England Biolabs), at an annealing temperature of 58°C and an extension time of 1 min at 72°C for 25 cycles.
Sequencing of the regions adjacent to the iro locus of pAPEC-1.
The iro gene cluster from strain χ7122 was previously characterized and sequenced. The flanking DNA regions of the iroBCDEN cluster were sequenced from APEC strain χ7122 by using custom primers. The sequencing was done at the Genome Québec facility (Montreal, Quebec, Canada). Sequences were analyzed with the ORF Finder and BLAST programs available online at http://www.ncbi.nlm.nih.gov.
Experimental infections of chickens via the air sacs.
For infection studies, eight groups of 3-week-old White Leghorn specific-pathogen-free chickens (Canadian Food Inspection Agency, Ottawa, Canada) were inoculated in the right thoracic air sac with 0.1 ml (107 CFU) of a bacterial inoculum grown overnight in brain heart infusion broth. Experimental infections and the lesion scoring were carried out as previously described (33). Chickens were euthanized at 48 h postinfection, and their spleens, livers, and lungs were removed; weighed; suspended in buffered saline with gelatin (BSG) (1 liter is 8.5 g NaCl, 0.3 g KH2PO4, 0.6g Na2HPO4, and 0.1 g gelatin); and homogenized with an Omnimixer homogenizer. Samples were diluted 1:3, 1:30, and 1:300 in BSG. Bacterial counts were performed by plating 100 μl of each diluted sample on MacConkey agar plates supplemented with the appropriate antibiotics.
Analysis of siderophores from culture supernatants and tissues of infected chickens.
Supernatants of 17-h cultures were obtained following centrifugation of bacterial cells at 3,200 × g for 15 min and addition of 5 mM FeCl3 to each supernatant. The precipitate was removed by centrifugation, and the supernatants were filtered on 0.2-μm membranes. Aliquots of 1 ml of supernatant were then prepared in 5% (vol/vol) formic acid, and 0.12 ng/ml of 5,6,7,8-tetradeutero-3,4-dihydroxy-2-heptylquinoline was added as an internal control (34). Each strain was cultured in triplicate, and a sample of each culture supernatant was analyzed by LC-MS/MS. All of the strains tested demonstrated similar growth curves to APEC wild-type strain χ7122 following growth in LB medium. For growth in iron-poor M63 medium, all strains tested grew similarly and attained an overnight (17 h) mean optical density at 600 nm (OD600) of from 1.2 to 1.4, except for strain χ7304(pIJ37 iroBDEN), which demonstrated a slight growth lag and an overnight mean OD600 of 1.0 (data not shown).
For detection of siderophores during infection, two groups of 20 chickens were infected as described above with a 0.1-ml inoculum containing either 107 or 108 CFU of strain χ7122. Twenty-four hours postinfection, pericardia, air sacs, livers, and blood were removed from surviving chickens, pooled, weighed, suspended in a solution of methanol and formic acid (19:1), and homogenized with an Omnimixer. Homogenates were centrifuged at 3,200 × g for 15 min. The supernatants were retained, and the insoluble pellets were then extracted three times with methanol-formic acid (19:1). The supernatants from each of the extractions were then combined and concentrated by evaporation. Five-hundred-microliter aliquots were then prepared with 5,6,7,8-tetradeutero-3,4-dihydroxy-2-heptylquinoline as an internal control. Samples were analyzed three times by LC-MS/MS.
LC-MS analyses.
Multiple reaction monitoring (MRM) analyses were performed using an Agilent HP 1100 high-performance liquid chromatograph (Agilent Canada, Mississauga, Ontario, Canada) coupled to a Micromass QuattroII spectrometer (Micromass Canada). Samples were injected onto a Zorbax Eclipse XDB-C8 4.6- by 150-mm column at a flow rate of 400 μl/min and a linear gradient of water-acetonitrile with 1% acetic acid. The high-performance liquid chromatography effluent was directed to the mass spectrometer through a Valco T splitter. The analyses were performed in positive electrospray ionization mode with a cone voltage of 30 V. Monitoring of daughter ions from specific pseudomolecular ions was performed by collision-induced dissociation with argon at different collision energies for each molecule ranging from 15 eV to 55 eV. Information concerning the different salmochelin- and enterobactin-related molecules detected by MRM MS analyses is presented in Fig. 2. The specific transition ions monitored from pseudomolecular ions to daughter ions of salmochelins SX, S1, linear diglucosyl-C-enterobactin (DGE) (S2), DGE (S4), S5, monoglucosyl-C-enterobactin (MGE), linear MGE, triglucosyl-C-enterobactin (TGE), and linear TGE were 404 > 299, 627 > 224, 1,012 > 224, 994 > 224, 789 > 386, 832 > 224, 850 > 224, 1,156 > 266 and 1,174 > 266 m/z, respectively. The transition ions monitored for enterobactin and its linear trimer [(DHBS)3], dimer [(DHBS)2], and monomer (DHBS) derivatives were 670 > 224, 688 > 224, 465 > 224, and 242 > 137 m/z, respectively. The transition ions monitored for the internal standard and aerobactin were 244 > 159 and 565 > 205 m/z, respectively. Quantification of each compound, with the exception of aerobactin, was determined from the response factor of enterobactin and corrected with the intensity of the signal of the internal standard.
FIG. 2.
Scheme showing molecular structures of each the catecholate siderophores that were analyzed. Enzymes predicted to play a role in processing of salmochelins, enterobactin, or DHBS derivatives based on biochemical studies of Lin et al. (36) and Zhu et al. (62) are indicated.
Purification of enterobactin.
Enterobactin was purified as described by Léveillé et al. (35), with additional purification by thin-layer chromatography on a 1-mm glass silica gel 60-Å plate (Whatman). A calibration curve was established from purified enterobactin, and this curve was used to estimate the quantities of all catecholate siderophores, on the basis that salmochelins and enterobactin-related molecules have a response factor similar to that of cyclic enterobactin.
