Abstract
Pili are putative virulence factors and promising vaccine candidates in Streptococcus agalactiae (group B Streptococcus [GBS]) infection, a leading cause of neonatal sepsis and meningitis. The genes necessary for pilus synthesis and assembly are clustered in pilus islands (PI). Each gene encodes three structural subunits (a backbone and two ancillary proteins) bearing a C-terminal LPXTG motif and two subfamily C sortases (SrtC) involved in covalent polymerization of the subunits. GBS strains also possess the conserved “housekeeping” sortase A (SrtA), but its role in pilus assembly is unclear. To address this issue, pilus expression and cell wall anchoring were analyzed in srtA deletion mutants. Loss of SrtA did not affect pilus polymerization. However, pilus expression on the cell surface was reduced, and pili accumulated in the culture supernatant. Furthermore, cell-associated pili could be readily released by detergent treatment, indicating that SrtA is involved in covalent anchoring of pili to the cell wall. When each of the genes comprising PI-2a was systematically deleted, only the absence of ancillary subunit GBS150 or the SrtC required for incorporation of GBS150 into pili mimicked the srtA mutant phenotype. Thus, from these data a model for GBS pilus assembly can be proposed in which PI sortases are responsible for polymerization of the pilus structure, while SrtA is required to covalently attach it to the cell wall, utilizing ancillary pilus subunit GBS150 as the anchor protein.
Streptococcus agalactiae (group B Streptococcus [GBS]) can be found as part of the normal flora in the gastrointestinal and genitourinary tracts of up to 50% of the healthy adult population (14, 31). However, as an opportunistic pathogen, it is the leading cause of neonatal sepsis, pneumonia, and meningitis in the industrialized world (6, 16, 31), and the incidence of GBS-mediated invasive disease is increasing in the elderly population (9).
Putative virulence factors for GBS include filamentous appendages that extend from the bacterial cell surface, termed pili. Such structures have been implicated in mediating attachment to human epithelial cells (8) and in the binding and invasion of brain microvascular endothelial cells (21). Pili have also been reported in other gram-positive bacteria, including Corynebacterium diphtheriae (11, 37, 40) and members of the oropharyngeal microflora, such as Streptococcus pyogenes (27), Streptococcus pneumoniae (3), and Actinomyces naeslundii (10, 13). Similar to GBS, these structures facilitate adhesion to a variety of host tissues (1, 3, 22, 23, 41) and/or mediate interbacterial coaggregation (25). Furthermore, pilus proteins of GBS, S. pyogenes, and S. pneumoniae have all been shown to elicit a protective immune response in mouse models and are, hence, vaccine candidates against these important pathogens (12, 18, 20, 27).
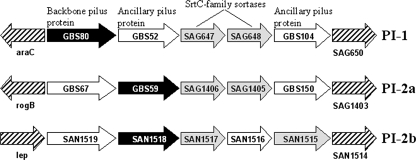
The genes encoding pili in GBS are located within two distinct loci, denoted pilus islands 1 and 2 (PI-1 and PI-2), and comparative analyses of available genomes revealed two variants of PI-2, designated PI-2a and PI-2b (29). All, however, conform to the same basic organization (Fig. 1). Each PI comprises three genes encoding LPXTG family proteins, which constitute the physical pilus structure, along with two genes encoding transpeptidase enzymes associated with polymerization of the pilus protein subunits. The three pilus proteins of GBS are expressed as precursor polypeptides with an N-terminal signal peptide and a C-terminal cell wall sorting signal (29). One of these proteins, termed the “backbone” subunit, forms the shaft of the pilus, while the other two “ancillary” subunits appear intermittently in the structure.
FIG. 1.
Schematic representation of GBS PIs. Genes encoding the three LPXTG proteins that comprise the pilus structure are represented by black (backbone subunit) and white (ancillary subunits) arrows. Subfamily SrtC transpeptidases that polymerize the protein subunits are shown in gray. Gene designations correspond to GBS strain 2603V/R, accession number AE009948 (PI-1 and PI-2a) or strain COH1, accession number AAJR00000000 (PI-2b).
Following translocation via the Sec system, pilus precursor proteins are transiently retained in the cell membrane by means of their C-terminal hydrophobic tails and become the target of membrane-associated transpeptidases of the sortase family (comprehensively reviewed by Marraffini et al. [24]). Based on phylogenetic analyses, two recent studies (5, 7) proposed the classification of sortases into either four (subfamilies A to D) or five (SrtA, SrtB, and families 3 to 5) subfamilies. GBS strains possess sortases from two of these subfamilies (5): sortase A (SrtA) and sortase C (SrtC or family 3). SrtA functions to anchor the majority of surface-exposed proteins in GBS that bear a C-terminal pentapeptide recognition sequence (LPXTG) to the bacterial cell wall. Specifically, SrtA cleaves the target protein between the threonyl and glycyl residues of the LPXTG motif to form an acyl enzyme intermediate. This is then resolved by the nucleophilic attack of amino groups, usually provided by the lipid II precursor of peptidoglycan, which is subsequently incorporated into the cell envelope. By contrast, the SrtC enzymes of GBS are located within the PI and function to polymerize only those LPXTG proteins located within the same PI (29). This process is not fully understood, but a mechanism proposed by Ton-That et al. (38) for pilus assembly in C. diphtheriae implies that SrtC transpeptidases serve to covalently join two protein subunits, utilizing the LPXTG motif of one and the conserved lysine residue of a canonical “pilin” motif of the other (reviewed in Telford et al. [34]). Mutating either of these motifs in the backbone subunit has been shown to abrogate pilus formation (37). Each PI of GBS contains two genes encoding SrtC transpeptidases. Both have been shown to be capable of polymerizing the backbone pilus subunit, but each preferentially incorporates one of the two ancillary proteins (29).
