Abstract
Typing of the porB variable region (VR) is an epidemiological tool that classifies gonococcal strains based on sequence differences in regions of the porB gene that encode surface-exposed loops. The frequent isolation of certain porB VR types suggests that some porin sequences confer a selective advantage during infection and/or transmission. Alternatively, certain porin types may be markers of strains that are successful due to factors unrelated to porin. In support of the first hypothesis, here we show urogenital tract isolates representing the most common PIA VR types identified in an urban clinic in Baltimore, MD, over a 10-year period belonged to several different clonal types, as determined by pulsed-field gel electrophoresis (PFGE). Serum resistance, which was confirmed by factor H and C4b-binding protein binding studies, was more often associated with gonococcal the most common VR types. In contrast, three porin-independent phenotypes, namely, lactoferrin utilization, β-lactamase production, and multiple transferable resistance (Mtr), were segregated with the PFGE cluster and not with the VR type. Data combined with another PIA strain collection showed a strong correlation between serum resistance and the most common VR types. A comparison of VR typing hybridization patterns and nucleotide sequences of 12 porB1a genes suggests that certain porin loop 1, 3, 6, and/or 7 sequences may play a role in the serum resistance phenotype. We conclude that some PorB PIA sequences confer a survival or transmission advantage in the urogenital tract, perhaps via increased resistance to complement-mediated killing. The capacity of some porin types to evade a porin-specific adaptive immune response must also be considered.
Neisseria gonorrhoeae is a highly successful pathogen that has an important social and economic impact on the world. An estimated 62.3 million infections occur worldwide (56). In 2006, an increase in gonorrhea rate occurred for the second consecutive year in the United States, with 358,366 cases reported (6). The major source of morbidity and mortality associated with gonorrhea is due to upper-reproductive tract infections in women, which can lead to involuntary infertility, ectopic pregnancy, and chronic pelvic pain. Over 77 million dollars was spent in the United States in the year 2000 on the diagnosis and treatment of acute gonorrhea and postinfection sequelae in patients 15 to 24 years of age (7). The fact that gonorrhea is a cofactor in human immunodeficiency virus (HIV) transmission (8) further emphasizes the need for improved therapeutic and prophylactic measures against gonococcal infections.
PorB is the predominant outer membrane protein of N. gonorrhoeae and is encoded by the porB gene. Gonococci possess only one of two porB alleles, porB1a or porB1b, which encodes the PIA or the PIB protein, respectively (10, 17). PorB assembles in the membrane as a trimeric protein in which each PIA or PIB monomer forms a β sheet barrel structure with eight predicted surface-exposed loops (4). PorB serves as an ion channel and is essential for cell survival. Several functions associated with gonococcal pathogenesis that may be selected on mucosal surfaces have also been attributed to porin, including serum resistance (41), induction (34, 35) or inhibition (3) of apoptosis, invasion (16), and antibiotic resistance (15). Among these porin-mediated functions, serum resistance is the function that is characterized best at the molecular level and one which Ram et al. (41, 42) showed is due to the binding of the complement regulatory proteins C4BP and/or fH to surface-exposed porin loops of serum-resistant P1A or P1B strains.
Unlike the majority of gonococcal surface structures, PorB is antigenically stable during infection. Porin diversity does exist among strains but primarily within the surface-exposed loops. These differences are exploited by the conventional N. gonorrhoeae serotyping method, which uses a panel of monoclonal antibodies to type strains (48), and sequence-based methods, such as multiple antigen sequence typing (31), porB sequencing (52), and porB variable region (VR) typing (50). porB typing methods are more discriminatory than serotyping and have revealed a high degree of porB genetic mosaicism in clinical isolates that is suggestive of horizontal exchange (14, 30). This observation is consistent with evidence that DNA uptake and integration play important roles in gonococcal adaptation (26, 37, 40, 47) and that there is positive selection that favors diversity of PorB surface loops, which is thought to result from immunity pressure (37, 40, 47). Gonococci are not purely panmictic populations, however; examples of extended clonal transmission have been reported (59).
In light of the strong evidence for PorB diversification, the identification of common porB types that persist over time raises the possibility that certain porin types may confer functional advantages. McKnew et al. (32) followed the recovery of urogenital isolates from two clinics in Baltimore, MD, over a 10-year period, with respect to porB VR type. Out of a total of 219 PIB isolates collected, 54 porB VR types were identified; 6 of these 54 VR types persisted over the length of the study. Lower diversity in VR type was detected among PIA isolates. Seven PIA porB VR types were identified among a total of 63 PIA isolates, and 61% of the strains expressed VR type PIA 1;2;1;1;1 or PIA 1;1;1;1/4;1. Based on these findings, these investigators hypothesized that a balance exists between function conservation and the ability to evade a porin-specific immune response.
Here, we further examine the hypothesis that certain more-“fit” porin types confer a functional phenotype that results in the persistence of certain PorB molecules among strains within a community. Our alternative hypothesis was that certain porin types are merely markers of successful strains that have an advantage due to factors other than porin. While PIA strains are isolated less often than PIB strains, here we focused only on PIA isolates as there is a more limited number of common PIA VR types and because the alternative hypothesis was more likely with PIA strains. We first measured the genetic relatedness of urogenital isolates from two P1A strain collections that were well characterized with regard to porB VR types. We then screened the isolates for serum resistance, a porin-dependent phenotype, and three porin-independent phenotypes that might influence fitness in vivo. We also determined the sequences of the porB1a genes of 12 isolates of representative PIA VR types to examine the conservation of porin sequences within a VR type that are not targeted by the VR typing method and to identify commonalities in the predicted amino acid sequence that might be involved in the serum resistance phenotypes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Two collections of urogenital isolates were examined in this study. The first set of isolates consisted of 25 PIA isolates from two different public health clinics in Baltimore, MD, that were typed by McKnew et al. (32). The isolates used were from the first 4 years of this 10-year study; isolates from 1991 and 1992 were collected from male patients as part of the Gonococcal Isolate Surveillance Program, and isolates from 1993 and 1994 were collected from both men and women for other studies. A second set of PIA isolates was obtained from a partner study that took place in Boston, MA, between September 1988 and April 1991. The laboratory strains FA1090 (PIB type, serum resistant [SR]), FA19 (PIA type, SR), MS11 (PIB type, serum intermediate [SI]), F62 (PIB type, serum sensitive [SS]) (41, 42), 15253 (P1A type, SR) (60), and FA6916, which is a transferrin receptor mutant of strain FA1090 (provided by P. F. Sparling) (9), were used as controls. All N. gonorrhoeae strains were cultured on GC agar supplemented with Kellogg's supplement I (25) and 12 μM Fe (NO3)3 and incubated at 37°C in 7% CO2.
Pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis (PFGE) was performed with a modification of the protocol described by Poh et al. (39). Briefly, each isolate was subcultured for 18 to 20 h on GC agar and then suspended in PIV buffer (0.2 M Tris-HCl, 5 M NaCl) to an initial optical density at 640 nm (OD640) of 0.80. Suspensions were centrifuged three times at 3,000 rpm for 15 min, and the bacterial pellet was resuspended in PIV buffer and then mixed with an equal volume of 1.2% (wt/vol) agarose (SeaKem Gold). This mixture was pipetted into molds and allowed to set at 4°C for 30 min. The plugs were treated with enhanced chemiluminescence lysis buffer (0.2 M Tris-HCl, 5 M NaCl, 0.5 M EDTA, 0.2% [wt/vol] sodium deoxycholate, 0.5% N-lauroylsarcosine, 1 mg/ml lysozyme, 20 μg/ml RNase) and incubated at 37°C overnight. The plugs were then treated with ESP lysis buffer (0.5 M EDTA, 1% [wt/vol] N-lauroylsarcosine, 1 mg/ml proteinase K) at 50°C overnight. Plugs were serially washed with Tris-EDTA (TE) buffer that contained 17.5 mg/ml phenylmethylsulfonyl fluoride, followed by washing with TE buffer alone and then with autoclaved H2O in preparation for digestion with NheI. Two-millimeter sections of each plug were incubated in enzyme buffer (NEB2; New England Biolabs) at 4°C for 15 min. Fresh NEB2 buffer was added to the plugs, and the plugs were incubated for an additional hour, followed by incubation with NheI in NEB2 buffer with bovine serum albumin at 37°C overnight. After TE buffer was added to stop the reaction, plugs were sealed into the wells of a 1% (wt/vol) agarose gel, and PFGE was performed at 1- to 20-s pulse times at 6 V for 18 h in 0.5× Tris-borate EDTA buffer at 4°C. Gels were stained with ethidium bromide and visualized with an AlphaImager system (Alpha Innotech). A low-range pulse-field gel marker that contains lambda DNA (New England Biolabs) was used as the molecular weight standard for all gels and for the universal standard for analysis. Images were analyzed, and dendrograms were created at the Center for Food Safety and Nutrition of the Food and Drug Administration, using Bionumerics software (Applied Maths). Dendrograms were constructed by using the dice coefficient with a pattern optimization of 1.5% and a band tolerance of 1.0%.
Bactericidal assays.
Clinical isolates were prescreened for their capacities to survive in 33% (vol/vol) pooled normal human serum (NHS) (Quidel), to identify the strains that were highly SR (>50% recovery) or SS (0 to 6% recovery) or that showed an SI level of serum resistance (6 to 50% recovery). Isolates were then retested with a microtiter assay to determine the titer at which 50% of organisms were killed (bactericidal50), using the appropriate range of NHS concentrations (0 to 50% for SR strains, 0 to 16% for SI strains, and 0 to 4% for SS strains). Briefly, bacteria were suspended in sterile phosphate-buffered saline (PBS) to an A600 of 0.07 and then diluted 1:1,000 in minimal essential medium (MEM). Thirty-microliter (100 to 400 CFU) portions of the diluted suspension were added to 60 μl of serially diluted NHS (1:2) in MEM or to MEM alone. After a 1-h incubation at 37°C, 30 μl of GC broth was added to each well, and 50-μl aliquots were cultured in duplicate on GC agar overnight. The average number of CFU recovered from each well was divided by the total number of CFU recovered from wells that lacked NHS and multiplied by 100; the resultant percentages were plotted against the concentration of NHS to obtain the bactericidal50 titer. Heat-inactivated NHS (HI-NHS) was prepared by incubating NHS at 56°C for 30 min and was tested in parallel with each assay. Prior to testing the clinical isolates, we standardized this assay in terms of the number of CFU input and the incubation time by using laboratory strains of known serum resistance phenotypes. When 100 to 400 CFU was used as the input number of F62 (SS) or MS11 (SI) bacteria, a linear decrease in survival was observed with increasing concentrations of NHS, and the bactericidal50 titers were reproducibly 0.5% NHS and 3.0 to 3.5% NHS, respectively. Based on this analysis, we defined SS and SI bacteria as those strains that had bactericidal50 titers of <3% and between 3 and 10%, respectively. The bactericidal50 titer against the SR strains FA1090 and FA19 ranged from 20 to 30% NHS and >66%, respectively, under these conditions, and was not dependent on the number of input CFU. Therefore, isolates were considered SR if more than 50% of the bacteria survived in more than 10% NHS, and no bactericidal50 titer was determined.
Human C4BP and fH binding.
The binding of C4b-binding protein (C4BP) and factor H (fH) to whole bacteria was measured by flow cytometry as described previously (41, 42). Briefly, bacteria cultured on chocolate agar plates were suspended in Hanks’ buffered saline solution containing 1 mM MgCl2 and 0.15 mM CaCl2. The reaction mixture contained 108 bacteria and either 0.5 μl of pooled NHS as a source of C4BP or 0.5 μg of pure human fH (CompTech, Tyler, TX) in a final volume of 100 μl. The mixture was incubated at 37°C for 10 min. C4BP was detected by using anti-C4BP monoclonal antibody 92 (21), and fH was detected by using monoclonal antibody 90X (24), both at a concentration of 5 μg/ml. Fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G (1:200 dilution; Sigma) was used to disclose the bound primary antibodies. Bacteria-bound C4BP and fH were detected by using flow cytometry (BD LSR II flow cytometer; Becton Dickinson). Analyses of fH and C4BP binding were performed by using FloJo fluorescence-activated cell sorter data analysis software (http://www.TreeStar.com). Strain F62 does not bind fH or C4BP and was used as a negative control. Strain 15253 binds both complement inhibitors and was used as a positive control.
Nucleotide sequence analysis.
The nucleotide sequences of selected porB1a and mtrR genes were determined as follows. The porB1a genes were PCR amplified using the primers PorBFouter (5′-TCGGCGGTAAATGCAAAGC-3′) and PorBRouter (5′-TGCAGATTAGAATTTGTGGCG-3′). The mtrR gene and its upstream promoter region were amplified by using the primers CXho and RXho (54). Reactions were performed in a 20-μl-volume mixture using 5 μl of genomic DNA with 1 μl of each primer, 0.5 μl of each 10 mM deoxynucleoside triphosphate, 2 μl of 10× reaction buffer, and 1 U of Deepvent DNA polymerase (New England Biolabs). PCR conditions were 30 cycles at 94°C for 45 s, 50°C for 45 s, and 72°C for 2 min (porB1a) and 30 cycles at 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min (mtrR). PCR products were analyzed on agarose gels and extracted using QIAquick spin columns (Qiagen). Sequencing reactions were performed with a BigDye sequencing reaction mixture using 100 to 300 ng of template DNA and 10 pmol of primers at 25 cycles at 94°C for 10 s, 50°C for 5 s, and 60°C for 4 min. The sequencing primer used for porB1a was PorBFouter, PorBRouter, PorBR502 (5′-CGCCGGTGTTTTTCAGG-3′), PorBF471 (5′-TTGAAAGGCGGCTTCGG-3′), or PorBF851 (5′-CTGCAAGTTCACCGTTTGG-3′); and the primers for the mtrR locus were CXho and RXho. Nucleotide sequence determination was performed by the USUHS Bioinstrumentation Center. DNA sequences were analyzed with Clone Manager software (Scientific & Educational Software).
Lactoferrin utilization.
The bacterium's capacity to utilize human lactoferrin as an iron source was assessed by a spot assay method. Briefly, bacteria were harvested from GC agar and suspended in PBS to an OD600 of 0.10. Bacterial suspensions were inoculated with a cotton swab onto GC media that contained Kellogg's supplement I and 50 μM of desferoxamine mesylate. Ten microliters of iron-loaded human transferrin or lactoferrin solution (20, 2, and 0.2 mg/ml) (Sigma), sterile H2O (negative control), and 12 μM Fe(NO3)3 (positive control) were dotted onto each plate. The presence or absence of growth on the spotted area was evaluated after overnight incubation. Strains MS11, which can utilize both transferrin and lactoferrin, FA1090, a natural lactoferrin receptor-deficient strain, and FA6916, which is incapable of using transferrin or lactoferrin, were used as controls.
MICs.