Statistical analyses.
Statistical analyses were performed using the Prism 4.0b software package (GraphPad Software, San Diego, CA).
Nucleotide sequence accession number.
The updated nucleotide sequence of the iroBCDEN cluster is available under GenBank accession no. AF449498.
RESULTS
Comparison of the iro-containing region of pAPEC-1 with other iro-containing regions.
A 21,019-bp region of pAPEC-1 containing the iro gene cluster and spanning from the iss gene to the cvaB gene was sequenced (accession no. AF449498). This region, or specific segments of it, is highly homologous to other sequences encoded on plasmids or genomic islands of other E. coli strains (5, 8, 28, 29, 56, 60). The regions immediately adjacent to the iro cluster on plasmid pAPEC-1 are also conserved in the pathogenicity island (PAI) carrying the iro cluster from CFT073 (Fig. 1), although an additional 24.5 kb (spanning open reading frames [ORFs] c1249 to c1221) upstream of the iroN gene codes for F1C fimbriae and microcin H47 (Fig. 1). The regions flanking the iro genes in PAIs from UPEC 536 (PAI III536) (5, 12), UTI89 (8), and probiotic strain Nissle 1917 (genomic island I-Nissle 1917) (26) are organized similarly to those in strain CFT073. An iro gene cluster is also present on virulence plasmid pLVKP from Klebsiella pneumoniae CG43 (9) (Fig. 1). The iro genes carried by pLVKP exhibit from 85 to 90% nucleotide identity (82 to 92% identity at the amino acid level) to those of APEC χ7122. However, in the iro cluster from K. pneumoniae CG43, the orientation of the iroN gene is inverted and the iroE gene is absent (Fig. 1). In addition, a DNA region which flanks iroB in pAPEC-1, corresponding to ORFs 3 and 4, exhibits 76% nucleotide identity to a putative integrase-encoding gene, LV233, upstream of the iro genes of pLVKP (Fig. 1). These comparative sequence analyses are in support of a lateral transfer of salmochelin-encoding iro gene clusters among PAIs and different plasmids in E. coli and K. pneumoniae.
The iro genes are associated with clinical isolates and virulence among avian E. coli isolates.
The presence of iro genes was investigated in avian E. coli isolates from clinical cases (APEC) and isolates from the feces of healthy poultry (environmental isolates). Two hundred ninety-eight APEC strains and 32 environmental isolates were previously classified into three lethality classes according to results from virulence assays in 1-day-old chicks. APEC strains classified as LC1 are highly virulent, LC2 strains are moderately virulent, and LC3 strains are of low virulence. PCR amplifications with primers specific to iroB and iroN genes determined the presence or absence of the iro cluster among strains. There was a 100% correlation between results obtained using primers specific to either the iroB or iroN genes. iro sequences were present in 244 of the 298 (82%) APEC isolates and were significantly associated (P < 0.0001) with APEC compared to environmental isolates, for which only 10 of 32 (31%) isolates contained iro sequences. Among APEC isolates, iro sequences were also significantly associated with the lethality of the strains, with 91% of the highly virulent (LC1) strains containing iro sequences, compared to only 45% of the low-virulence (LC3) strains (Table 2). Taken together, the results demonstrate an association of iro genes with both clinical origin and increased virulence of avian E. coli.
TABLE 2.
Distribution of iro genes among APEC and avian fecal commensal isolates according to lethality class
| Lethality classa | No. (%) of E. coli isolates
|
|||
|---|---|---|---|---|
| APEC
|
Avian fecal commensal
|
|||
| Total | iro positiveb | Total | iro positiveb | |
| LC1 | 222 | 203 (91)c | 1 | 1 (100) |
| LC2 | 38 | 24 (63) | 12 | 6 (50) |
| LC3 | 38 | 17 (45) | 19 | 3 (16) |
| Total | 298 | 244 (82)d | 32 | 10 (31) |
Lethality classes were defined by lethality in 1-day-old chicks as follows: LC1, 50% lethal dose (LD50) of <108 CFU; LC2, LD50 of ≥108 CFU; LC3, not lethal at ≥108 CFU (14).
Positive PCR amplification of the iroB and iroN genes.
iro sequences were significantly associated with APEC isolates from LC1 as compared to APEC isolates from LC2 or LC3 (P < 0.0001) using Fisher's exact test.
iro sequences were significantly associated with APEC isolates relative to environmental isolates (P < 0.0001) using Fisher's exact test.
Role of iro genes in APEC virulence.
To characterize the importance of specific iro genes in APEC virulence, we determined the capacities of different mutants to infect chickens. Groups of 3-week-old birds were inoculated in the right caudal thoracic air sac with χ7122 or its isogenic mutant derivatives. From the inoculation site, virulent strains are typically able to invade and infect deeper tissues, generate gross lesions, and cause a systemic infection (43). However, in this model, attenuated strains are impaired in their capacity to colonize deeper tissues (4, 13, 33).
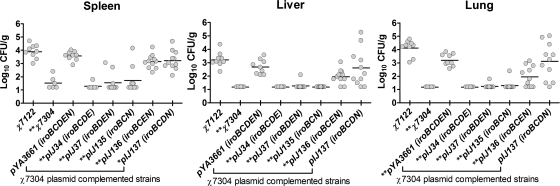
Bacterial counts from internal organs at 48 h postinfection (Fig. 3) demonstrated a systemic infection in chickens infected with virulent strains. Wild-type APEC χ7122 colonized the lung, liver, and spleen, whereas the Δiro ΔiucABC iutA mutant, χ7304, was not isolated from lungs or livers and was present in very low numbers from the spleens of infected chickens (Fig. 3). Complementation of χ7304 with the complete iro gene cluster (pYA3661) restored the capacity to colonize organs, although bacterial numbers in the lungs were significantly reduced (P = 0.003) compared to those in the wild-type parent strain, χ7122 (Fig. 3). The introduction of plasmids lacking either iroN (pIJ34 iroBCDE), iroC (pIJ37 iroBDEN), or iroDE (pIJ135 iroBCN) was not effective at complementing strain χ7304, and very few bacteria were isolated from the tissues of chickens infected with these strains (Fig. 3). Compared to complementation with the complete iro cluster (pYA3661), the absence of iroD (pIJ136) conferred a significantly reduced capacity to colonize the liver (P = 0.007), spleen (P = 0.03), and lungs (P = 0.003), although levels were markedly higher than those observed in the absence of iroN, iroC, or both iroD and -E (Fig. 3). In contrast, when χ7304 was complemented with pIJ137 (iroBCDN), which lacks iroE, complementation was as effective as complementation with the complete iro gene cluster. In fact, χ7304(pIJ137 iroBCDN) demonstrated no significant difference in colonization of tissues compared to χ7304(pYA3661). In addition, χ7304(pIJ137) demonstrated no significant difference in colonization of the lungs or liver compared to wild-type APEC strain χ7122.