There is growing evidence that, in addition to the SrtC transpeptidases, the housekeeping SrtA may play some role in GBS pilus assembly. Nevertheless, the mechanism by which this might occur remained unclear. By comparing mutants defective in PI-associated sortases to those lacking SrtA, this study confirmed that, while not involved in pilus polymerization, SrtA is essential for the permanent anchoring of GBS pili to the cell wall. Moreover, a detailed analysis of PI-2a identified ancillary protein GBS150 as the substrate for SrtA and thus the anchor protein of these pilus structures. The work presented here, therefore, provides some of the first direct evidence as to the mechanism by which SrtA-mediated anchoring of pili can occur in GBS.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Streptococci (listed in Table 1) were routinely grown in Todd Hewitt broth (THB) or chemically defined synthetic medium (FMC) (35, 36) at 37°C in 5% CO2. Escherichia coli cells were grown aerobically at 37°C in Luria-Bertani medium. When required, antibiotics were added to the medium at the following concentrations: erythromycin, 5 μg ml−1 (S. agalactiae) or 400 μg ml−1 (E. coli); chloramphenicol, 10 μg ml−1 (S. agalactiae) or 20 μg ml−1 (E. coli).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Invitrogen |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG/pMON14272/pMON7124 | Invitrogen |
| S. agalactiae | ||
| 515 | Wild type | D. Kasper, Harvard Medical School, Boston, MA |
| 515 (pAMp) | Wild-type complemented with pAM401/gbs80P+T | This study |
| 515ΔsrtA | ΔsrtA | This study |
| 515ΔsrtA (pAMp) | ΔsrtA complemented with pAM401/gbs80P+T | This study |
| 515ΔsrtA+ | ΔsrtA complemented with pAM401-srtA | This study |
| Δ59 | Δgbs59 | 29 |
| Δ67 | Δgbs67 | 29 |
| Δ150 | Δgbs150 | 29 |
| Δ1405 | ΔSAG1405 | 29 |
| Δ1406 | ΔSAG1406 | 29 |
| Plasmids | ||
| pJRS233 | 6.0 kb; Emr; ColE1 ori; temperature-sensitive E. coli-streptococcal shuttle vector | 28 |
| pJRS233-ΔsrtA | pJRS233-derived containing overlapping flanking sequences of srtA gene | This study |
| pAM401/gbs80P+T | 11.5 kb; Cmr; ColE1 ori; E. coli-streptococcal shuttle vector pAM401 containing promoter of gbs80 | 29 |
| pAM401-srtA | pAM401/gbs80P+T-derived containing entire srtA coding sequence | This study |
Antisera.
LPXTG family and PI proteins were expressed as His-tagged fusion proteins and purified by affinity chromatography, as reported previously (26). Specific antisera were then generated by immunizing CD1 mice with each of these recombinant proteins (20).
Construction of in-frame deletion mutants.
Standard recombinant DNA techniques were employed as described by Sambrook et al. (30). Plasmids (Table 1) were purified from E. coli cells using a Wizard Plus SV Miniprep System (Promega, Madison, WI). Oligonucleotides (Table 2) were synthesized in-house or by Invitrogen (Carlsbad, CA). Chromosomal DNA was prepared from mutanolysin-treated streptococcal cells using a Nucleospin Tissue Kit (Clontech, Mountain View, CA). PCRs were performed using GoTaq DNA polymerase as recommended by the manufacturer (Promega). PCR products were purified using the Wizard SV Gel/PCR Clean-Up System (Promega). DNA restriction and modification enzymes were used under the conditions specified by the manufacturer (NEB, Ipswich, MA).
TABLE 2.
Primers used in this study
| Primer | Sequencea | Reference |
|---|---|---|
| srtA.F1D | CTGTGCCTCGAGGGAGCGTCAGAGTCAAGC | This study |
| srtA.F2B | ATTGTTGGGTTGGCTCGTATTATTGTGCATGCTGAA | This study |
| srtA.R1B | ATGCACAATAATACGAGCCAACCCAACAATAATTAG | This study |
| srtA.R2 | GCGTTGCTGCAGCAGCATTGCGATACATATCTT | This study |
| srtAcomp.F | AGAACAGCGGCCGCAAAGAATAGGAAGTTATGCGTAAT | This study |
| srtAcomp.R | GTCGGGAGATCTTTCTAACTACCTTCTAGAGATTAATTTG | This study |
| srttrans.F | CTGCTCAAACGAAATCACAT | This study |
| srttrans.R | CTTGCCAGGTGTATCATCAA | This study |
| gyrA.F | GTCATGGAAACTTTGGTTCA | This study |
| gyrA.R | GCTCTTTCCCATTTGAAGTT | This study |
| srtAhyp.F | TATTCGTGAAAACCATCGTC | This study |
| srtAhyp.R | CAATTCGGCCTACCTATTCT | This study |
| srtAunk.F | ATTTACCCTAGCGAGTCCAG | This study |
| srtAunk.R | AGAGCACTCTCCCCAGTTAC | This study |
Restriction enzyme sites are underlined.
The srtA gene of S. agalactiae 515 was inactivated using the PCR method of splicing by overlap extension, as described previously (15). PCR amplification with primers srtA.F1D/srtA.R1B and srtA.F2B/srtA.R2 from genomic DNA template generated two fragments comprising the flanking sequences of the srtA gene (1,309 bp and 1,095 bp), with an overlap of 30 bp. These were ligated via the overlap and cloned into the temperature-sensitive allelic exchange vector pJRS233 (gift of June Scott, Emory University, Atlanta, GA) (28), generating plasmid pJRS233-ΔsrtA. Transformation and allelic exchange were then performed as described previously (29). Confirmation of predicted insertions was obtained by PCR amplification and sequencing.
To complement the srtA deletion mutant, a DNA fragment (796 bp) incorporating the entire srtA gene was PCR amplified from genomic DNA using primers srtAcomp.F and srtAcomp.R. This product was cloned into E. coli-streptococcal shuttle vector pAM401/gbs80P+T (29), generating plasmid pAM401-srtA. This construct was purified and used to transform S. agalactiae srtA mutants by electroporation. Complementation was confirmed by detection of srtA RNA transcript using the primers srttrans.F and srttrans.R. RNA extraction and cDNA synthesis were performed as described below. Empty pAM401/gbs80P+T without the srtA coding sequence was also used to transform wild-type and srtA mutant strains as controls for effects induced by vector alone.
RNA extraction and cDNA synthesis.