The MICs of penicillin G (Pen G), ampicillin (Amp), erythromycin (Erm), tetracycline (Tet), azithromycin (Az), and Triton X-100 (TX-100) were determined by agar dilution (http://www.cdc.gov/STD/Gonorrhea/lab/agar.htm). The MIC was defined as the lowest concentration of antibiotic or TX-100 on which the isolate did not grow. For analysis of the mtr locus, we used a 1.4-kb DNA fragment that contained the mtrR gene and its upstream promoter region that was amplified from isolates with MICs of >0.5 μg/ml (Erm and Az) and ≥1,000 μg/ml (TX-100) and sequenced using the primers CXho and RXho, as described above. PCR fragments were transformed into the gonococcal strain FA19, and transformants were selected on GC agar with 0.5 μg/ml Erm (54). The mtrR locus from mutant strain JF1, which carries an inactivated mtrR gene (12), was also amplified and used as a positive control. Production of β-lactamase was detected by using a rapid acidometric method (Hardy Diagnostics).
RESULTS
Analysis of genetic relatedness.
A schematic that illustrates the VR typing system, which targets loops 1, 2, 3, 6, and 7 of the porB1a gene, is shown in Fig. 1. To explore the genetic relatedness of the P1A strains of commonly isolated VR types, we analyzed 25 PIA urogenital tract isolates obtained over the first 4 years of the McKnew study by using PFGE. The number of isolates of each VR type was selected to resemble the distribution of VR types that were observed over the first 3 to 4 years of the initial study. Isolates of the VR types PIA 1;2;1;1;1 and PIA 1;1;1;1/4;1 were recovered for every year of the 10-year study or for 5 consecutive years and 2 years later, respectively. The VR types PIA 1;2;1;1;1 and PIA 1;1;1;1/4;1 represented 42% and 19% of the total number of PIA isolates recovered over the period of the study, and we define these VR types as common here, based on their high recovery rate and persistence over time. The VR type PIA 2;4;3;3;3 was highly represented among PIA isolates collected over the first 3 years of the McKnew study, but no isolates of this VR type were recovered during the remaining 6 years of the study. One isolate each was of the VR type PIA 1;3;1;1/4;1 and the VR type PIA 1;nt;1;1;1, which are VR types that were identified either in a single year or over 2 consecutive years.
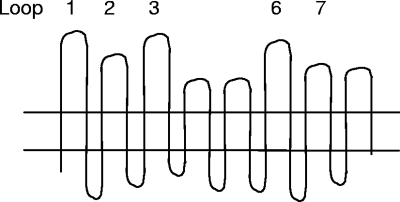
FIG. 1.
PIA loops targeted by the VR typing system. The PIA VR typing system is based on differences in the nucleotide sequences that encode five surface-exposed loops (loops 1, 2, 3, 6, and 7). The exact regions of each loop that correspond to the nucleotide sequences targeted by the oligonucleotide probes are shown in the report by McKnew et al. (32). As examples, VR type PIA 1;2;1;1;1 indicates hybridization to loop 1, probe 1 (probe 1-1), loop 2, probe 2 (probe 2-1), loop 3, probe 1 (probe 3-1), loop 6, probe 1 (probe 6-1), and loop 7, probe 1 (probe 7-1). VR type PIA 1;1;1;1/4;1 is very similar to PIA 1;2;1;1;1, with the exception of the loop 2 probe and minor sequence differences in loop 6. Numbers that are separated by a slash refer to loop sequences that hybridized to two typing probes. A designation of nt, instead of a number, refers to a loop sequence that did not hybridize to any probe and thus is considered nontypeable.
The 25 PIA isolates exhibited eight distinct PFGE pattern groups, based on a cutoff value of 85% band similarity (Fig. 2). The 10 isolates of the most common VR type, PIA 1;2;1;1;1, were segregated to four PFGE clusters (A1, A2, B, and G); the 6 isolates of the next most common type, PIA 1;1;1;1/4;1, were segregated to two different PFGE clusters (C and E). These results show that genetically unrelated strains can express highly related porin sequences, and they are consistent with the hypothesis that porin variants may therefore confer an advantage. Seven isolates expressed the VR type PIA 2;4;3;3;3. These isolates, which were collected in 1991 (1 isolate), 1992 (3 isolates), and 1993 (3 isolates) were highly related and fell into a single PFGE cluster (cluster F). Isolates of the VR types PIA 1;3;1;1/4;1 and PIA 1;nt;1;1;1 had unique PFGE patterns.
FIG. 2.
Relatedness of the N. gonorrhoeae isolates (LG1 to LG26) used in this study. A composite of gels on which NheI-restricted genomic DNA from the 26 PIA Baltimore isolates was fractionated by PFGE is shown; normalization to a universal standard is described in Materials and Methods. A dendrogram that groups patterns into eight distinct clusters (A to H) based on the percentage of band similarity is also shown. Isolates that produced patterns that were <85% similar (see scale in the upper left corner) were considered different strains. Isolates of VR type PIA 1;2;1;1;1 belonged to four distinct PFGE patterns, which ranged from 90 to 96% similarity, while isolates of VR type PIA 1;1;1;1/4;1 fell into two clusters that showed 92 to 95% similarity.
Associations among VR type, lactoferrin utilization, and antibiotic resistance.
The capacity to utilize human lactoferrin as an iron source (LF+) is a porin-independent phenotype that occurs via a specific outer membrane receptor (44). Approximately 50% of gonococcal strains are LF+ (33). In our study, 10 of the 25 P1A isolates (40%) were LF+ (Table 1). No association was seen between the LF+ phenotype and possession of a more common VR type. Six of 10 isolates of the VR type PIA 1;2;1;1;1 and 2 of 6 isolates of the VR type PIA 1;1;1;1/4;1 were LF+. All isolates of the VR type PIA 2;4;3;3;3 were LF−. Importantly, with one exception (cluster A1), isolates within a PFGE cluster were uniform with respect to LF phenotype.
TABLE 1.
Serum resistance and LF phenotypes of PIA isolates from the Baltimore study
| VR type | Isolate | PFGE cluster | Serum resistance | LF use |
|---|---|---|---|---|
| 1;2;1;1;1 | LG14 | A1 | SR | + |
| LG12 | A1 | SR | − | |
| LG23 | A2 | SR | − | |
| LG25 | A2 | SR | − | |
| LG22 | A2 | SR | − | |
| LG11 | B | SI | + | |
| LG13 | B | SI | + | |
| LG15 | B | SR | + | |
| LG7 | B | SR | + | |
| LG24 | G | SR | + | |
| 1;1;1;1/4;1 | LG19 | C | SR | + |
| LG20 | C | SR | + | |
| LG4 | E | SR | − | |
| LG5 | E | SR | − | |
| LG6 | E | SR | − | |
| LG21 | E | SR | − | |
| 2;4;3;3;3 | LG2 | F | SS | − |
| LG8 | F | SS | − | |
| LG10 | F | SI | − | |
| LG17 | F | SS | − | |
| LG9 | F | SR | − | |
| LG16 | F | SR | − | |
| LG18 | F | SR | − | |
| 1;3;1;1/4;1 | LG26 | H | SR | + |
| 1;nt;1;1;1 | LG70 | D | SR | + |
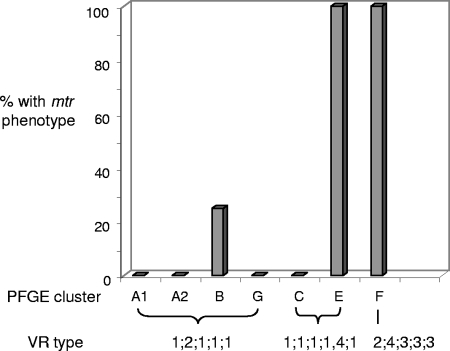
The susceptibility of each isolate to antibiotics was also examined to determine if certain resistance patterns correlated with the more frequently isolated VR types. We were particularly interested in the multitransferable resistance (Mtr) phenotype, which results from mutations in the mtrR repressor gene or its promoter region. The consequence of mutations in the mtrR locus is derepression of the mtrCDE operon (19). The mtr mutants display increased resistance to host-derived antimicrobial substrates of the MtrC-MtrD-MtrE pump (46) and increased fitness in vivo (54). Twelve isolates exhibited the Mtr phenotype, which is defined as increased resistance to the macrolide antibiotics Erm and Az (49, 61), Pen G (53), and the nonionic detergent TX-100 (18). With one exception, the Mtr phenotype segregated with the PFGE cluster, and not all strains of the VR types PIA 1;1;1;1/4;1 and PIA 1;2;1;1;1 had this phenotype, as would be predicted should the Mtr phenotype be responsible for the frequent isolation of certain VR types (Fig. 3). The remaining strains of VR type PIA 1;2;1;1;1 were susceptible or moderately susceptible to all antibiotics tested, with MICs in the susceptible range for Pen G (0.25 to 1.0 μg/ml), Amp (0.125 to 0.5 μg/ml), Tet (0.25 to 1.0 μg/ml), Erm (0.125 to 0.25 μg/ml), and Az (0.125 μg/ml), and MICs of ≤500 μg/ml for TX-100. In addition to exhibiting the Mtr phenotype, all seven isolates of VR type PIA 2;4;3;3;3 (PFGE cluster F) exhibited markedly elevated MICs for Pen G (>4.0 μg/ml). Upon subsequent testing, each of these isolates expressed high levels of β-lactamase activity.