FIG. 3.
Bacterial numbers present in the lungs, spleens, and livers of chickens infected with wild-type APEC strain χ7122, isogenic mutant derivatives, and iro plasmid-complemented strains. Data points represent bacterial counts from tissues isolated from different chickens (n = 5 to 11) 48 h postinfection. Horizontal bars represent the median bacterial CFU. Statistical differences compared with the wild-type strain are noted: * and **, P < 0.05 and P < 0.01, respectively, using the two-tailed Mann-Whitney test.
Gross lesions of colibacillosis present in the air sacs, livers, and pericardia of infected chickens were also in accordance with the bacterial levels observed in different tissues (Table 3). Gross lesions were present in the air sacs, livers, and hearts of chickens infected with APEC strain χ7122 or χ7304 complemented with the complete iro gene cluster (pYA3661) (Table 3). In contrast, lesions were minimal or absent in chickens infected with strain χ7304 or χ7304 complemented with plasmids lacking either iroN (pIJ34), iroC (pIJ37), or iroDE (pIJ135). Strain χ7304(pIJ136 iroBCEN), which lacks iroD, generated lesions of airsacculitis that were somewhat fewer than those induced by χ7304(pYA3661), although this reduction was not significant (P = 0.07). However, χ7304(pIJ136) generated lesions of pericarditis/perihepatitis that were significantly decreased compared to those generated by strain χ7304(pYA3661) (P = 0.003). In contrast, strain χ7304(pIJ137 iroBCDN), which lacks iroE, demonstrated no significant difference in generation of lesions of airsacculitis or pericarditis/perihepatitis compared to χ7304(pYA3661).
TABLE 3.
Score-based evaluation of gross lesions in organs of infected chickens
| E. coli strain | Mean lesion score ± SEMa
|
|
|---|---|---|
| Air sacsb | Liver and heartc | |
| χ7122 | 2.7 ± 0.2 | 3.2 ± 0.3 |
| χ7304 (Δiro ΔiucABC iutA) | 1.5 ± 0.4* | 0.8 ± 0.3** |
| χ7304(pYA3661 iroBCDEN) | 2.8 ± 0.3 | 3.2 ± 0.2 |
| χ7304(pIJ34 iroBCDE) | 1.6 ± 0.4* | 1.3 ± 0.5* |
| χ7304(pIJ37 iroBDEN) | 1.4 ± 0.3** | 1.0 ± 0.2** |
| χ7304(pIJ135 iroBCN) | 1.4 ± 0.3** | 0.8 ± 0.1** |
| χ7304(pIJ136 iroBCEN) | 1.9 ± 0.3 | 1.4 ± 0.1** |
| χ7304(pIJ137 iroBCDN) | 2.7 ± 0.3 | 2.4 ± 0.4 |
Lesion scores are presented as the mean (± standard error of the mean [SEM]) of values attributed to gross lesions of airsacculitis and combined lesions of perihepatitis/and perihepatitis as described in reference 33.
Mean lesion scores for airsacculitis in both caudal thoracic air sacs.
Combined lesion scoring values for pericarditis and perihepatitis. Statistical differences compared with the wild-type strain using the two-tailed Mann-Whitney test are noted: * and **, P < 0.05 and P < 0.001, respectively.
Detection of siderophores produced by APEC χ7122 in vitro and in vivo.
Relative levels of siderophores were initially determined from culture supernatants and from tissues of chickens infected with APEC strain χ7122 by LC-MS/MS using an MRM approach. Relative quantities of specific siderophores detected from samples are detailed in Fig. 3. Siderophores were detected from the air sacs and pericardia of chickens infected with 108 CFU. Attempts to detect siderophores from blood or liver or at an infective dose of 107 CFU were not successful. Culture supernatants following overnight growth in iron-poor M63 medium comprised a mean of 77.1% ± 3.3% of nonglucosylated enterobactin derivatives, 21.1% ± 2.6% of salmochelins, and only 1.9% ± 0.65% of aerobactin (Fig. 4). In contrast, in extracts from air sac tissues, aerobactin comprised 58.9% ± 3.4% of the siderophores, enterobactin derivatives comprised 27.1% ± 1.5%, and salmochelins comprised 14.0% ± 4.9%. In extracts from pericardial tissues, aerobactin, salmochelins, and enterobactin derivatives, respectively, comprised 48.0% ± 4.5%, 27.1% ± 5.1%, and 24.9% ± 0.59% of the siderophores detected. Overall, there was a major shift in the relative quantities of aerobactin and enterobactin and DHBS molecules produced in vitro compared to in vivo, whereas salmochelin levels were similar from either culture supernatants or tissues of infected chickens.
FIG. 4.
Mean distribution of siderophores produced by wild-type APEC χ7122 in iron-poor M63 medium (A) and in air sacs (B) and pericardia (C) of infected chickens. The error values are standard errors of the means. Bold values indicate significant differences for the same molecule compared to levels present in iron-poor M63 medium. The underlined value indicates a significant difference between the air sacs and the pericardial membrane. (D) Relative total percent distribution of siderophores belonging to each siderophore group. *, significant difference for the same group of siderophores between in vivo and in vitro conditions (P < 0.05) using the unpaired t test.
