Abstract
The Francisella tularensis live vaccine strain (LVS), in contrast to its iglC mutant, replicates in the cytoplasm of macrophages. We studied the outcome of infection of the murine macrophagelike cell line J774A.1 with LVS and with iglC, iglD, and mglA mutants, the latter of which is deficient in a global regulator. Compared to LVS, all of the mutants showed impaired intracellular replication up to 72 h, and the number of the mglA mutant bacteria even decreased. Colocalization with LAMP-1 was significantly increased for all mutants compared to LVS, indicating an impaired ability to escape into the cytoplasm. A lysosomal acidity-dependent dye accumulated in approximately 40% of the vacuoles containing mutant bacteria but not at all in vacuoles containing LVS. Preactivation of the macrophages with gamma interferon inhibited the intracellular growth of all strains and significantly increased acidification of phagosomes containing the mutants, but it only slightly increased the LAMP-1 colocalization. The intracellular replication and phagosomal escape of the iglC and iglD mutants were restored by complementation in trans. In conclusion, the IglC, IglD, and MglA proteins each directly or indirectly critically contribute to the virulence of F. tularensis LVS, including its intracellular replication, cytoplasmic escape, and inhibition of acidification of the phagosomes.
Macrophages perform a sentinel function aimed at phagocytosing and killing invading pathogens. Internalization is followed by a complex remodeling of the phagosome, during which the interactions of the phagosome with organelles of the endocytic pathway and ultimately fusion with lysosomes lead to formation of a hostile environment that can degrade ingested bacteria (13, 32). Phagocytosis has been exploited by certain microbes in order to form a symbiotic relationship with a host or by microbes that have developed an ability to parasitize the phagocytic cells and use them as a principal habitat for replication. Some pathogenic microbes, such as Mycobacterium, Salmonella, Legionella, and Chlamydia spp., subvert the composition of phagosomal compartments and obtain nutrients from the cytoplasm to support their replication, whereas other pathogenic microbes, such as Listeria, Shigella, and Rickettsia spp., manage to escape from the confined intracellular compartments and directly use the cytoplasm as their habitat (17).
Francisella tularensis is a highly virulent, facultative intracellular bacterium and the etiological agent of the zoonotic disease tularemia. This disease affects predominantly lagomorphs, rodents, and humans but has been found in more than 200 mammalian species (23). The bacterium appears to be very well adapted to the intracellular environment. Two of the F. tularensis subspecies, F. tularensis subsp. tularensis and F. tularensis subsp. holarctica, are highly contagious and virulent, even for humans. Moreover, Francisella novicida, a low-virulence species, is of interest since it has often been used in experimental models of tularemia (18). The F. tularensis live vaccine strain (LVS) became attenuated by repeated passage in vitro of a virulent strain of F. tularensis subsp. holarctica (10). This strain is currently used as a human vaccine strain and provides good, albeit not complete, protection against laboratory-acquired tularemia. The behavior of LVS in murine and human macrophages appears to resemble that of virulent F. tularensis strains, and although attenuated for mice, this strain causes a disease that closely resembles tularemia in wild rodents (10). Thus, in many respects the LVS strain appears to be a useful model for elucidating the intracellular virulence mechanisms of F. tularensis.
Uptake of F. tularensis by human monocytic cells occurs via a process that requires complement component C3 and actin-dependent pseudopod formation (8) Additionally, entry of Francisella into host cells has been reported to depend upon the mannose receptor, the Fc-γ receptor, and class A scavenger receptors in both human and murine host cells (3, 25, 30). Bacterium-containing phagosomes soon interact with early and late endosomes, and within hours F. tularensis escapes from the phagosomal compartment of the monocytes and replicates in the cytosol (9, 14, 22).
Despite the fact that several F. tularensis genomes have been sequenced (20, 24, 27), the successful adaptation of this organism to the intracellular habitat is not well understood. Features necessary for expression of full virulence of other intracellular pathogens, such as type III or IV secretion systems, are not present. Our current knowledge indicates that F. tularensis utilizes novel mechanisms in order to survive and multiply intracellularly. Escape from the phagosome of phagocytic cells is required for intracellular replication, and this escape is dependent on factors of the intracellular growth locus (Igl) operon (22, 29). The functions of the four proteins which are encoded by the Igl operon, IglA, IglB, IglC, and IglD, are elusive. The transcription of these proteins is regulated by the global regulator MglA (4, 21). An iglC mutant of F. tularensis LVS does not replicate intracellularly and lacks the ability of wild-type strains to cause apoptosis of infected cells (15, 19). Also, the LVS mutant and an iglC mutant of Schu S4 are both essentially avirulent in mice (15, 31). Together, the Igl proteins appear to be essential for F. tularensis to be a successful intracellular pathogen. A bioinformatic analysis of F. novicida has revealed that IglA and IglB show strong similarity to members of a recently described type VI secretion system in Vibrio cholerae (11, 26), but there is no experimental evidence that Francisella possesses such a secretion system.