FIG. 3.
Percentage of isolates with the Mtr phenotype and the distribution with respect to VR type. The Mtr phenotype did not segregate by VR type. Isolates within a PFGE cluster were uniform with respect to the Mtr phenotype, with the exception of one isolate in cluster B. One of four isolates within cluster B (isolate LG7; VR type PIA 1;2;1;1;1), all four isolates in cluster E (VR type PIA 1;1;1;1/4;1), and all seven isolates within cluster F (VR type PIA 2;4;3;3;3) exhibited an antibiotic resistance pattern suggestive of the Mtr phenotype.
To confirm the genetic linkage between the mtrR locus and the Mtr resistance pattern, we PCR amplified the mtrR gene and the intergenic region between mtrR and the mtrCDE operon that contains the mtrR promoter and the MtrR binding site (23, 29) from strains with the Mtr phenotype. Consistent with the fact that mtrR loci cause derepression of the mtrCDE operon, transformation of the PCR products into strain FA19 yielded transformants that grew on GC agar with Erm (0.5 μg/ml). The nucleotide sequence of each mtrR amplicon was also determined. Strains LG2 and LG7, which fall into different PFGE clusters, harbor a G>A substitution at nucleotide 115 in the mtrR structural gene, which is predicted to cause an Ala39>Tyr substitution in the MtrR protein. Isolates LG4, LG5, LG6, and LG21, which belong to the same PFGE cluster, harbor a G>A substitution at nucleotide 134 of the mtrR gene that is predicted to cause a Gly45>Asp substitution in the MtrR protein. Both the Ala39>Tyr (11) and the Gly45>Asp (49) substitutions were described previously and are hypothesized to be located in the DNA-binding α-helix motif of the MtrR protein.
In summary, both the lactoferrin and the Mtr phenotypes were strongly associated with the PFGE type but not the porB VR type. These results confirm the presence of genetically distinct strains within persistent VR types as demonstrated by PFGE and indicate that neither lactoferrin usage nor overexpression of the MtrC-MtrD-MtrE efflux pump system is responsible for the frequent isolation of certain porB types.
Association between VR type and serum resistance.
Having established that the most successful PIA VR types are expressed by genetically and phenotypically different strains, we next tested the sensitivity of the strains to the bactericidal activity of NHS. As is typical of PIA strains, the majority of isolates (19 of 25; 76%) were SR, with more than 50% of the bacteria withstanding exposure to 10% NHS for 1 h (Table 1). The majority of isolates of the VR types PIA 1;2;1;1;1 and PIA 1;1;1;1/4;1 were SR (14 of 16; 87%), and the remaining two isolates demonstrated an SI phenotype with bactericidal50 titers of 7.2 and 8.0% NHS. These SI isolates (strains LG11 and LG13) belonged to PFGE cluster B, which also had two SR isolates. In contrast to the more common VR types, a high percentage of SS isolates was detected among isolates of the VR type PIA 2;4;3;3;3 (PFGE cluster F), with three of seven isolates (43%) having bactericidal50 titers of less than 3% NHS. One 2;4;3;3;3 isolate, LG10, was SI (bactericidal50 titer, 5%), and three were SR. None of the SS or SI isolates was sensitive to HI-NHS, which ruled out the possibility that factors in human serum besides complement might contribute to strain susceptibility.
To further examine the link between serum resistance and certain VR types, we screened a second set of clinical isolates for serum resistance levels (2). These isolates originally came from a partner study that consisted of male subjects who tested positive for N. gonorrhoeae infection at a Boston clinic and their female partners who were subsequently contacted and tested (28). Twelve of the 19 isolates (63%) were of the VR type PIA 1;2;1;1;1, and 7 of the isolates had VR types that were not detected in the Baltimore study (VR types PIA 3;1;2;2;2 and PIA 3;1;1/2;2/3;2). Six different clonal types were identified by PFGE analysis of the Boston isolates (Table 2). Similar to the Baltimore isolates, the 12 Boston isolates of VR type 1;2;1;1;1 were distributed over four clusters (I, J, K, and N), and a majority (9 of 12; 75%) were SR. One isolate was SI (bactericidal50 titer, 4.0%), and unlike the PIA 1;2;1;1;1 isolates in the Baltimore collection, two isolates of this common VR type had an SS phenotype. These isolates (LG66F and LG65F) fell into the same PFGE cluster and were from the same sexual network. A higher percentage of isolates of VR types PIA 3;1;2;2;2 and PIA 3;1;1/2;2/3;2 (4 of 7; 57%) were SS. The remaining three isolates exhibited an SI phenotype, with bactericidal50 titers ranging from 3.4 to 6.0%. Statistical analysis of combined data from the Baltimore and the Boston strain collections showed significant differences between the number of PIA 1;2;1;1;1 and PIA 1;1;1;1/4;1 isolates with SR and SI phenotypes (26 of 28 isolates) and the number of SR and SI isolates of the less frequently isolated VR types PIA 1;3;1;1/4;1, PIA 1;nt;1;1;1, PIA 2;4;3;3;3, PIA 3;1;2;2;2, and PIA 3;1;1/2;2/3;2 (9 of 16 isolates) (P < 0.006; Fisher's exact test, two-tailed).
TABLE 2.
Serum resistance phenotypes of P1A isolates from the Boston partner study
| VR type | Isolate | Partner networka | PFGE cluster | Serum resistance |
|---|---|---|---|---|
| 1;2;1;1;1 | LG66F | 1 | I | SS |
| LG65F | 1 | I | SS | |
| LG69F | N/A | J | SR | |
| LG52M | 2 | K | SR | |
| LG53F | 2 | K | SR | |
| LG48F | N/A | K | SR | |
| LG61M | 3 | N1 | SR | |
| LG62F | 3 | N1 | SR | |
| LG54M | 5 | N1 | SR | |
| LG55F | 5 | N2 | SR | |
| LG64F | 4 | N2 | SR | |
| LG63M | 4 | N3 | SI | |
| 3;1;1/2;2/3;2 | LG67F | 6 | L | SS |
| LG68F | 6 | L | SS | |
| 3;1;2;2;2 | LG58M | 7 | M1 | SI |
| LG59F | 7 | M1 | SI | |
| LG60F | 7 | M1 | SI | |
| LG49M | 8 | M2 | SS | |
| LG50F | 8 | M2 | SS |
N/A, only one isolate from these partner networks was obtained.