Bacterial mid-exponential-phase cultures (6 ml) were stabilized using RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany), according to manufacturer's instructions, and the cells were subsequently harvested (3,000 × g for 20 min at 4°C). Cells were incubated at 37°C for 10 min in Tris-EDTA buffer containing 15 mg ml−1 lysozyme and 100 U of mutanolysin and mixed with 350 μl of buffer RLT (RNeasy Mini Kit; Qiagen). RNA was then prepared using an RNeasy Mini Kit (Qiagen), according to manufacturer's instructions. The integrity of the RNA was confirmed by gel electrophoresis, and the RNA was then treated with DNase I (Promega) for 2 h at 37°C. The concentration of RNA was determined by measuring the A260 in a spectrophotometer. RNA (2 μg) was reverse transcribed into cDNA with random hexamer primers, as described previously (43). For each RNA sample, a control cDNA reaction in the absence of reverse transcriptase was performed to check for DNA contamination.
Immunoblotting.
S. agalactiae strains were maintained at 37°C and 5% CO2 in either THB or chemically defined FMC medium. For total protein extracts, mid-exponential-phase cells were harvested, washed in phosphate-buffered saline (PBS) and resuspended in 50 mM Tris-HCl containing 400 U of mutanolysin. Cell suspensions were incubated at 37°C for 2 h and lysed by three cycles of freeze-thawing, and the cellular debris was removed (12,000 × g for 10 min). Supernatants were collected, and protein concentration was determined using a Bio-Rad protein assay (Hercules, CA). Proteins (20 μg) were resolved on 3 to 8% or 4 to 12% NuPage Novex sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Invitrogen) and transferred to nitrocellulose. Membranes were probed with mouse antiserum directed against LPXTG family or PI proteins (1:1,000 dilution), followed by a rabbit anti-mouse horseradish peroxidase-conjugated secondary antibody (Dako, Glostrup, Denmark). Bands were then visualized using an Opti-4CN substrate kit (Bio-Rad) or SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
To visualize proteins released during growth, culture supernatants were harvested (3,000 × g for 20 min), dialyzed extensively against distilled H2O and concentrated by lyophilization, before being subjected to SDS-PAGE, with 2 ml of supernatant equivalent loaded per well. Corresponding cell pellets were digested with mutanolysin, as described above, prior to dialysis and lyophilization.
Protein solubility in detergent.
Bacterial cultures (10 ml) were grown to an optical density at 600 nm of 1.0, harvested (3,000 × g for 20 min), washed with PBS, and then adjusted to 1 × 109 cells ml−1 in 0.5 M Tris-HCl (pH 8), with or without 0.5% (wt/vol) SDS. Cell suspensions were incubated for 1.5 h at 25°C with gentle agitation. Supernatants were collected (3,000 × g for 20 min), dialyzed extensively against distilled H2O, and concentrated by lyophilization, before being subjected to SDS-PAGE, with 2 ml of supernatant equivalent loaded per well. Corresponding cell pellets were digested with mutanolysin prior to visualization by immunoblotting, as described above.
Flow cytometry.
Following SDS treatment, as described above, harvested bacterial cells were resuspended in PBS containing 0.1% (wt/vol) paraformaldehyde. Cell suspensions were incubated at 37°C for 1 h, followed by an additional incubation at 25°C for 1 h. Fixed cells were then washed in PBS and incubated at 25°C for 20 min in newborn calf serum (Sigma, St. Louis, MO). Bacteria were then incubated for 1 h at 4°C with preimmune or immune serum diluted 1:200 in dilution buffer (PBS, 0.1% [wt/vol] bovine serum albumin, 20% [vol/vol] newborn calf serum). Cells were washed in PBS-0.1% (wt/vol) bovine serum albumin and incubated for a further 1 h with R-phycoerythrin-conjugated F(ab)2 goat anti-mouse immunoglobulin G (1:100 dilution) (Jackson ImmunoResearch Laboratories, West Grove, PA). Cells were washed again, resuspended in PBS, and analyzed with a FACSCalibur apparatus (Becton Dickinson, Franklin Lakes, NJ), using FlowJo software (Tree Star, Ashland, OR). The difference in mean fluorescence levels between preimmune and immune sera was calculated for each pilus protein.
RESULTS
Generation of S. agalactiae srtA mutants.
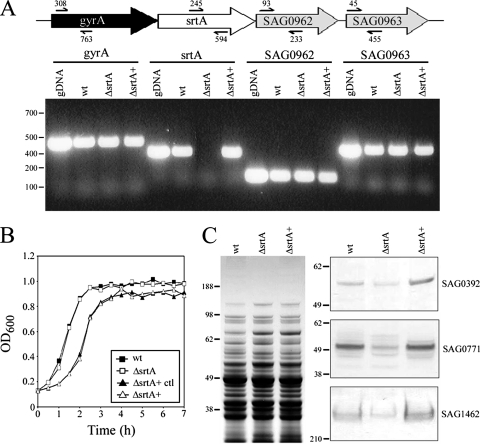
To characterize the role of SrtA in S. agalactiae pilus assembly, an in-frame deletion was made in the srtA gene of strain 515 (possessing PI-2a) using the PCR method of splicing by overlap extension. This resulted in the deletion of SrtA amino acid residues 29 to 213, which include the catalytic domain (17). Confirmation of the deletion was obtained by sequence analysis and loss of the srtA transcript, as detected by reverse transcription-PCR (RT-PCR) (Fig. 2A). On the S. agalactiae genome, srtA is closely flanked by gyrA and two genes encoding downstream proteins SAG0962 and SAG0963 (Fig. 2A). Thus, to ensure that any phenotype associated with the srtA mutation did not arise from loss of expression of these neighboring genes, RT-PCR was used to confirm that transcripts for gyrA, SAG0962, and SAG0963 were present in each of the srtA mutants (Fig. 2A). To further exclude the possibility of polar effects, complementation of the srtA mutation was performed using expression vector pAM401 carrying the complete srtA coding sequence under the control of a GBS-specific promoter. Restoration of srtA expression was confirmed by RT-PCR (Fig. 2A).
FIG. 2.