The present study aimed to examine the phenotypes of F. tularensis mutants lacking expression of the IglC, IglD, and MglA proteins with regard to the fate of the organisms in murine macrophages. We demonstrated that LVS ΔiglD and ΔmglA strains, like an LVS ΔiglC mutant, are unable to escape from phagosomes, showing that each of the investigated Igl proteins and MglA have important roles.
MATERIALS AND METHODS
Bacterial strains.
F. tularensis LVS (ATCC 29684) was supplied by the U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, Frederick, MD. It was grown to logarithmic phase in modified Mueller-Hinton broth, harvested, and stored frozen at −70°C. A mutant of F. tularensis LVS, designated the ΔiglC mutant, was generated by allelic replacement of the two gene copies encoding the 23-kDa protein IglC, as described elsewhere (15), and mglA, and iglD mutants were constructed using the same strategy. The regions located upstream or downstream of each gene were amplified by PCR and, for the mglA mutant, were treated with restriction enzymes, ligated, and cloned directly into XbaI/SalI-digested pPV (15). To generate the iglD mutant, a second, overlapping PCR was performed using purified fragments complementary to the flanking regions as templates (all primer sequences used to generate mutants are available upon request). After restriction enzyme digestion and purification, the PCR fragments were cloned into XbaI/SalI-cleaved pPV. The resulting plasmids were designated pPV-ΔmglA and pPV-ΔiglD, respectively. These plasmids were first introduced into Escherichia coli S17-1 by electroporation and then transferred to LVS by conjugation (15). Clones with pPV-ΔmglA or pPV-ΔiglD integrated into the LVS chromosome by a single recombination event were selected on plates containing chloramphenicol and polymyxin B, and integration was verified by PCR (15). Clones with integrated plasmids were then subjected to sucrose selection. This procedure selected for a second crossover event, in which the integrated plasmid, carrying sacB, was excised from the chromosome. Chloramphenicol-sensitive clones were examined by PCR, which confirmed deletion of the genes. Since there are two copies of the iglD gene in the chromosome, conjugation and sucrose selection were repeated with the mutant from which the first copy was deleted. This procedure resulted in removal of the ribosome-binding site and nucleotides 1 to 374 of the iglD gene, and in the mglA mutant nucleotides 37 to 564 were removed. All deletions were in frame and thus did not affect transcription downstream.
For complementation in trans of the iglC mutant, plasmid pKK214iglC, carrying the iglC gene under control of the LVS groESL promoter, was used (19). The plasmid used for complementation of the iglD mutant was created by amplification of the iglD gene by PCR and cloning it into pKK214gfp (1) after removal of the gfp gene, resulting in pKK289iglD. Using a similar strategy, the mglA gene was amplified by PCR and cloned into the pKK289Km-gfp plasmid, a derivative of pKK214gfp that expresses a Tn5-derived kanamycin resistance gene, resulting in plasmid pKK289Km-mglA. All plasmids used for complementation were then introduced into the corresponding mutants by cryotransformation (15).
For construction of green fluorescent protein (GFP)-expressing bacteria, the LVS, ΔiglC, and ΔiglD mutants were transformed with pKK214gfp (1), and the ΔmglA mutant was transformed with pKK289Km-gfp. To express an igl gene and gfp on the same plasmid, the genes and their ribosome-binding sites were cloned directly into PstI-digested pKK214gfp, resulting in plasmids pKK289iglC/gfp and pKK289iglD/gfp. Finally, the plasmids were introduced into the corresponding mutants by cryotransformation.
Formalin-killed LVS (FK-LVS) was prepared by incubating GFP-expressing LVS in 4% paraformaldehyde for 45 min at 37°C, followed by three washes in phosphate-buffered saline (PBS).
Western blot analysis.