Correlation of serum resistance with fH- and C4BP-binding and the porB1a sequence.
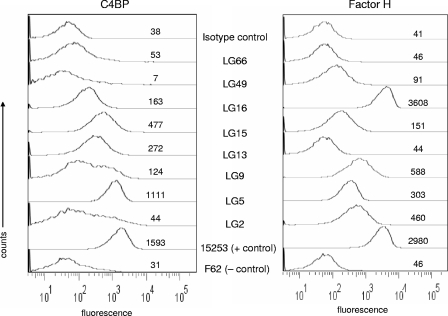
To further analyze the relationship between serum resistance and VR type, we also tested the degree to which isolates bind the complement regulatory proteins C4BP and fH. We were particularly interested in isolates of the same VR type that differed in serum resistance phenotype. Cluster F contained SR, SI, and SS isolates (Table 1). Consistent with an SR phenotype, isolates LG9 and LG16 from this cluster bound both C4BP and fH; the SS isolate LG2, in contrast, bound fH, but only minimal amounts of C4BP (Fig. 4). Among the 22 isolates of VR type PIA 1;2;1;1;1, LG66 was one of two SS isolates, and this strain did not bind either inhibitor. Isolate LG13, an SI isolate of VR type PIA 1;2;1;1;1 that showed 50% survival in 7.2% NHS, bound C4BP but not fH. In contrast, LG15, which is an SR isolate of VR type PIA 1;2;1;1;1, bound C4BP and fH. We also examined an SS strain of the VR type PIA 3;1;2;2;2 (isolate LG49), and found that it bound low amounts of fH but did not bind C4BP. Consistent with a serum resistance phenotype, isolate LG5 (VR type 1;1;1;1/4;1) bound both inhibitors well. We conclude that the capacity of these strains to bind either or both inhibitors correlated well with the serum resistance phenotype, and therefore, the differences in serum resistance phenotypes among isolates of the same porB VR type was not due to errors or limitations of the bactericidal assay used.
FIG. 4.
C4BP and fH binding to N. gonorrhoeae. Flow cytometry was used to measure the binding of C4BP (left panel) and fH (right panel) to bacterial strains. The SS strain F62 (does not bind either inhibitor) and strain 15253 (binds both regulators) were used as negative and positive controls, respectively. A representative isotype control with strain LG2, in which either NHS or fH was omitted from the reaction mixture, is shown. The x axis represents fluorescence on a log10 scale, and the y axis the number of events (counts). The numbers alongside each trace indicate the median fluorescence of the entire population. One representative experiment of two reproducible repeats is shown.
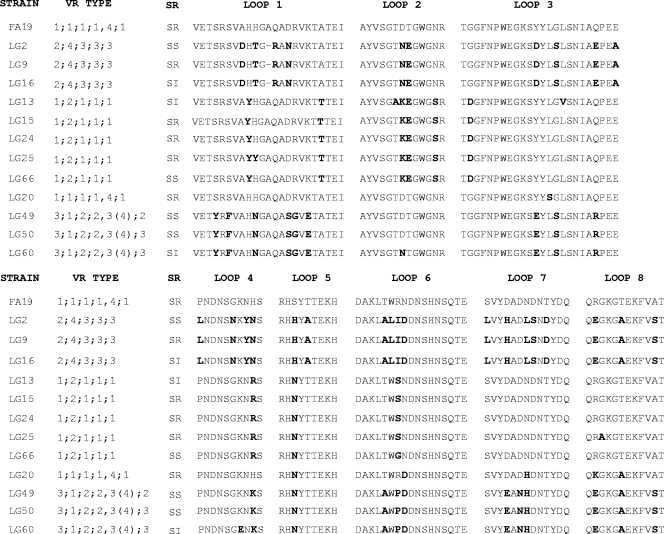
The identification of closely related isolates that shared the same VR type but differed in serum resistance and C4BP/fH binding phenotypes prompted us to determine the nucleotide sequence of the porB1a genes from representative SS and SR/SI isolates. Additionally, we sequenced the porB1a genes of three closely related isolates of VR type PIA 3;1;2;2;2 that showed SS or SI phenotypes and one of the 6 SR VR type PIA 1;1;1;1/4;1 isolates to see if commonalities in the predicted amino acid sequences could be identified that might be involved in the serum resistance phenotypes. Among SR strains of VR type PIA 1;2;1;1;1, the sequences were identical, with the exception of a one-amino-acid change in loops 2 and 3 (isolate LG13; PFGE cluster B) and loops 1 and 8 (isolate LG25; PFGE cluster A2) (Fig. 5). As predicted from the VR types, isolates of VR type PIA 3;1;2;2;2 showed a high degree of diversity in loop 1 (six different amino acids), and three differences in loops 3, 6, and 7 compared to the corresponding sequences of isolates of the other VR types. These isolates (LG49, LG50, and LG60) were SS or SI.
FIG. 5.
Predicted amino acid sequence of surface-exposed loops of selected P1A strains. The porB1a genes from 12 isolates were sequenced as described in Materials and Methods, and the predicted amino acid sequences of the surface-exposed loops are shown. Bold indicates residues that are different from the FA19 predicted amino acid sequence. In general, sequences were segregated with the VR type, and only a few or no differences are seen in the predicted amino acid sequence of SR, SI, and SS isolates within the same VR type.
Importantly, the predicted loop sequences of the SS PIA 1;2;1;1;1 isolate LG66 were identical to those of the SR PIA 1;2;1;1;1 isolates LG15 and LG24, with the exception of a tyrosine instead of a histidine in loop 1 and a glycine instead of a serine in loop 6 (Fig. 5). The heterogeneity observed for serum resistance phenotypes for isolates of VR type PIA 2;4;3;3;3 also did not correspond to differences in porin sequence based on the fact that the predicted amino acid loop sequences of the SS isolate LG2 were identical to those of the highly SR isolates LG9 and LG16. We conclude from these results that the serum resistance exhibited by the less common VR types and by occasional isolates of the more common VR types is most likely due to a factor other than porin.
We also found that the predicted amino acid sequences of the surface-exposed loops were identical or very similar for isolates within a VR type and were not segregated with the PFGE cluster, including loops that are not targeted by the VR typing probes (Fig. 5). When the predicted amino acid sequences of porins of different VR types were compared, differences were present in all loops, with the exception of loop 5. PIA loop 5, which is likely to be the least surface exposed, was relatively conserved. Consistent with the sequences predicted by the probe hybridization patterns, there were only one or two differences between the predicted amino acid sequence of loops 1, 3, 6, and 7 of five different PIA 1;2;1;1;1 isolates. These strains had three to four different residues predicted to occur in loop 2 compared to those of strains of PIA 1;1;1;1/4;1.
In summary, among strains that are not highly related by PFGE but have the same VR type, the entire porin sequence segregates by VR type and not by PFGE type. This result is consistent with the fact that certain porins are selectively maintained. We found no evidence that sequence differences are responsible for differences in serum resistance phenotypes among isolates with the same VR type. We also confirmed the reliability of the VR typing system to group strains with the same predicted P1A sequence.