Confirmation of S. agalactiae srtA mutant generation. (A) Transcription of srtA and flanking genes (gyrA, SAG0962, and SAG0963) in wild-type and srtA mutant strains. RNA was extracted from wild-type (wt), srtA deletion (ΔsrtA) and complemented (ΔsrtA+) strains, cDNA was synthesized, and the presence of transcripts was detected by PCR. Wild-type genomic DNA (50 ng) was used as a positive control, as indicated. DNA markers (bp) are given on the left-hand side. Schematic indicates positioning of primer sites. (B) Comparative growth of wild-type (filled squares) and srtA deletion (empty squares) and complemented strains (empty triangles), along with strain 515 (pAMp) as a complementation vector control (filled triangles). Bacteria were grown for 7 h at 37°C in 5% CO2 in THB medium, and the optical density at 600 nm was measured at the indicated times. (C) Western immunoblot analyses of wild-type and srtA mutant strains with antiserum against LPXTG family surface proteins. Total protein extracts (20 μg) were collected as described in Materials and Methods, blotted onto nitrocellulose, and probed with antisera directed against SAG0392, SAG0771, and SAG1462 (right panel). Extracts were also stained with Coomassie blue as a protein loading control (left panel). Molecular size markers (kDa) are indicated.
No difference in growth rates was seen for the srtA deletion mutant compared to wild type (Fig. 2B). The complemented strain showed a slightly reduced rate of growth, but a similar reduction in growth upon transformation of the wild-type with empty pAM401 vector confirmed that this was due to the presence of the expression vector (Fig. 2B). A functional effect of the srtA mutation was also confirmed by examining the expression of three surface-exposed proteins bearing classic SrtA subfamily LPXTG motifs: SAG0392, SAG0771, and SAG1462. As in previous studies of the effects of mutating SrtA on surface protein expression in streptococci (2, 17, 19, 39), immunoblot analyses of total protein extracts confirmed that levels of all three proteins were reduced in the srtA mutant compared to the wild type (Fig. 2C). Levels were restored or exceeded those of the wild type in the complemented strain (Fig. 2C).
Effect of srtA mutation on pilus expression.
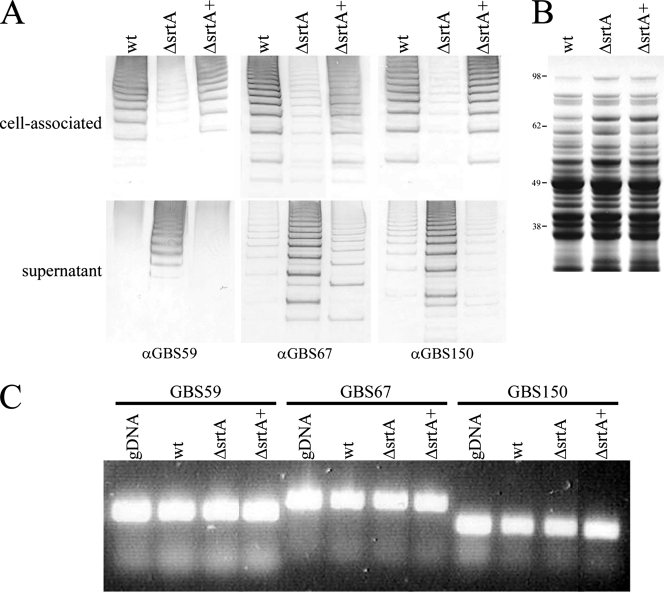
To investigate the role of SrtA in pilus assembly, the first step was to examine overall levels of pilus expression in the srtA mutant compared to the wild type. Immunoblot analyses of total protein extracts revealed the expected high-molecular-weight laddering indicative of pilus-like structures (18). Comparison of these protein profiles indicated that whole-cell extracts of the srtA mutant possessed fewer pili than the wild type (Fig. 3A, upper panel). Using antiserum directed against each of the pilus proteins, pilus loss was found to involve the entire pilus structure, affecting the backbone protein, GBS59, and also the two ancillary proteins, GBS67 and GBS150. Complementation of the srtA mutation restored expression of all three pilus structural proteins to levels comparable to those of the wild type (Fig. 3A, upper panel). This was not seen for the srtA mutant transformed with an empty expression vector alone (data not shown). Thus, while not affecting the process of pilus polymerization, loss of SrtA appeared to result in a lower level of pili associated with the bacterial cells.
FIG. 3.
Relative abundance of pilus proteins associated with wild-type 515 and srtA mutant strains. (A) Proteins were collected from FMC culture supernatants or harvested cell pellets of wild-type 515 (wt) and srtA deletion (ΔsrtA) and complemented (ΔsrtA+) strains, blotted onto nitrocellulose, and probed with antiserum directed against each of the PI-2a pilus proteins (GBS59, GBS67, and GBS150). (B) Total protein extracts were also stained with Coomassie blue as protein loading controls. Molecular size markers (kDa) are indicated. (C) Transcription of PI-2a pilus protein subunits in wild-type and srtA mutant strains. RNA was extracted from wild-type (wt) and srtA deletion (ΔsrtA) and complemented (ΔsrtA+) strains; cDNA was synthesized, and the presence of backbone (GBS59) and ancillary (GBS67/GBS150) protein transcripts was detected by PCR. Wild-type genomic DNA (50 ng) was used as a positive control, as indicated. α, anti.
To determine if the lower levels of pili resulted from a reduction in pilus protein gene expression or from pilus loss from the cell surface, bacterial cells were cultured in chemically defined medium so that levels of pilus found in the extracellular and cell-associated fractions could be compared directly. As before, wild-type strain 515 had significantly higher levels of pilus protein in the cell-associated fraction than the srtA mutant (Fig. 3A, upper panel). However, whereas only low levels of pilus protein were detected in the extracellular fraction of the wild-type strain, much greater quantities were present for the srtA mutant (Fig. 3A, lower panel). Protein profiles comparable to those of the wild type were restored upon complementation of the srtA mutation (Fig. 3A). In accordance with the cell-associated fractions, pili lost from the bacterial cell surface into the extracellular environment were shown to comprise all three pilus proteins. These data indicated, therefore, that in the absence of SrtA, pili were produced at levels comparable to those of the wild type but were then lost from the bacterial cell surface. No significant difference was seen in transcript levels for any of the three pilus proteins between the wild type and srtA mutants, as detected by RT-PCR (Fig. 3C). This added further support to the hypothesis that lower levels of pili in the absence of SrtA resulted from pilus loss, as opposed to a reduction in gene expression.
Anchoring of pili by SrtA.