Bacterial lysates in Laemmli sample buffer were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel. Each lane was loaded with lysate corresponding to 3 × 107 bacteria. Proteins were then transferred onto nitrocellulose membranes using a semidry blotter (EBU 4000; C.B.S. Scientific Co., Del Mar, CA). The membranes were then incubated overnight at 4°C with the following primary antibodies: mouse monoclonal anti-F. tularensis IglC diluted 1:3,000 (kindly provided by Jiri Stulik, Institute of Molecular Pathology, Faculty of Military Health Sciences, University of Defense, Trebesska, Hradec Kralove, Czech Republic); chicken IgY anti-F. tularensis IglD diluted 1: 20,000 (Agrisera, Vännäs, Sweden); and monoclonal anti-F. tularensis intracellular growth locus subunit B (IglB) protein clone IglB1 NR-3195 diluted 1:8,000 (obtained through the NIH Biodefense and Emerging Infections Research Repository, NIAID, NIH). Secondary horseradish peroxidase (HRP)-conjugated antibodies were used at the following dilutions: goat anti-mouse HRP-conjugated antibody, 1:2,000 (Santa Cruz Biotechnology, CA); and rabbit anti-chicken IgY HRP-conjugated antibody, 1:20,000 (Sigma-Aldrich, St. Louis, MO).
Infection of macrophages.
For all infections, mouse macrophagelike J774A.1 cells were seeded in tissue culture plates the day before infection in Dulbecco modified Eagle medium (DMEM) (GIBCO BRL, Grand Island, NY) with 10% heat-inactivated fetal bovine serum (FBS). After adhesion, the monolayers were washed and reconstituted with prewarmed fresh medium and then incubated overnight at 37°C in the presence of 5% CO2. To some wells, 100 to 1,000 U/ml of gamma interferon (IFN-γ) (Peprotech, London, United Kingdom) was added 12 h before infection and was present throughout the infection. Following incubation overnight, the wells were washed and reconstituted with fresh culture medium. Bacteria were grown overnight and resuspended in ice-cold PBS to a density of approximately 2.5 × 109 bacteria/ml. Bacteria were added to each well at the indicated multiplicity of infection (MOI) in DMEM with 10% noninactivated human serum from a nonimmune donor, and bacterial uptake was allowed to occur for 60 min at 37°C, as described previously (9). After they were infected, the monolayers were washed three times and then incubated for the indicated periods of time in prewarmed DMEM with FBS supplemented with 5 μg/ml of gentamicin. The time point after 30 min of gentamicin treatment was defined as zero time.
At 0, 24, 48, and 72 h, supernatants were collected for further analysis, and after washing, the macrophage monolayers were lysed in 0.1% deoxycholate. One-hundred-microliter portions of serial dilutions in PBS of the lysates were plated on plates containing GC II agar base (BD Diagnostic Systems, Maryland) with hemoglobin and IsoVitaleX (BD Diagnostic Systems) and incubated at 37°C until colonies could be enumerated. The total number of bacteria/well was calculated. The assay was performed in triplicate and was repeated three times.
Assay of cell death.
Culture supernatants (50 μl) from the bacterial viability assay were transferred to a 96-well plate and mixed with 50 μl of a substrate mixture prepared according to the manufacturer's instructions (lactate dehydrogenase [LDH] assay kit; Promega, Madison, WI). The plates were incubated in the dark at room temperature for 30 min, and then 50 μl of a stop solution was added. Absorbance at 492 nm was determined with a Tecan Sunrise plate reader (Tecan Systems, San Jose, CA) 1 h later. Uninfected J774 cells lysed in 0.1% deoxycholate served as a positive control, and the value for this control was arbitrarily considered 100% cell lysis. Sample absorbance was expressed as a percentage of the positive control value. The assay was performed with triplicate samples and was repeated three times.
Intracellular immunofluorescence assay.