Quantification of siderophores from APEC χ7122 and mutant derivatives.
As iron-poor M63 medium was suitable for production of salmochelins from supernatants of strain χ7122, salmochelins, enterobactin, and linear DHBS molecules were quantified directly from the supernatants of strain χ7122, χ7304, and complemented strains grown in this medium. As expected, deletion of the iro gene cluster eliminated production of salmochelins in isogenic strain χ7304 (Fig. 5 and Table 4). Loss of the iro gene cluster in strain χ7304 also resulted in a mean overall 2.9-fold increase in enterobactin and DHBS molecules compared to the level of the wild-type strain χ7122 (Fig. 5).
FIG. 5.
Mean concentrations of enterobactin and DHBS molecules and salmochelins detected in culture supernatants. The amount of each molecule was normalized with the internal standard, and quantities were summed up according to the related siderophore group: salmochelins or enterobactin and nonglucosylated DHBS derivatives (Enterobactin/DHBS). Error bars represent the standard errors of the means. ¤, significant difference between the total enterobactin and DHBS molecules produced by strain χ7304 compared to χ7122; ¤¤, significant difference between the total enterobactin and DHBS molecules and total salmochelins produced by strain χ7304(pYA3661) compared to χ7122. *, significant difference between the total enterobactin and DHBS molecules produced compared to χ7304(pYA3661); **, significant difference between the total enterobactin and DHBS molecules and salmochelins produced compared to χ7304(pYA3661) (P < 0.05).
TABLE 4.
Mean concentrations of salmochelins detected in culture supernatants
| Strain (genotype) | Concn (μM) of salmochelina
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Monoglucosylated molecules
|
Diglucosylated molecules
|
Triglucosylated molecules
|
|||||||
| MGE | Linear MGE | S1 | SX | DGE (S4) | Linear DGE (S2) | S5 | TGE | Linear TGE | |
| χ7122 | 1.7 ± 0.28 | 1.1 ± 0.11 | 5.1 ± 0.12 | 9.8 ± 0.33 | 0.31 ± 0.03 | 1.1 ± 0.02 | 0.45 ± 0.02 | ND | ND |
| χ7304b | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| χ7304(pYA3661 iroBCDEN)b | 0.22 ± 0.02 | 0.14 ± 0.03 | 9.2 ± 0.94 | 31.0 ± 4.4 | 0.06 ± 0.004 | 2.9 ± 0.07 | 1.7 ± 0.18 | ND | ND |
| χ7304(pIJ34 iroBCDE)c | 0.09 ± 0.01 | 0.07 ± 0.01 | 8.6 ± 1.1 | 26.1 ± 3.9 | 0.04 ± 0.005 | 8.8 ± 0.98 | 1.5 ± 0.23 | ND | 0.06 ± 0.01 |
| χ7304(pIJ137 iroBCDN)c | 0.17 ± 0.001 | 0.22 ± 0.01 | 9.9 ± 0.55 | 20.4 ± 1.1 | 0.42 ± 0.02 | 22.7 ± 1.1 | 1.5 ± 0.05 | 0.04 ± 0.001 | 0.19 ± 0.01 |
| χ7304(pIJ136 iroBCEN)c | 0.09 ± 0.01 | 0.06 ± 0.01 | 4.0 ± 0.33 | 19.6 ± 1.8 | 0.23 ± 0.02 | 5.6 ± 0.94 | 1.7 ± 0.09 | ND | 0.04 ± 0.004 |
| χ7304(pIJ53 iroB)c | ND | ND | 0.04 ± 0.001 | ND | ND | ND | ND | ND | ND |
| χ7304(pIJ121 iroBC)c | 0.09 ± 0.01 | ND | 1.3 ± 0.02 | 3.2 ± 0.26 | 0.52 ± 0.02 | 5.4 ± 0.12 | 0.43 ± 0.02 | 0.25 ± 0.01 | 0.13 ± 0.01 |
| χ7304(pIJ135 iroBCN)c | 0.13 ± 0.01 | 0.04 ± 0.002 | 1.1 ± 0.04 | 3.5 ± 0.40 | 0.46 ± 0.01 | 5.1 ± 0.03 | 0.46 ± 0.04 | 0.29 ± 0.01 | 0.15 ± 0.01 |
| χ7304(pIJ37 iroBDEN)c | ND | ND | 1.3 ± 0.1 | 31.7 ± 3.5 | ND | 0.71 ± 0.07 | 0.05 ± 0.005 | ND | ND |
Values represent means ±standard errors of the means of at least three independently grown cultures. Bold values indicate significant differences (P < 0.05) using a two-tailed unpaired t test. ND, molecule not detected in samples.
Statistical differences were determined by using χ7122 as the reference strain.
Statistical differences were determined by using χ7304(pYA3661 iroBCDEN) as the reference.
Complementation of strain χ7304 with plasmid pYA3661 (iroBCDEN) increased salmochelin levels by a mean of 2.3-fold overall compared to APEC χ7122 and reduced the overall level of enterobactin-related molecules by 2.1-fold (Fig. 5). Specifically, introduction of the complete iro gene cluster conferred an increase in glucosylation of enterobactin derivatives and an increased hydrolysis of these products into linear DGE (S2), S1, S5, and SX compared to levels produced by wild-type strain χ7122 (Tables 4 and 5). Complementation of χ7304 with plasmid pIJ34 (iroBCDE), which lacks the iroN gene encoding the salmochelin receptor, resulted in catecholate siderophore levels that were similar to those observed with pYA3661 (iroBCDEN) (Fig. 5). However, the lack of iroN resulted in an increase in linear DGE (S2) and a reduction in MGE, linear MGE, enterobactin, and (DHBS)3 compared to pYA3661 (Tables 4 and 5).