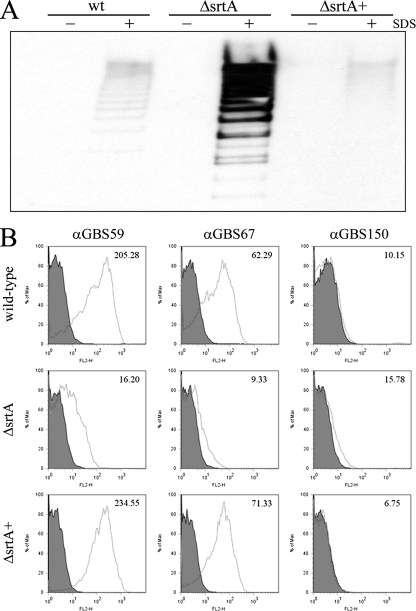
Given the role of SrtA as a transpeptidase, studies progressed to determine if the pilus loss seen with the srtA mutants resulted from a failure of pili to be securely anchored to peptidoglycan of the bacterial cell wall. Proteins that are covalently attached to the cell wall will be unaffected by mild detergent treatment. By contrast, proteins that are held transiently within the cell membrane, as would be predicted for noncovalently bound pili in srtA mutant cells, will be susceptible to solubilization (17, 37, 38). Consequently, wild-type and srtA mutant cells were incubated with 0.5% SDS, and pilus loss was compared. No pilus loss occurred in the absence of SDS for any of the strains tested. Incubation of wild-type strain 515 with SDS resulted in barely detectable levels of pilus loss into the extracellular environment, possibly reflecting a very low-level lysis during the incubation step. Similarly, this background level of pilus release was seen for the srtA complemented mutant (Fig. 4A). By contrast, a pilus protein ladder was clearly detectable in the supernatant following SDS treatment of the srtA mutant (Fig. 4A). This pilus loss was further confirmed by fluorescence-activated cell sorting (FACS) analysis. A comparison of mean fluorescence values (comparing preimmune and immune sera) for backbone protein (GBS59) indicated a greater than 90% reduction in staining for the srtA mutant cells compared to the wild type (Fig. 4B). Similarly, staining of srtA mutant cells for ancillary protein 1 (GBS67) was 85% lower than that of wild-type cells (Fig. 4B). Levels were restored to those of the wild type for both protein subunits in the srtA complemented strain (Fig. 4B). As reported previously (29), surface staining was negative for ancillary protein 2 (GBS150). Taken together, these data imply that in the presence of SrtA, pili are tightly anchored to the cell wall of the bacteria, whereas in its absence, pili are only transiently held within the membrane and are therefore susceptible to release by detergent. The small fraction of cell-associated pili that are still detectable in the srtA mutant following SDS treatment may represent pili that have failed to be solubilized by the detergent or that are retained through interactions with other membrane components such as the PI-associated sortases.
FIG. 4.
Effects of mild detergent treatment on pilus anchoring by wild-type (wt) and srtA mutant strains. (A) Bacterial cells were incubated with (+) or without (−) 0.5% SDS for 1.5 h, and the supernatants were harvested. Proteins were then extracted as described in Materials and Methods, blotted onto nitrocellulose, and probed with antiserum directed against the pilus backbone protein (GBS59). (B) Flow cytometry analysis of bacterial cells. Cells were incubated with 0.5% SDS for 1.5 h, paraformaldehyde fixed, and then stained with antiserum against each of the three pilus proteins followed by an R-phycoerythrin secondary antibody. Filled histograms correspond to staining of bacteria with preimmune serum, while black histograms indicate staining with specific antiserum. The change in mean fluorescence is indicated in the top right-hand corner of each plot. wt, wild-type; ΔsrtA, srtA deletion mutant; ΔsrtA+, srtA complemented mutant; α, anti.
Identification of the pilus anchor protein.
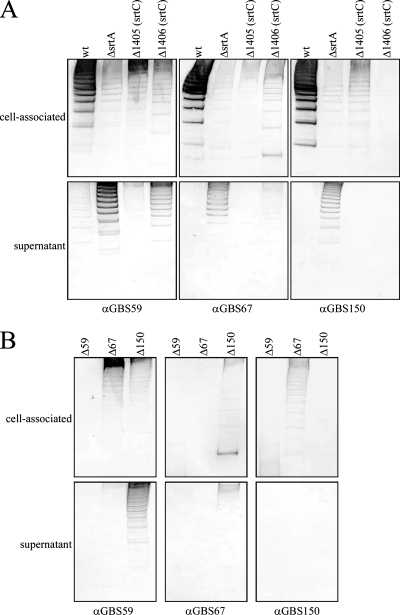
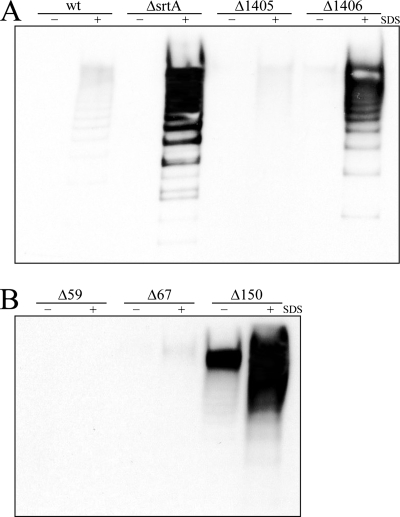
Having established a role for SrtA in the cell wall anchoring of pili, the next stage was to identify the protein upon which SrtA was acting. As a component(s) of the PI was a likely candidate, the amount of cell-associated pili was analyzed for a panel of deletion mutants in which expression of each of the PI-2a components was abrogated: GBS59, GBS67, GBS150 (GBS proteins comprising pilus structure), and sortases SAG1405 and SAG1406 (sortase subfamily C enzymes). This analysis showed that, as had been reported previously (29), deletion of the backbone protein GBS59 completely eliminated pilus polymerization (Fig. 5B). The roles of sortases SAG1405 and SAG1406 in preferentially incorporating ancillary proteins GBS67 and GBS150 into the pilus structure, respectively (29), were also confirmed (Fig. 5A). Staining of total protein extracts with antiserum specific for the backbone protein, which can be polymerized by SAG1405 or SAG1406 with equal efficiency, showed that mutants in SAG1406 and GBS150 possessed the lowest levels of pili among the PI mutants (Fig. 5, upper panels). This implied that GBS150 is the natural substrate of SrtA and that when GBS150 is absent from pili, due to deletion or lack of incorporation by SAG1406, pili cannot be covalently bound to the cell wall and are shed. To confirm this hypothesis, levels of pilus protein released into the extracellular environment during growth by each mutant were investigated. Culture supernatants from wild-type strain 515 and a deletion mutant of the SAG1405 sortase contained barely detectable levels of pilus protein (Fig. 5A, lower panel). By contrast, significantly higher levels of pilus protein were released by the GBS150-specific SAG1406 sortase mutant, similar to the profile seen in the absence of SrtA (Fig. 5A, lower panel). Furthermore, analyses of mutants in each of the three pilus proteins revealed that only the absence of GBS150 resulted in a loss of pilus that mimicked the effects seen for the srtA mutant (Fig. 5B, lower panel), albeit at lower levels. That this pilus loss resulted from a failure of pili to be covalently anchored was further confirmed by SDS treatment (Fig. 6). In contrast to wild-type 515 or mutants in SAG1405 or GBS67, significant pilus loss was observed only in the presence of SDS from the cell surface of mutants in SAG1406 and GBS150.