J774 cells (2.5 × 105 cells/well) in DMEM with 10% FBS were seeded onto glass coverslips in 24-well plates. The following day, the cells were infected with GFP-expressing F. tularensis LVS or mutant strains at an MOI of 30 or with green fluorescent latex beads (Sigma-Aldrich, St. Louis, MO) at an MOI of 10 for 1 h; after this the cells were washed three times and incubated for the indicated periods of time. For incubation times between 20 and 72 h, 1 × 105 J774 cells were infected at an MOI of 5. After infection, the cells were washed with PBS, fixed in 4% paraformaldehyde for 15 min, washed again with PBS, permeabilized with 0.15% saponin, blocked in 2% bovine serum albumin-saponin, and incubated in blocking buffer with primary antibodies against the LAMP-1 glycoprotein diluted 1:500 (BD Pharmingen, San Jose, CA) or against cathepsin D (CatD) diluted 1:200 (Dako Cytomation, Glostrup, Denmark). Then an anti-rat or anti-rabbit immunoglobulin G antibody conjugated to Alexa 555 (Molecular Probes, Eugene, OR) was added at a dilution of 1:1,000. After three washes in PBS, coverslips were mounted using Dako Cytomation mounting medium.
To assay the acidity of the phagosomes, Lysotracker Red DND99 (Molecular Probes) was added to culture supernatants 30 min before the indicated time points at a 1:10,000 dilution to allow accumulation of the dye in acidic organelles. The monolayers were then washed once with DMEM and three times with PBS and fixed in 4% paraformaldehyde. The Lysotracker Red slides were analyzed immediately after fixation, washing, and mounting.
Glass slides were analyzed by using an epifluorescence microscope (Zeiss Axioskop2; Carl Zeiss MicroImaging GmbH, Germany) or a confocal microscope (Leica SP2; Leica Microsystems, Bensheim, Germany) for determination of the degree of colocalization of GFP-labeled F. tularensis and LAMP-1, CatD, or Lysotracker Red staining. A total of 100 bacteria on each coverslip were scored, each stain was analyzed in triplicate, and experiments were repeated two or three times.
Electron microscopy.
J774 cells (2 × 106 cells) in 6 ml of DMEM with FBS were seeded in tissue culture plates that were 6 cm in diameter. The following day the monolayers were infected at an MOI of 50 for 1 h and then washed three times and incubated in medium containing 5 μg/ml of gentamicin. After incubation, the monolayers were washed with sodium cacodylate buffer (0.1 M, pH 7.4) before fixation in 2.5% glutaraldehyde for 2 h in the same buffer. Then the cells were washed, scraped from the dishes, and postfixed for 1 h with 1% osmium tetroxide. After washing, the cell pellet was dissolved in 2% agarose and centrifuged in a warm centrifuge. The embedded pellet was cut into small cubes and stained with uranyl acetate (1% solution in methanol) overnight. Following dehydration using an acetone series, the specimens were embedded in Vestopal. Ultrathin sections (70 nm) were cut and contrasted with uranyl acetate and lead citrate before they were viewed with a JEOL 1200 EX-II electron microscope (JEOL Ltd., Tokyo, Japan). To examine membrane integrity, at least 50 bacteria from different sections of duplicate samples were analyzed for each time point and categorized as (i) bacteria with an intact or slightly damaged phagosomal membrane or (ii) bacteria with a mostly degraded membrane or no membrane.
Statistical analysis.
All values were expressed as means ± standard errors of the means. One-way analysis of variance (ANOVA), followed by a post hoc test (Bonferroni) and a one-tailed Student t test, was used to identify differences between groups. All statistical analyses were carried out using the SPSS software (version 15.0).
RESULTS
Characterization of Igl protein expression of mglA and igl mutants.
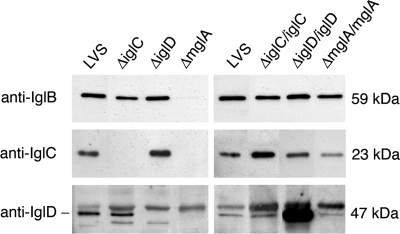
All strains were assayed for protein expression by Western blotting with anti-IglB, anti-IglC, or anti-IglD antibodies. The levels of the IglB protein were intact in the ΔiglC and ΔiglD strains, the levels of the IglC protein were intact in the ΔiglD strain, and the levels of the IglD protein were intact in the ΔiglC strain, thus showing that the deletions did not have any polar effects. The IglB, IglC, and IglD proteins were absent or the levels were very low in the ΔmglA strain (Fig. 1). The complemented igl mutants showed restored expression of the corresponding Igl proteins. The complemented ΔmglA strain had intact levels of IglB, while the levels of IglC and IglD were slightly lower than the levels in LVS (Fig. 1).
FIG. 1.