TABLE 5.
Mean concentrations of enterobactin-related molecules detected in culture supernatants
| Strain (genotype) | Concn (μM) of enterobactin-related moleculea:
|
|||
|---|---|---|---|---|
| Enterobactin | (DHBS)3 | (DHBS)2 | DHBS | |
| χ7122 | 12.1 ± 3.4 | 25.3 ± 4.6 | 33.1 ± 4.7 | 11.6 ± 1.2 |
| χ7304b | 62.9 ± 7.2 | 62.7 ± 6.2 | 86.6 ± 9.3 | 25.9 ± 3.9 |
| χ7304(pYA3661 iroBCDEN)b | 1.5 ± 0.16 | 1.9 ± 0.25 | 9.6 ± 1.01 | 27.1 ± 3.7 |
| χ7304(pIJ34 iroBCDE)c | 0.9 ± 0.07 | 0.9 ± 0.11 | 10.9 ± 1.9 | 20.8 ± 3.1 |
| χ7304(pIJ137 iroBCDN)c | 3.8 ± 0.14 | 5.1 ± 0.23 | 22.2 ± 0.9 | 13.9 ± 0.8 |
| χ7304(pIJ136 iroBCEN)c | 0.6 ± 0.06 | 0.8 ± 0.12 | 8.6 ± 0.57 | 15.0 ± 1.3 |
| χ7304(pIJ53 iroB)c | 1.6 ± 0.35 | 0.6 ± 0.10 | 0.5 ± 0.01 | ND |
| χ7304(pIJ121 iroBC)c | 3.7 ± 0.20 | 2.9 ± 0.09 | 10.7 ± 0.4 | 1.6 ± 0.10 |
| χ7304(pIJ135 iroBCN)c | 4.6 ± 0.36 | 3.9 ± 0.29 | 11.8 ± 0.5 | 2.6 ± 0.15 |
| χ7304(pIJ37 iroBDEN)c | 2.7 ± 0.25 | 3.7 ± 0.35 | 15.3 ± 1.5 | 8.2 ± 1.12 |
Values represent means ± the standard errors of the means of at least three independently grown cultures. Bold values indicate significant differences (P < 0.05) using a two-tailed unpaired t test. ND, molecule not detected in samples.
Statistical differences were determined by using χ7122 as the reference strain.
Statistical differences were determined by using χ7304(pYA3661 iroBCDEN) as the reference strain.
Complementation of χ7304 with pIJ137 (iroBCDN), which lacks the iroE gene encoding a periplasmic hydrolase, did not alter the overall production of enterobactin or salmochelin molecules, compared to χ7304(pYA3661) (Fig. 5), although profiles for specific catecholate molecules differed (Tables 4 and 5). In contrast, complementation with pIJ136 (iroBCEN), which lacks the iroD gene encoding a cytoplasmic esterase, resulted in global 1.4-fold and 1.6-fold decreases in salmochelins and enterobactin and DHBS derivatives, respectively, compared to χ7304(pYA3661) (Fig. 5).
Complementation of mutant χ7304 with a plasmid carrying only iroB (pIJ53) resulted in a marked overall decrease of enterobactin-related molecules and salmochelins (Fig. 5). Traces of S1 were detected in the supernatants of this strain (Table 4), and although enterobactin levels in χ7304(pIJ53 iroB) supernatants were similar to those from χ7304(pYA3661), all linear DHBS products were greatly reduced (Table 5). Complementation of strain χ7304 with either pIJ121 (iroBC) or pIJ135 (iroBCN) restored production and secretion of salmochelins to some extent, although overall salmochelin levels were 4.0-fold lower than those in supernatants of χ7304(pYA3661) (Fig. 5). Despite the overall decrease in salmochelins, introduction of either pIJ121 (iroBC) or pIJ135 (iroBCN) to χ7304 resulted in high levels of DGE (S4) and TGE and reduced levels of SX, S1, and S5 (Table 4). χ7304(pIJ121 iroBC) and χ7304(pIJ135 iroBCN) also generated higher levels of enterobactin and (DHBS)3, whereas levels of DHBS were greatly reduced compared to those of χ7304(pYA3661) (Table 5). Taken together, these results suggest that enterobactin molecules underwent increased glucosylation in the presence of the IroB glucosyltransferase, but the tricyclic salmochelins could not be efficiently secreted in the absence of the IroC transporter and were poorly processed in the absence of the IroD and IroE hydrolases.
Complementation of χ7304 with plasmid pIJ37 (iroBDEN), which lacks the iroC gene, further demonstrated the importance of this gene for secretion of salmochelins, as only the monomeric SX salmochelin was present at appreciable levels in the supernatant of χ7304(pIJ37 iroBDEN) (Table 4). However, the overall level of enterobactin and DHBS molecules detected was similar to that of χ7304(pYA3661) (Fig. 5), and in fact levels of enterobactin, (DHBS)3, and (DHBS)2 were increased compared to those of χ7304(pYA3661) (Table 5). Hence, overall iroC was important for secretion of salmochelins in the supernatant but was not important for secretion of enterobactin.
DISCUSSION
The iro genes encoding the salmochelin siderophore system were initially identified as an iron- and pH-regulated locus (named the iroA locus) in Salmonella enterica serovar Typhimurium (20, 21). Further studies determined that the iroA locus comprised two divergently transcribed sets of genes (iroBCDE and iroN) (2). The iro sequences which are present in all phylogenetic lineages of S. enterica (2), are associated with the Fels-2 prophage region inserted at the tmRNA (ssrA) site (61), and were likely acquired through horizontal gene transfer. iro gene clusters were also identified within PAIs located at different tRNA sites on the chromosomes of several ExPEC strains (12, 51, 60). In addition, some E. coli and Klebsiella pneumoniae strains contain iro sequences localized to plasmids (9, 13, 28, 29, 56). The presence of common sequences bordering iro genes in different strains suggests a possible common ancestry of these horizontally acquired genes (Fig. 1).