FIG. 5.
Relative abundance of pilus proteins associated with sortase mutants (A) or pilus protein mutants (B) of PI-2a. Proteins were collected from FMC culture supernatants (lower panels) or harvested cell pellets (upper panels), as described in Materials and Methods, blotted onto nitrocellulose, and probed with antiserum directed against each of the pilus proteins (GBS59, GBS67, and GBS150). wt, wild-type; ΔsrtA, srtA deletion mutant; Δ1405 and Δ1406, pilus-associated sortase C family deletion mutants; Δ59, -67, and -150, pilus protein deletion mutants; α, anti.
FIG. 6.
Effects of mild detergent treatment on sortase mutants (A) or pilus protein mutants (B) of PI-2a. Bacterial cells were incubated with (+) or without (−) 0.5% SDS for 1.5 h, and the supernatants were harvested. Proteins were then extracted as described in Materials and Methods, blotted onto nitrocellulose, and probed with antiserum directed against the pilus backbone protein (GBS59). wt, wild-type; ΔsrtA, srtA deletion mutant; Δ1405 and Δ1406, pilus-associated sortase C family deletion mutants; Δ59, -67, and -150, pilus protein deletion mutants.
Fate of the pilus anchor protein.
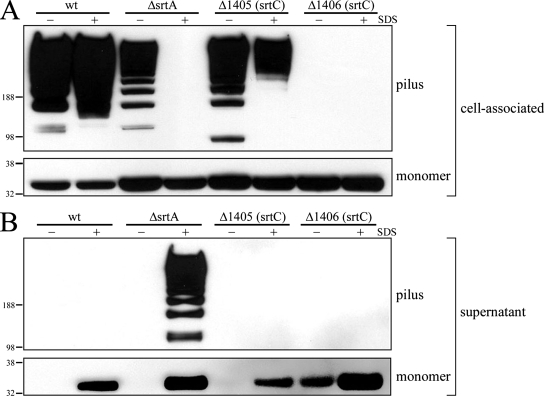
These data implied that GBS150 could serve as the target for both the PI-associated SrtC transpeptidases and for SrtA, with differing outcomes. SrtC enzymes act upon GBS150 for its incorporation into the pilus structure, while SrtA targets GBS150 for the purpose of pilus anchoring. To investigate the nature of these interactions in greater detail, the fate of GBS150, both as a monomer and as part of the polymerized pilus, was monitored in the absence of SrtA, SAG1405, or SAG1406. No polymerized structures were lost from wild-type cells into the extracellular fraction upon treatment with SDS, indicating that these pili were securely anchored to the bacterial cell wall (Fig. 7, compare top blots of both panels). This was also seen for cells lacking GBS67-specific sortase SAG1405 (Fig. 7, compare top blots of both panels). By contrast, SDS treatment of srtA mutant cells resulted in the release of all detectable polymerized GBS150 from the cell surface (Fig. 7A, upper panel) and its concomitant appearance in the extracellular fraction (Fig. 7B, upper blot). No polymerized GBS150 was detected in the absence of SAG1406, thereby confirming the role of sortase SAG1406 in principally incorporating GBS150 into pili (Fig. 7A, upper blot).
FIG. 7.
Fate of ancillary protein GBS150 in the absence of SrtA or the pilus-associated SrtC transpeptidases. Bacterial cells were incubated with 0.5% SDS for 1.5 h, and the suspensions were subsequently separated into cell-associated (A) and extracellular (B) fractions. Proteins were extracted as described in Materials and Methods, blotted onto nitrocellulose, and probed with antiserum directed against ancillary protein GBS150. wt, wild-type; ΔsrtA, srtA deletion mutant; Δ1405 and Δ1406, pilus-associated sortase C family deletion mutants.
Surprisingly, monomeric GBS150 was released by SDS from all of the strains tested, including the wild-type strain (Fig. 7B, lower blot). This indicated that, in contrast to polymerized GBS150, a substantial fraction of the monomer was not covalently linked to the cell wall. This fraction may represent nascent GBS150 that is not yet incorporated into pili and is still tethered to the membrane through its C-terminal transmembrane region. Some monomeric GBS150 remained associated with the cells following SDS treatment (Fig. 7A, lower blot). This occurred even in the absence of the SrtA and SrtC transpeptidases, implying that either an additional component is interacting with the monomer or that the SDS treatment is not 100% effective at disrupting the monomer-cell membrane interactions. Interestingly, some loss of monomeric GBS150 occurred from the SAG1406 mutant, even in the absence of SDS (Fig. 7B, lower blot). This supports the notion that SAG1406 interacts with the monomeric form of GBS150 to incorporate it into the pilus structure. Taken together, these data imply that a significant proportion of GBS150 monomer is not anchored to the cell wall and so is susceptible to release by SDS, whereas once incorporated into the pilus structure, GBS150 is anchored and is thus SDS resistant.
DISCUSSION
Current models of pilus assembly (24, 32-34), based predominantly on the prototype PI of C. diphtheriae, propose that this process comprises two phases: polymerization and anchoring. Since the formation of high-molecular-weight structures was seen with the srtA deletion mutants generated in this study and at levels comparable to those of the wild-type strain, it was clear that pilus formation is not abrogated by the loss of SrtA. This is in direct contrast to the effects of deleting both SrtC transpeptidases, which eliminates the generation of all polymerized structures (8, 29). Thus, it can be concluded that any role for SrtA is distinct from that of the PI-associated SrtC function of subunit polymerization. Nevertheless, the observed shedding of these structures from the cell surface of the srtA mutants implied that SrtA does indeed play some role in pilus assembly, specifically, in the anchoring phase.