DISCUSSION
N. gonorrhoeae induces no or limited immunity to reinfection. This trait, combined with the propensity to cause asymptomatic infection, allows this organism to be well sustained in community outbreaks. The high incidence of mixed infections and the capacity of the gonococcus to diversify its genetic information via natural transformation and an inherently high mutation rate result in the nonclonal, panmictic nature of this pathogen (20, 26, 36). The more frequently isolated porB1a types over extended periods of time suggest a positive selection for specific PIA porin sequences. Selective pressure may be influenced by structural or functional constraints and opposed by the need for freedom in sequence diversity that is sufficient to facilitate evasion of the host response. This hypothesis was also proposed by Fudyk et al. (14), who compared the porB genes of gonococcal isolates from female sex workers in Kenya to sequences from strains from other regions. Gonococcal strains of different geographic origins appeared to have the identical porB genes or segments of the same gene. Our demonstration that N. gonorrhoeae isolates with the most common PIA VR types fall into several genetically unrelated PFGE clusters and, thus, presumably have different ancestral backgrounds further supports this hypothesis. We found that PFGE, which has been used by others to investigate outbreaks and the emergence of antibiotic-resistant strains (51, 58), was sufficiently discriminatory to distinguish between LF+ and LF− clonal types within a VR type, with one exception, and to confirm partner relationships, which were based on interview data. Isolates from male and female partners fell into the same clusters, the majority of which had almost identical band patterns. The use of a second enzyme may have provided a higher level of discrimination of isolates within a cluster. However, even if these isolates are less related than indicated by measurement, our conclusion that porin types can be expressed by isolates of different ancestral backgrounds would not change. We therefore did not pursue further measures of genomic relatedness.
Several functions have been attributed to gonococcal porin, including serum resistance, due to downregulation of complement activation via the binding of complement regulatory proteins (41, 42). We found that isolates of the porB VR types PIA 3;1;1/2;2/3;2 and PIA 3;1;2;2;2, which were not isolated with high frequencies in the Baltimore study, and VR type PIA 2;4;3;3;3 were more often sensitive to the bactericidal activity of NHS. In contrast, significantly more isolates of the most common VR types, PIA 1;2;1;1;1 and PIA 1;1;1;1/4;1, showed high or intermediate levels of serum resistance; isolates of these VR types hybridize to the same oligonucleotide probes used to type loops 1, 3, 6, and 7 but to different probes for the loop 2-encoding region, as do isolates of VR type 1;3;1;1/4;1, which were not tested in this study but which were isolated for 5 of the 10 years of the Baltimore study (32). Interestingly, isolates that hybridized to probe 1 for loop 1 were more often serum resistant than those that hybridized to probe 2 or 3 for loop 1. Using hybrid porin strains, Ram et al. found that P1A loop 1 was required, but alone not sufficient, for C4BP binding (41), and that loop 5 of certain P1A strains such as FA19 bound to fH (42). The exact binding motifs on P1A for these complement inhibitors are not known. The data from our study appear to be consistent with the finding that loop 1 is important in serum resistance. Sequence data for loop 5, which is not targeted by the VR typing probe, suggest that the fH binding site may be conformational and involve other loops.
We note that serum resistance did not always segregate strictly by VR type or by the predicted amino acid loop sequences. Several other factors can influence serum resistance in N. gonorrhoeae, including surface molecules that undergo phase-variable expression, which might explain why otherwise genetically related isolates can differ in serum resistance phenotypes. Specifically, phase-variable expression of the glycosyl transferase genes (lgtA, lgtC, and lgtG) involved in lipooligosaccharide (LOS) biosynthesis can lead to a marked difference in the levels of serum resistance (45), and opacity protein-mediated serum resistance was reported for an SI strain and shown to be phenotypically recessive to LOS phenotype (5). Interestingly, the LOS structure can modulate the binding of C4BP to N. gonorrhoeae (43), and we speculate that differences in complement inhibitor binding and serum resistance phenotypes observed among strains with identical P1A sequences (such as LG2, LG9, and LG16) may be, at least in part, related to differences in LOS glycan substitutions.
One interesting finding of this project came from the analysis of isolates of VR type PIA 2;4;3;3;3. This group of isolates appeared only during the first 3 years of the 10-year period analyzed by McKnew et al. (32); whether these isolates were present prior to the initiation of the study is not known. We have identified this porB type infrequently in other strain collections, and we have observed these loop sequences only with this specific combination (data not shown). None of the individual loop sequences of this PIA type has been observed in mosaic combination with other VRs, a result which suggests that the horizontal exchange of portions of this porB gene has not been successful. It is possible that immunity to porin or other epitopes expressed by these isolates may affect its capacity to persist within the communities studied. None of these isolates could use lactoferrin as an iron source, and all of these isolates demonstrated the Mtr phenotype and had β-lactamase activity. Whether these three phenotypes influence the fitness of this clone is not clear. Mutation of the lactoferrin receptor did not attenuate N. gonorrhoeae in the human urethritis model, unless the transferrin receptor was also not expressed (1), and while data from a murine infection model suggest that the mtr mutation may confer increased survival during infection (54), the consequence of β-lactamase-mediated antibiotic resistance on gonococcal fitness has not been examined. It is possible that β-lactamase production may be accompanied by a fitness loss based on the traditional inverse association between resistance to antibiotics and fitness (27).
PIA strains are historically more often associated with disseminated gonococcal infections than with PIB strains. However, our data suggest that resistance to complement may be important to PIA strains during urogenital tract infections, since the concentration of serum used to define serum resistance, specifically 10%, is likely to reflect concentrations of complement encountered during mucosal infections rather than in the bloodstream. Other porin-mediated functions that could confer a selective advantage include potential differences in nutrient acquisition, invasion (16), the capacity to induce (34, 35, 55) or inhibit (3) apoptosis, and changes in permeability that might result in higher levels of antibiotic resistance. It is not known if heterogeneity in porin molecules confers differences in porin-related apoptotic properties, and porin-mediated differences in antibiotic resistance have been demonstrated only for PIB strains (15, 57). The capacity of some porin types to evade a porin-specific adaptive immune response must also be considered. Plummer et al. (38) reported that women with gonococcal pelvic inflammatory disease and female commercial sex workers with a history of gonococcal infection were at increased risk for reinfection with an isolate of a different serovar. From these studies, one might conclude that these women developed some form of PorB-specific immunity. However, no evidence of serovar-specific immunity was found in studies of predominantly male urethral infections in rural populations in North Carolina (13, 22). The role of immune evasion in PorB diversification deserves further investigation, specifically with regard to gender, type of infection, and relationship to mixed and asymptomatic infections.
In summary, we conclude that some PIA molecules confer a selective advantage to strains that results in the persistence of certain PorB types over time and among geographically distinct strains and that this advantage may be the increased resistance to complement-mediated killing in the urogenital tract. Whether similar or additional selective pressures affect the isolation of P1B strains, which are more common and also more often SS, is not yet known. More diversity also exists among PIB VR types, although a subset of VR types also appears to be more common (32). An examination of these hypotheses with respect to P1B diversity and conservation is under way. The data presented in the current work should help delineate the PIA porin sequences that are involved in serum resistance through fH and C4BP binding. Our results may also provide new focus to the design of porin-based vaccines and novel immunoprophylactic strategies for this highly antigenically diverse pathogen.
Acknowledgments
This work was supported by National Institutes of Health STI/TM CRC grant U19 AI31496 (A.E.J.), NIH grant RO1-AI062755 (W.M.S.), and NIH grants AI54544 and AI032725 (S.R.).
W.M.S. was supported in part by a senior research career scientist award from the VA Medical Research Service.
Editor: A. Camilli
Footnotes
Published ahead of print on 9 June 2008.