Among E. coli isolates, iro sequences were shown to be associated with ExPEC isolated from neonatal meningitis (38), UTIs, and prostatitis in humans (1, 30, 51) as well as APEC (15, 48). In accordance with these reports, in our study iro sequences were highly associated with the virulence of APEC strains in 1-day-old chicks, and iro sequences were also significantly associated with APEC compared to fecal commensal E. coli isolates from poultry (Table 2). The increased association of iro sequences among ExPEC and APEC isolates also correlates with the importance of iro genes for the virulence of E. coli in different infection models (13, 19, 38, 54). However, prior to our current study, aside from the IroN salmochelin receptor, the individual roles of iro-containing genes for E. coli virulence had not been investigated.
To our knowledge, this is the first study to compare relative levels of catecholate siderophores (salmochelins, enterobactin, and derivatives) and aerobactin produced by E. coli during infection of host tissues as well as following growth in iron-poor culture medium. The proportions of salmochelins produced in tissues and in vitro were quite similar. In contrast, clearly there were major differences between the relative amounts of aerobactin and enterobactin siderophores present in vivo and in vitro, and there were a marked increase in aerobactin and a decrease in enterobactin observed from host extraintestinal tissues (Fig. 4). The production of different siderophores by ExPEC is likely required to provide an adaptive advantage, as different siderophores may exhibit optimal activity under various conditions, including pH, carbon availability, and growth temperature. From this standpoint, the increased level of aerobactin detected in vivo may be due to the fact that aerobactin acts as an effective siderophore under the physiological conditions present in these tissues, and therefore its expression may be upregulated in vivo. In vitro, aerobactin was better produced under slightly acid conditions, whereas the catecholate siderophores were more abundant at neutral or slightly alkaline pH by strain Nissle 1917 (59). These findings suggest that important regulatory mechanisms, in addition to iron availability itself, can alter production of siderophores in vitro as well as in vivo and further emphasize the role aerobactin may contribute to E. coli extraintestinal virulence. In addition to regulatory changes that may occur in vivo, it is also possible that the decreased detection of enterobactin from tissues may in part be due to association of this siderophore with host proteins such as albumin or NGAL orthologs.
As the presence of multiple iron uptake systems may hinder the analysis of the roles of specific iron transport systems or their individual components in extraintestinal E. coli infections (13, 54, 57), we used plasmid complementation of an attenuated salmochelin- and aerobactin-negative APEC strain to assess the specific importance of individual iro genes for APEC virulence and for the production of different types of salmochelin molecules. Complementation of the attenuated mutant with the complete iro gene cluster on a medium-copy plasmid restored lesion scores to levels similar to those of the wild-type parent strain, χ7122 (Table 3), and considerably increased colonization of systemic organs even in the absence of aerobactin (Fig. 3). Compensation for the lack of aerobactin and the regaining of virulence are likely due to an increased capacity to produce and process salmochelins. Introduction of the iro genes on pYA3661 resulted in over twice the production of salmochelins and a greater level of glucosylated dimers and monomers compared to those of the wild-type parent (Table 5 and Fig. 4).
Complementation of attenuated strain χ7304 with plasmids containing different combinations of iro genes demonstrated that, in particular, the iroN and iroC genes, as well as the combination of iroD and iroE genes, were critical for virulence. The IroN salmochelin receptor-encoding gene did not effect salmochelin production, although it was required for virulence. These results are in line with the attenuation of ExPEC iroN mutants (38, 54). Recently, IroN has also been shown to demonstrate a cell invasion phenotype, which could further contribute to its importance for ExPEC virulence (16). In contrast to ExPEC, loss of IroN, together with FepA, in Salmonella enterica serovar Typhimurium and S. enterica serovar Enteritidis had little effect on systemic spread in mouse and chicken models, respectively, indicating that salmochelin uptake and enterobactin uptake are not critical for Salmonella virulence (45). However, since the combined loss of IroN, FepA, and Cir resulted in attenuation, uptake of DHBS and enterobactin breakdown products appears to contribute to Salmonella virulence (45). Thus, although ExPEC possess numerous catecholate siderophore receptors, including FepA, Fiu, Cir, Iha, and other uncharacterized putative siderophore receptors, it is likely that only IroN functions as the salmochelin receptor and that the uptake of salmochelins is a prerequisite for full virulence.
χ7304(pIJ37 iroBDEN), which lacks only iroC, secreted no detectable cyclic glucosylated salmochelins, very little glucosylated linear trimers and dimers, and a substantial level of SX monomer (Table 4). However, in the absence of iroC, enterobactin, (DHBS)3, and (DHBS)2 were abundant. These results support a role for the iroC gene product in the export of salmochelins, with the exception of the SX monomer, which may possibly be exported by another pathway. Furthermore, the attenuation of this mutant indicates that enterobactin and its breakdown products could not serve as efficient siderophores in vivo. Recently, Crouch et al. (10) also demonstrated the role of iroC for secretion of DGE (S4) and linear DGE (S2) as well as the importance of this gene for virulence of Salmonella enterica serovar Typhimurium in the mouse.
χ7304(pIJ135 iroBCN), which lacks both the iroD and iroE genes encoding salmochelin hydrolases (62), secreted small amounts of SX monomer and S1 and S5 dimers and some of the largest amounts of TGE and DGE (S4) detected in supernatants. Hence, the lack of virulence of this strain is likely due to a reduced processing of cyclic salmochelins for efficient iron acquisition during an infection. The IroD hydrolase appeared to play a predominant role in virulence compared to the IroE hydrolase (Fig. 3 and Table 3). These results are in accordance with the enzymatic activities of these hydrolases (36). Purified IroE demonstrated higher selectivity for apo-siderophores and mostly only cleaved cyclic compounds into linear trimers (36), suggesting IroE may be more important for export or processing prior to release of siderophores for iron scavenging. In contrast, IroD demonstrated higher affinity for Fe3+-loaded siderophores and efficiently processed cyclic salmochelins and enterobactin into trimers, dimers, and monomers, favoring its role in cytoplasmic release of iron (36). The lack of IroE in the iro gene cluster of K. pneumoniae strain CG3, a bacteremia isolate that is highly virulent in mice (7), further supports that iroE may be of minimal importance for a fully functional salmochelin system. Although the importance of salmochelins for virulence of K. pneumoniae remains to be established, iroE may have been lost by attrition, at least in this K. pneumoniae strain, if it provided no selective advantage. However, since iro gene clusters have been shown to be present in multiple copies in certain E. coli strains, the possibility that there are additional copies of iro genes, including an iroE ortholog, within certain K. pneumoniae strains cannot be excluded.