Loss of pili from the srtA mutant cell surface correlates well with the previous findings of Dramsi et al. (8), in which an srtA mutant in GBS strain NEM316 was shown by transmission electron microscopy to bear fewer pili on its surface than the wild type. However, in that study this observation was attributed to a down-regulation in transcription of the pilus protein genes and therefore failure to express protein subunits in the absence of SrtA. The present work, however, found no evidence to support this finding. By contrast, the extracellular release of significant quantities of pili by srtA mutants and the visible expression of protein monomers clearly demonstrated that pilus structures were formed. Based on these data, therefore, it would be predicted that the presence of fewer pili on the surface resulted from shedding into the extracellular environment. To further support a role for SrtA in the anchor phase of pilus assembly, cells were subjected to mild detergent treatment. Such a method is often employed with gram-positive bacteria to distinguish between proteins that are membrane associated (SDS soluble) and those that are covalently anchored to the peptidoglycan of the cell wall (SDS resistant) (17, 37, 38). In this way it was shown that pili fail to be covalently attached to the cell wall in the absence of SrtA but, rather, are held transiently in the cell membrane before being secreted into the extracellular environment. A similar finding has also been reported in C. diphtheriae, for which it was shown that in the absence of the SrtA homologue, SrtF, pili were shed into the culture medium and were more readily solubilized by boiling in SDS (33, 37).
Only the absence of ancillary subunit GBS150, either due to deletion of the gene itself or of the gene encoding the SrtC transpeptidase (SAG1406) required for GBS150 incorporation into the pilus, resulted in a profile of pilus loss that mimicked that of the srtA mutation. This implied, therefore, that GBS150 is the target of SrtA activity in pilus anchoring. Such a role could explain why surface staining for GBS150 by FACS is consistently negative (Fig. 4B) (29). Although at present we cannot say conclusively if this negative staining indicates that GBS150 is absent from the pilus shaft or that it is present but hidden from antibodies, one might predict that the anchor protein would be less accessible throughout the pilus structure than the other pilus subunits. Importantly, however, based on the model proposed here, inclusion of GBS150 within the pilus shaft and its role as the anchor are not mutually exclusive. Interestingly, FACS staining is also negative for GBS150 homologues from PI-1 and PI-2b (29), suggesting that these other PIs of GBS might also utilize an ancillary protein anchor. Furthermore, these data provide an explanation for the observation made by Dramsi et al. (8) that the GBS150 homologue in strain NEM316, GBS1474, was localized predominantly at the base of the pilus structure, as detected by immunogold electron microscopy.
The role of GBS150 as the pilus anchor is also supported by the sequence data. Based on the classification system proposed by Comfort and Clubb (5), subunits GBS59 and GBS67 both possess sortase recognition motifs that fit the classic family 3 (SrtC) motif (IPXTGG), while the motif of GBS150 is closer to that of SrtA (LPKTGM). Nevertheless, it is clear that GBS150 is also recognized by the SrtC transpeptidases as its incorporation into the pilus structure occurs even in the absence of SrtA. This implies, therefore, that elements in addition to the LPXTG motif may determine the specificity of sortase substrates. Data presented here suggest that a significant fraction of GBS150 monomer is not anchored to the bacterial cell wall, while in its polymerized form as a component of the pilus, GBS150 is securely attached. This implies that efficient SrtA anchoring of GBS150 to the cell wall only occurs following SrtC-mediated incorporation of GBS150 into the pilus structure. One might speculate, therefore, that it is a conformational change in GBS150 following polymerization that enables it to be recognized by SrtA. Alternatively, the monomeric form of GBS150 might be protected from SrtA by other proteins, such as the SrtC transpeptidases themselves, or by a chaperone protein, as was recently proposed for pilus assembly in group A Streptococcus (42). Such possibilities are currently under investigation.
The processes that govern the order for the uptake and inclusion of each pilus component remain unclear. However, observations that pilus length is greatly extended upon overexpression of the backbone subunit (18, 29, 37) imply that subunit stoichiometry may play at least some part in regulating overall pilus composition. As such, one might expect to find the anchor protein in lower abundance than those proteins that constitute the pilus shaft. That a pool of monomeric GBS150 was found within the cells, therefore, may support the notion that GBS150 can occur as both a pilus shaft component and as the pilus anchor. However, since it is not possible to quantify the relative abundance of each pilus protein by Western immunoblotting, this remains a question for future studies.
Taken together, these data can be used to expand on the recent models proposed for pilus assembly (24, 32-34). This process initiates with translocation of the pilus precursor subunits via the Sec system to the bacterial cell membrane, where they are retained by means of their C-terminal hydrophobic tails. Here, they are brought into close proximity with membrane-associated transpeptidases of both the SrtA and SrtC subfamilies. In contrast to C. diphtheriae, no initiating tip subunit has yet been found in GBS, but this and previous reports (29) have shown that it is the backbone subunit, GBS59, and, to a lesser extent, ancillary protein GBS67 that form the bulk of the pilus structure. SrtC enzymes, SAG1405 and SAG1406, therefore function to polymerize predominantly GBS59 and GBS67 subunits and in this way extend the pilus structure away from the bacterial cell surface. During this phase the growing pili interact with the membrane-associated SrtC enzymes, but these structures are not, however, covalently attached to the bacterial cell wall. For this to occur, SrtA is required. For SrtA to recognize and act upon pili, subunit GBS150 must be incorporated into the structure via its canonical pilin motif, predominantly by the action of sortase SAG1406. SrtA then cleaves GBS150 at its LPXTG motif and catalyzes its covalent attachment to peptidoglycan of the cell wall, thereby securely anchoring the pilus structures to the bacterial surface.
The model reported here relates specifically to PI-2a of GBS, and studies will now extend to PI-1 and PI-2b. Nevertheless, recent reports of SrtA-mediated pilus anchoring in C. diphtheriae (33) and Bacillus cereus (4) imply that this may be a common mechanism among gram-positive bacteria. Given the potential importance of pili to bacterial pathogenesis and vaccine strategies, elucidating these models in even greater detail will remain a priority.