REFERENCES
- 1.Anderson, J. E., M. Hobbs, G. Biswas, and P. F. Sparling. 2003. Opposing selective forces for expression of the gonococcal lactoferrin receptor. Mol. Microbiol. 481325-1337. [DOI] [PubMed] [Google Scholar]
- 2.Bash, M. C., P. Zhu, S. Gulati, D. McKnew, P. A. Rice, and F. Lynn. 2005. por variable-region typing by DNA probe hybridization is broadly applicable to epidemiologic studies of Neisseria gonorrhoeae. J. Clin. Microbiol. 431522-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binnicker, M., R. Williams, and M. Apicella. 2004. Gonococcal porin IB activates NF-κB in human urethral epithelium and increases the expression of host antiapoptotic factors. Infect. Immun. 726408-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blake, M. S., and E. C. Gotschlich. 1986. Functional and immunological properties of pathogenic neisserial surface proteins, p. 377-400. In M. Inouye (ed.), Bacterial outer membranes as model systems. John Wiley & Sons, Inc., New York, NY.
- 5.Bos, M. P., D. Hogan, and R. J. Belland. 1997. Selection of Opa+ Neisseria gonorrhoeae by limited availability of normal human serum. Infect. Immun. 65645-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. 2006. Trends in reportable sexually transmitted diseases in the United States, 2006. National surveillance data for chlamydia, gonorrhea, and syphilis. Centers for Disease Control and Prevention, Atlanta, GA.
- 7.Chesson, H. W., J. M. Blandford, T. L. Gift, G. Tao, and K. L. Irwin. 2004. The estimated direct medical cost of sexually transmitted diseases among American youth, 2000. Perspect. Sex. Reprod. Health 3611-19. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, M. S., I. F. Hoffman, R. A. Royce, P. Kazembe, J. R. Dyer, C. C. Daly, D. Zimba, P. L. Vernazza, M. Maida, S. A. Fiscus, and J. J. Eron, Jr., for the AIDSCAP Malawi Research Group. 1997. Reduction of concentration of HIV-1 in semen after treatment of urethritis: implications for prevention of sexual transmission of HIV-1. Lancet 3491868-1873. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen, C. N., M. Kelley, M. M. Hobbs, J. E. Anderson, J. G. Cannon, M. S. Cohen, and P. F. Sparling. 1998. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol. Microbiol. 27611-616. [DOI] [PubMed] [Google Scholar]
- 10.Derrick, J. P., R. Urwin, J. Suker, I. M. Feavers, and M. C. Maiden. 1999. Structural and evolutionary inference from molecular variation in Neisseria porins. Infect. Immun. 672406-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewi, B. E., S. Akira, H. Hayashi, and W. Ba-Thein. 2004. High occurrence of simultaneous mutations in target enzymes and MtrRCDE efflux system in quinolone-resistant Neisseria gonorrhoeae. Sex. Transm. Dis. 31353-359. [DOI] [PubMed] [Google Scholar]
- 12.Folster, J. P., and W. M. Shafer. 2005. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J. Bacteriol. 1873713-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, K. K., J. C. Thomas, D. H. Weiner, R. H. Davis, P. F. Sparling, and M. S. Cohen. 1999. Longitudinal evaluation of serovar-specific immunity to Neisseria gonorrhoeae. Am. J. Epidemiol. 149353-358. [DOI] [PubMed] [Google Scholar]
- 14.Fudyk, T., I. W. Maclean, J. N. Simonsen, E. Njagi, J. Kimani, R. C. Brunham, and F. A. Plummer. 1999. Genetic diversity and mosaicism at the por locus of Neisseria gonorrhoeae. J. Bacteriol. 1815591-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill, M. J., S. Simjee, K. Al-Hattawi, B. D. Robertson, C. S. Easmon, and C. A. Ison. 1998. Gonococcal resistance to β-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob. Agents Chemother. 422799-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorby, G. L., A. F. Ehrhardt, M. A. Apicella, and C. Elkins. 2001. Invasion of human fallopian tube epithelium by Escherichia coli expressing combinations of a gonococcal porin, opacity-associated protein, and chimeric lipo-oligosaccharide. J. Infect. Dis. 184460-472. [DOI] [PubMed] [Google Scholar]
- 17.Gotschlich, E. C., M. E. Seiff, M. S. Blake, and M. Koomey. 1987. Porin protein of Neisseria gonorrhoeae: cloning and gene structure. Proc. Natl. Acad. Sci. USA 848135-8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141611-622. [DOI] [PubMed] [Google Scholar]
- 19.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 1774162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton, H. L., and J. P. Dillard. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59376-385. [DOI] [PubMed] [Google Scholar]
- 21.Hardig, Y., A. Hillarp, and B. Dahlback. 1997. The amino-terminal module of the C4b-binding protein alpha-chain is crucial for C4b binding and factor I-cofactor function. Biochem. J. 323469-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobbs, M., T. M. Alcorn, R. H. Davis, W. Fischer, J. C. Thomas, I. Martin, C. Ison, P. F. Sparling, and M. S. Cohen. 1998. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J. Infect. Dis. 179371-381. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann, K. M., D. Williams, W. M. Shafer, and R. G. Brennan. 2005. Characterization of the multiple transferable resistance repressor, MtrR, from Neisseria gonorrhoeae. J. Bacteriol. 1875008-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jokiranta, T. S., P. F. Zipfel, J. Hakulinen, S. Kuhn, M. K. Pangburn, J. D. Tamerius, and S. Meri. 1996. Analysis of the recognition mechanism of the alternative pathway of complement by monoclonal anti-factor H antibodies: evidence for multiple interactions between H and surface bound C3b. FEBS Lett. 393297-302. [DOI] [PubMed] [Google Scholar]
- 25.Kellogg, D. S., Jr., W. L. Peacock, Jr., W. E. Deacon, L. Brown, and D. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 851274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kline, K. A., E. V. Sechman, E. P. Skaar, and H. S. Seifert. 2003. Recombination, repair and replication in the pathogenic Neisseriae: the 3 R's of molecular genetics of two human-specific bacterial pathogens. Mol. Microbiol. 503-13. [DOI] [PubMed] [Google Scholar]
- 27.Lenski, R. E. 1997. The cost of antibiotic resistance-from the perspective of a bacterium. Ciba Found. Symp. 207131-151. [DOI] [PubMed] [Google Scholar]
- 28.Lin, J. S., S. P. Donegan, T. C. Heeren, M. Greenberg, E. E. Flaherty, R. Haivanis, X. H. Su, D. Dean, W. J. Newhall, J. S. Knapp, S. K. Sarafian, R. J. Rice, S. A. Morse, and P. A. Rice. 1998. Transmission of Chlamydia trachomatis and Neisseria gonorrhoeae among men with urethritis and their female sex partners. J. Infect. Dis. 1781707-1712. [DOI] [PubMed] [Google Scholar]
- 29.Lucas, C. E., J. T. Balthazar, K. E. Hagman, and W. M. Shafer. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 1794123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynn, F., M. M. Hobbs, J. M. Zenilman, F. M. Behets, K. Van Damme, A. Rasamindrakotroka, and M. C. Bash. 2005. Genetic typing of the porin protein of Neisseria gonorrhoeae from clinical noncultured samples for strain characterization and identification of mixed gonococcal infections. J. Clin. Microbiol. 43368-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, I. M., C. A. Ison, D. M. Aanensen, K. A. Fenton, and B. G. Spratt. 2004. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 1891497-1505. [DOI] [PubMed] [Google Scholar]