The importance of salmochelins as newly discovered siderophores that are able to evade the mammalian innate immune response protein NGAL (lipocalin 2 and siderocalin) has been revelatory for furthering our understanding of the role of the enterobactin pathway as a precursor required for the generation of these glucosylated virulence factors (18, 27) and the importance of NGAL as a host response protein that plays a critical role in host protection from certain bacterial infections and possibly detection of enterobactin (19, 22, 40). In the avian host, it is clear from our study that the enterobactin system alone is insufficient for APEC virulence, and it is thus probable that an avian host defense protein akin to NGAL plays a similar role in protection against enterobactin-mediated iron acquisition. A number of different chicken lipocalins which have been shown to be upregulated during inflammation or in response to bacterial signaling molecules have been identified (41), and one of these may have a protective role against enterobactin-mediated bacterial iron acquisition. However, as demonstrated in the present work, the salmochelin-siderophore system, comprised of functional synthesis, export, import, and processing components encoded on the iro gene cluster, is efficient at circumventing any avian innate host defenses that may respond to or sequester enterobactin.
Acknowledgments
Kind thanks to R. Curtiss III (Biodesign Institute, Arizona State University) and H. Y. Kang (Pusan National University, Korea) for helpful discussions and support.
M.C. was funded by a Fondation Armand-Frappier scholarship. Funding for this project was provided by the Natural Sciences and Engineering Research Council of Canada, the Canadian Foundation for Innovation, and the Canada Research Chairs programs.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 9 June 2008.
REFERENCES
- 1.Bauer, R. J., L. Zhang, B. Foxman, A. Siitonen, M. E. Jantunen, H. Saxen, and C. F. Marrs. 2002. Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection—usp, iha, and iroNE. coli. J. Infect. Dis. 1851521-1524. [DOI] [PubMed] [Google Scholar]
- 2.Bäumler, A. J., T. L. Norris, T. Lasco, W. Voight, R. Reissbrodt, W. Rabsch, and F. Heffron. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 1801446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bister, B., D. Bischoff, G. J. Nicholson, M. Valdebenito, K. Schneider, G. Winkelmann, K. Hantke, and R. D. Sussmuth. 2004. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17471-481. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. K., and R. Curtiss III. 1996. Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 9311149-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzuszkiewicz, E., H. Bruggemann, H. Liesegang, M. Emmerth, T. Olschlager, G. Nagy, K. Albermann, C. Wagner, C. Buchrieser, L. Emody, G. Gottschalk, J. Hacker, and U. Dobrindt. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. USA 10312879-12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1341141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, H. Y., J. H. Lee, W. L. Deng, T. F. Fu, and H. L. Peng. 1996. Virulence and outer membrane properties of a galU mutant of Klebsiella pneumoniae CG43. Microb. Pathog. 20255-261. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 1035977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y. T., H. Y. Chang, Y. C. Lai, C. C. Pan, S. F. Tsai, and H. L. Peng. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337189-198. [DOI] [PubMed] [Google Scholar]
- 10.Crouch, M. L., M. Castor, J. E. Karlinsey, T. Kalhorn, and F. C. Fang. 2008. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 67971-983. [DOI] [PubMed] [Google Scholar]
- 11.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30299-316. [PubMed] [Google Scholar]
- 12.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536) to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 706365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dozois, C. M., M. Dho-Moulin, A. Brée, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the tsh genetic region. Infect. Immun. 684145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewers, C., G. Li, H. Wilking, S. Kiessling, K. Alt, E. M. Antao, C. Laturnus, I. Diehl, S. Glodde, T. Homeier, U. Bohnke, H. Steinruck, H. C. Philipp, and L. H. Wieler. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297163-176. [DOI] [PubMed] [Google Scholar]
- 16.Feldmann, F., L. J. Sorsa, K. Hildinger, and S. Schubert. 2007. The salmochelin siderophore receptor IroN contributes to invasion of urothelial cells by extraintestinal pathogenic Escherichia coli in vitro. Infect. Immun. 753183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischbach, M. A., H. Lin, D. R. Liu, and C. T. Walsh. 2006. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2132-138. [DOI] [PubMed] [Google Scholar]
- 18.Fischbach, M. A., H. Lin, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl. Acad. Sci. USA 102571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischbach, M. A., H. Lin, L. Zhou, Y. Yu, R. J. Abergel, D. R. Liu, K. N. Raymond, B. L. Wanner, R. K. Strong, C. T. Walsh, A. Aderem, and K. D. Smith. 2006. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. USA 10316502-16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 1744317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, J. W., Y. K. Park, I. S. Bang, K. Karem, H. Betts, H. K. Hall, and E. Shaw. 1994. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology 140341-352. [DOI] [PubMed] [Google Scholar]
- 22.Goetz, D. H., M. A. Holmes, N. Borregaard, M. E. Bluhm, K. N. Raymond, and R. K. Strong. 2002. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 101033-1043. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths, E. 1999. Iron in biological systems, p. 1-26. In J. J. Bullen and E. Griffiths (ed.), Iron and infection: molecular, physiological and clinical aspects, 2nd ed. John Wiley & Sons, Hoboken, NJ.