Acknowledgments
We thank Andrew Edwards for critical reading of the manuscript.
This work was supported by Marie Curie Transfer of Knowledge Fellowship MTKD-CT-2004-509261.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 9 June 2008.
REFERENCES
- 1.Abbot, E. L., W. D. Smith, G. P. S. Siou, C. Chiriboga, R. J. Smith, J. A. Wilson, B. H. Hirst, and M. A. Kehoe. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 91822-1833. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 1842181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 1032857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budzik, J. M., L. A. Marraffini, and O. Schneewind. 2007. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66495-510. [DOI] [PubMed] [Google Scholar]
- 5.Comfort, D., and R. T. Clubb. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 722710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dermer, P., C. Lee, J. Eggert, and B. Few. 2004. A history of neonatal group B streptococcus with its related morbidity and mortality rates in the United States. J. Pediatr. Nurs. 19357-363. [DOI] [PubMed] [Google Scholar]
- 7.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of gram-positive bacteria. Res. Microbiol. 156289-297. [DOI] [PubMed] [Google Scholar]
- 8.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 601401-1413. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, M. S., and C. J. Baker. 2005. Group B streptococcal infections in elderly adults. Clin. Infect. Dis. 41839-847. [DOI] [PubMed] [Google Scholar]
- 10.Ellen, R. P., D. L. Walker, and K. H. Chan. 1978. Association of long surface appendages with adherence-related functions of the gram-positive species Actinomyces naeslundii. J. Bacteriol. 1341171-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaspar, A. H., and H. Ton-That. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 1881526-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gianfaldoni, C., S. Censini, M. Hilleringmann, M. Maschioni, C. Facciotti, W. Pansegrau, V. Masignani, A. Covacci, R. Rappuoli, M. A. Barocchi, and P. Ruggiero. 2007. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 751059-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girard, A. E., and B. H. Jacius. 1974. Ultrastructure of Actinomyces viscosus and Actinomyces naeslundii. Arch. Oral Biol. 1971-79. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, S. M., N. Uldbjerg, M. Kilian, and U. B. S. Sorensen. 2004. Dynamics of Streptococcus agalactiae colonization in women during and after pregnancy and in their infants. J. Clin. Microbiol. 4283-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8528-535. [PubMed] [Google Scholar]
- 16.Johri, A. K., L. C. Paoletti, P. Glaser, M. Dua, P. K. Sharma, G. Grandi, and R. Rappuoli. 2006. Group B Streptococcus: global incidence and vaccine development. Nat. Rev. Microbiol. 4932-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalioui, L., E. Pellegrini, S. Dramsi, M. Baptista, N. Bourgeois, F. Doucet-Populaire, C. Rusiok, M. Zouine, P. Glaser, F. Kunst, C. Poyart, and P. Trieu-Cuot. 2005. The SrtA sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect. Immun. 733342-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauer, P., C. D. Rinaudo, M. Soriani, I. Magarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B Streptococcus. Science 309105. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S. F., and T. L. Boran. 2003. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 71676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. Nardi Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B Streptococcus vaccine by multiple genome screen. Science 309148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maisey, H. C., M. Hensler, V. Nizet, and K. S. Doran. 2007. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 1891464-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2007. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 64111-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manetti, A. G. O., C. Zingaretti, F. Falugi, S. Capo, M. Bombaci, F. Bagnoli, G. Gambellini, G. Bensi, M. Mora, A. M. Edwards, J. M. Musser, E. A. Graviss, J. L. Telford, G. Grandi, and I. Margarit. 2007. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64968-983. [DOI] [PubMed] [Google Scholar]
- 24.Marraffini, L. A., A. C. DeDent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McIntire, F. C., A. E. Vatter, J. Baros, and J. Arnold. 1978. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infect. Immun. 21978-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montigiani, S., F. Falugi, M. Scarselli, O. Finco, R. Petracca, G. Galli, M. Mariani, R. Manetti, M. Agnusdei, R. Cevenini, M. Donati, R. Nogarotto, N. Norais, I. Garaguso, S. Nuti, G. Saletti, D. Rosa, G. Ratti, and G. Grandi. 2002. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect. Immun. 70368-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. O. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA 10215641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8809-819. [DOI] [PubMed] [Google Scholar]
- 29.Rosini, R., C. D. Rinaudo, M. Soriani, P. Lauer, M. Mora, D. Maione, A. Taddei, I. Santi, C. Ghezzo, C. Brettoni, S. Buccato, I. Margarit, G. Grandi, and J. L. Telford. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61126-141. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott, J. R., and D. Zahner. 2006. Pili with strong attachments: gram-positive bacteria do it differently. Mol. Microbiol. 62320-330. [DOI] [PubMed] [Google Scholar]
- 33.Swaminathan, A., A. Mandlik, A. Swierczynski, A. Gaspar, A. Das, and H. Ton-That. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 66961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4509-519. [DOI] [PubMed] [Google Scholar]
- 35.Terleckyj, B., and G. D. Shockman. 1975. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terleckyj, B., N. P. Willett, and G. D. Shockman. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 501429-1438. [DOI] [PubMed] [Google Scholar]
- 38.Ton-That, H., L. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53251-261. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi, M., Y. Terao, T. Ogawa, T. Takahashi, S. Hamada, and S. Kawabata. 2006. Role of Streptococcus sanguinis sortase A in bacterial colonization. Microbes Infect. 82791-2796. [DOI] [PubMed] [Google Scholar]
- 40.Yanagawa, R., and E. Honda. 1976. Presence of pili in species of human and animal parasites and pathogens of the genus Corynebacterium. Infect. Immun. 131293-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeung, M. K. 1999. Molecular and genetic analyses of Actinomyces spp. Crit. Rev. Oral Biol. Med. 10120-138. [DOI] [PubMed] [Google Scholar]
- 42.Zahner, D., and J. R. Scott. 9 November 2007. SipA is required for pilus formation in Streptococcus pyogenes serotype M3. J. Bacteriol. doi: 10.1128/JB.01520-07. (Subsequently published, J. Bacteriol. 190:527-535, 2008.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Y., Y. Lei, A. Khammanivong, and M. C. Herzberg. 2004. Identification of a novel two-component system in Streptococcus gordonii V288 involved in biofilm formation. Infect. Immun. 723489-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]