- 32.McKnew, D. L., F. Lynn, J. M. Zenilman, and M. C. Bash. 2003. Porin variation among clinical isolates of Neisseria gonorrhoeae over a 10-year period, as determined by Por variable region typing. J. Infect. Dis. 1871213-1222. [DOI] [PubMed] [Google Scholar]
- 33.Mickelsen, P. A., E. Blackman, and P. F. Sparling. 1982. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from lactoferrin. Infect. Immun. 35915-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller, A. D. G., F. Dux, M. Naumann, T. F. Meyer, and T. Rudel. 1999. Neisserial porin (PorB) causes rapid calcium influx in target cells and induces apoptosis by the activation of cysteine proteases. EMBO J. 18339-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naumann, M., T. Rudel, and T. Meyer. 1999. Host cell interactions and signalling with Neisseria gonorrhoeae. Curr. Opin. Microbiol. 262-70. [DOI] [PubMed] [Google Scholar]
- 36.O'Rourke, M., and B. G. Spratt. 1994. Further evidence for the non-clonal population structure of Neisseria gonorrhoeae: extensive genetic diversity within isolates of the same electrophoretic type. Microbiology 1401285-1290. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Losada, M., R. P. Viscidi, J. C. Demma, J. Zenilman, and K. A. Crandall. 2005. Population genetics of Neisseria gonorrhoeae in a high-prevalence community using a hypervariable outer membrane porB and 13 slowly evolving housekeeping genes. Mol. Biol. Evol. 221887-1902. [DOI] [PubMed] [Google Scholar]
- 38.Plummer, F. A., J. N. Simonsen, H. Chubb, L. Slaney, J. Kimata, M. Bosire, J. O. Ndinya-Achola, and E. N. Ngugi. 1989. Epidemiologic evidence for the development of serovar-specific immunity after gonococcal infection. J. Clin. Investig. 831472-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poh, C. L., G. K. Loh, and J. W. Tapsall. 1995. Resolution of clonal subgroups among Neisseria gonorrhoeae IB-2 and IB-6 serovars by pulsed-field gel electrophoresis. Genitourin. Med. 71145-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Posada, D., K. A. Crandall, M. Nguyen, J. C. Demma, and R. P. Viscidi. 2000. Population genetics of the porB gene of Neisseria gonorrhoeae: different dynamics in different homology groups. Mol. Biol. Evol. 17423-436. [DOI] [PubMed] [Google Scholar]
- 41.Ram, S., M. Cullinane, A. M. Blom, S. Gulati, D. P. McQuillen, B. G. Monks, C. O'Connell, R. Boden, C. Elkins, M. K. Pangburn, B. Dahlback, and P. A. Rice. 2001. Binding of C4b-binding protein to porin: a molecular mechanism of serum resistance of Neisseria gonorrhoeae. J. Exp. Med. 193281-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ram, S., D. P. McQuillen, S. Gulati, C. Elkins, M. K. Pangburn, and P. A. Rice. 1998. Binding of complement factor H to loop 5 of porin protein 1A: a molecular mechanism of serum resistance of nonsialylated Neisseria gonorrhoeae. J. Exp. Med. 188671-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ram, S., J. Ngampasutadol, A. D. Cox, A. M. Blom, L. A. Lewis, F. St Michael, J. Stupak, S. Gulati, and P. A. Rice. 2007. Heptose I glycan substitutions on Neisseria gonorrhoeae lipooligosaccharide influence C4b-binding protein binding and serum resistance. Infect. Immun. 754071-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 321117-1123. [DOI] [PubMed] [Google Scholar]
- 45.Shafer, W. M., A. Datta, V. S. Kolli, M. M. Rahman, J. T. Balthazar, L. E. Martin, W. L. Veal, D. S. Stephens, and R. Carlson. 2002. Phase variable changes in genes lgtA and lgtC within the lgtABCDE operon of Neisseria gonorrhoeae can modulate gonococcal susceptibility to normal human serum. J. Endotoxin Res. 847-58. [PubMed] [Google Scholar]
- 46.Shafer, W. M., X. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 951829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, N. H., J. M. Smith, and B. G. Spratt. 1995. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: evidence of positive Darwinian selection. Mol. Biol. Evol. 12363-370. [DOI] [PubMed] [Google Scholar]
- 48.Tam, M. R., T. M. Buchanan, E. G. Sandstrom, K. K. Holmes, J. S. Knapp, A. W. Siadak, and R. C. Nowinski. 1982. Serological classification of Neisseria gonorrhoeae with monoclonal antibodies. Infect. Immun. 361042-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka, M., N. H. Huruya, K. Konomi, I. Irie, S. Kanayama, A. Saika, and I. Kobayashi. 2006. Analysis of mutations within multiple genes associated with resistance in a clinical isolate of Neisseria gonorrhoeae with reduced ceftriaxone susceptibility that shows a multidrug-resistant phenotype. Int. J. Antimicrob. Agents 277. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, D. K., C. D. Deal, C. A. Ison, J. M. Zenilman, and M. C. Bash. 2000. A typing system for Neisseria gonorrhoeae based on biotinylated oligonucleotide probes to PIB gene variable regions. J. Infect. Dis. 1811652-1660. [DOI] [PubMed] [Google Scholar]
- 51.Unemo, M., T. Berglund, P. Olcén, and H. Fredlund. 2002. Pulsed-field gel electrophoresis as an epidemiologic tool for Neisseria gonorrhoeae: identification of clusters within serovars. Sex. Transm. Dis. 2925-31. [DOI] [PubMed] [Google Scholar]
- 52.Unemo, M., P. Olcén, J. Albert, and H. Fredlund. 2003. Comparison of serologic and genetic porB based typing of Neisseria gonorrhoeae: consequences for future characterization. J. Clin. Microbiol. 414141-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veal, W. L., R. A. Nicholas, and W. M. Shafer. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 1845619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warner, D. M., J. P. Folster, W. M. Shafer, and A. E. Jerse. 2007. Regulation of the MtrC-MtrD-MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J. Infect. Dis. 1961804-1812. [DOI] [PubMed] [Google Scholar]
- 55.Weel, J. F., and J. P. van Putten. 1991. Fate of the major outer membrane protein P.IA in early and late events of gonococcal infection of epithelial cells. Res. Microbiol. 142985-993. [DOI] [PubMed] [Google Scholar]
- 56.WHO. 1999. Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates. World Health Organization, Geneva, Switzerland.
- 57.Woodford, N., K. M. Bindayna, C. S. Easmon, and C. A. Ison. 1989. Associations between serotype and susceptibility to antibiotics of Neisseria gonorrhoeae. Genitourin. Med. 6586-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia, M., W. L. Whittington, K. K. Holmes, F. A. Plummer, and M. C. Roberts. 1995. Pulsed-field gel electrophoresis for genomic analysis of Neisseria gonorrhoeae. J. Infect. Dis. 171455-458. [DOI] [PubMed] [Google Scholar]
- 59.Xia, M., W. L. Whittington, K. K. Holmes, and M. C. Roberts. 1997. Genomic homogeneity of the AHU/IA-1,2 phenotype of Neisseria gonorrhoeae during its disappearance from an urban population. Sex. Transm. Dis. 24561-566. [DOI] [PubMed] [Google Scholar]
- 60.Yamasaki, R., D. E. Kerwood, H. Schneider, K. P. Quinn, J. M. Griffiss, and R. E. Mandrell. 1994. The structure of lipooligosaccharide produced by Neisseria gonorrhoeae, strain 15253, isolated from a patient with disseminated infection. Evidence for a new glycosylation pathway of the gonococcal lipooligosaccharide. J. Biol. Chem. 26930345-30351. [PubMed] [Google Scholar]
- 61.Zarantonelli, L., G. Borthagaray, E. H. Lee, and W. M. Shafer. 1999. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob. Agents Chemother. 432468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]