- 24.Griffiths, E., and P. Williams. 1999. The iron uptake systems of pathogenic bacteria, fungi and protozoa, p. 87-212. In J. J. Bullen and E. Griffiths (ed.), Iron and infection: molecular, physiological and clinical aspects, 2nd ed. John Wiley & Sons, Hoboken, NJ.
- 25.Gross, W. G. 1994. Diseases due to Escherichia coli in poultry, p. 237-259. In C. L. Gyles (ed.), E. coli in domestic animals & humans. CABI, Wallingford, United Kingdom.
- 26.Grozdanov, L., C. Raasch, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, and U. Dobrindt. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 1865432-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 1003677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, T. J., S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 1885975-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanamaru, S., H. Kurazono, S. Ishitoya, A. Terai, T. Habuchi, M. Nakano, O. Ogawa, and S. Yamamoto. 2003. Distribution and genetic association of putative uropathogenic virulence factors iroN, iha, kpsMT, ompT and usp in Escherichia coli isolated from urinary tract infections in Japan. J. Urol. 1702490-2493. [DOI] [PubMed] [Google Scholar]
- 31.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 32.Konopka, K., and J. B. Neilands. 1984. Effect of serum albumin on siderophore-mediated utilization of transferrin iron. Biochemistry 232122-2127. [DOI] [PubMed] [Google Scholar]
- 33.Lamarche, M. G., C. M. Dozois, F. Daigle, M. Caza, R. Curtiss III, J. D. Dubreuil, and J. Harel. 2005. Inactivation of the Pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect. Immun. 734138-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lepine, F., E. Deziel, S. Milot, and L. G. Rahme. 2003. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta 162236-41. [DOI] [PubMed] [Google Scholar]
- 35.Léveillé, S., M. Caza, J. R. Johnson, C. Clabots, M. Sabri, and C. M. Dozois. 2006. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect. Immun. 743427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, H., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 12711075-11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marrs, C. F., L. Zhang, and B. Foxman. 2005. Escherichia coli mediated urinary tract infections: are there distinct uropathogenic E. coli (UPEC) pathotypes? FEMS Microbiol. Lett. 252183-190. [DOI] [PubMed] [Google Scholar]
- 38.Negre, V. L., S. Bonacorsi, S. Schubert, P. Bidet, X. Nassif, and E. Bingen. 2004. The siderophore receptor IroN, but not the high-pathogenicity island or the hemin receptor ChuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect. Immun. 721216-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson, A. L., J. M. Barasch, R. M. Bunte, and J. N. Weiser. 2005. Bacterial colonization of nasal mucosa induces expression of siderocalin, an iron-sequestering component of innate immunity. Cell. Microbiol. 71404-1417. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, A. L., A. J. Ratner, J. Barasch, and J. N. Weiser. 2007. Interleukin-8 secretion in response to aferric enterobactin is potentiated by siderocalin. Infect. Immun. 753160-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pagano, A., P. Giannoni, A. Zambotti, D. Sanchez, M. D. Ganfornina, G. Gutierrez, N. Randazzo, R. Cancedda, and B. Dozin. 2004. Phylogeny and regulation of four lipocalin genes clustered in the chicken genome: evidence of a functional diversification after gene duplication. Gene 33195-106. [DOI] [PubMed] [Google Scholar]
- 42.Payne, S. M., and A. R. Mey. 2004. Pathogenic Escherichia coli, Shigella, and Salmonella, p. 199-218. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 43.Pourbakhsh, S. A., M. Boulianne, B. Martineau-Doize, C. M. Dozois, C. Desautels, and J. M. Fairbrother. 1997. Dynamics of Escherichia coli infection in experimentally inoculated chickens. Avian Dis. 41221-233. [PubMed] [Google Scholar]
- 44.Provence, D. L., and R. Curtiss III. 1992. Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or curli production. Infect. Immun. 604460-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabsch, W., U. Methner, W. Voigt, H. Tschäpe, R. Reissbrodt, and P. H. Williams. 2003. Role of receptor proteins for enterobactin and 2,3-dihydroxybenzoylserine in virulence of Salmonella enterica. Infect. Immun. 716953-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54881-941. [DOI] [PubMed] [Google Scholar]
- 47.Restieri, C., G. Garriss, M.-C. Locas, and C. M. Dozois. 2007. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl. Environ. Microbiol. 731553-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 1512097-2110. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36241-256. [DOI] [PubMed] [Google Scholar]
- 50.Ron, E. Z. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 928-32. [DOI] [PubMed] [Google Scholar]
- 51.Russo, T. A., U. B. Carlino, A. Mong, and S. T. Jodush. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect. Immun. 675306-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russo, T. A., and J. R. Johnson. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5449-456. [DOI] [PubMed] [Google Scholar]
- 53.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 1811753-1754. [DOI] [PubMed] [Google Scholar]
- 54.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, T. J. Barnard, and J. R. Johnson. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 707156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schubert, S., B. Picard, S. Gouriou, J. Heesemann, and E. Denamur. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 705335-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sorsa, L. J., S. Dufke, J. Heesemann, and S. Schubert. 2003. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 713285-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 696179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valdebenito, M., B. Bister, R. Reissbrodt, K. Hantke, and G. Winkelmann. 2005. The detection of salmochelin and yersiniabactin in uropathogenic Escherichia coli strains by a novel hydrolysis-fluorescence-detection (HFD) method. Int. J. Med. Microbiol. 29599-107. [DOI] [PubMed] [Google Scholar]
- 59.Valdebenito, M., A. L. Crumbliss, G. Winkelmann, and K. Hantke. 2006. Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int. J. Med. Microbiol. 296513-520. [DOI] [PubMed] [Google Scholar]
- 60.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams, K. P. 2003. Traffic at the tmRNA gene. J. Bacteriol. 1851059-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, M., M. Valdebenito, G. Winkelmann, and K. Hantke. 2005. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 1512363-2372. [DOI] [PubMed] [Google Scholar]