Western blot showing the levels of IglB, IglC, and IglD proteins in the igl and mglA mutants and in the complemented igl and mglA mutants. The presence of the three proteins in the LVS, ΔiglC, ΔiglD, and ΔmglA strains is shown in the blot on the left, and the presence in the complemented mutant strains is shown in the blot on the right. Each lane was loaded with a lysate prepared from 3 × 107 bacteria.
The mglA and igl mutants of F. tularensis LVS all have reduced replication rates in J774 cells.
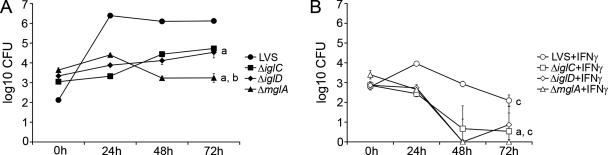
The iglC mutant of F. tularensis LVS has previously been reported to have an impaired ability to replicate intracellularly in J774 cells (15, 19, 22). Here, we monitored the numbers of bacteria in J774 cells infected with LVS or the iglC, iglD, or mglA mutant. The cells were cultured in the absence or presence of IFN-γ, a cytokine which is required for activation of mechanisms controlling the intracellular growth of F. tularensis (2, 22). Intracellular bacterial counts were assessed up to 72 h. After this, all cell cultures, including noninfected macrophages, showed signs of severe cytopathogenicity and detachment of cells. During the time period assessed, the number of intracellular LVS bacteria increased by 4.0 log10 (Fig. 2). The number of the ΔiglC and ΔiglD bacteria increased by 1.7 and 1.2 log10, respectively, whereas the number of ΔmglA bacteria decreased by 0.4 log10. Thus, the igl mutants were capable of intracellular replication, albeit at a rate lower than that of F. tularensis LVS. These results are in agreement with our previous findings concerning the critical role of IglC in F. tularensis LVS (15) and additionally indicate that IglD and MglA also must be expressed for effective intracellular replication of F. tularensis LVS. The decreased intracellular replication rates are similar to what has been observed for iglC, iglD, and mglA mutants of F. novicida (16, 21, 28, 29).
FIG. 2.
Numbers of F. tularensis bacteria in J774 cells. J774 cells were infected for 1 h with the indicated F. tularensis strains at an MOI of 5 without (A) or with (B) IFN-γ and then incubated for the indicated periods. The numbers of bacteria at 0, 24, 48, and 72 h are expressed as the mean ± standard error of the mean log10 CFU for triplicate wells in one of the three experiments performed. The letter “a” indicates that the number of bacteria was significantly lower than the number of LVS bacteria at 24, 48, and 72 h (P < 0.05, as determined by a one-way ANOVA). The exception was iglD with IFN-γ at 72 h (P < 0.24). The comparisons were performed separately for IFN-γ-treated cultures. The letter “b” indicates that the log10 number of CFU of the ΔmglA mutant was significantly less (P < 0.001) than the values for the igl mutants at 48 and 72 h. The letter “c” indicates that IFN-γ treatment significantly inhibited the growth of each strain at 24, 48, and 72 h (P < 0.01).
Macrophages activated with IFN-γ were capable of restricting the replication of all strains; the number of LVS bacteria decreased by 0.7 log10, the numbers of ΔiglC and ΔiglD bacteria decreased by 2.3 and 2.0 log10, respectively, and the ΔmglA strain was completely eliminated during the 72 h of incubation (Fig. 2).
The mglA and igl mutants of F. tularensis LVS exhibit reduced cytopathogenicity in J774 cells.
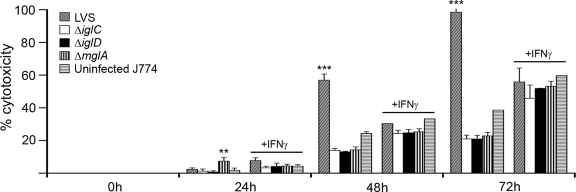
Upon infection of J774 cells, LVS causes cytopathogenic effects, a virulence trait which has not been observed for the ΔiglC strain (15, 19). Here the cytopathogenicity of each of the mutants was studied by assaying the LDH released into the cell medium. The results were expressed as percentages of the level recorded in the medium of lysed, noninfected cells. When cells were infected with F. tularensis LVS, the LDH levels increased from 2 to 98% in the interval from 24 to 72 h (Fig. 3). Compared to F. tularensis LVS, all mutants showed an impaired ability to cause cytopathogenicity, and the LDH levels at 72 h after infection were only ∼20% of the levels in noninfected lysed cells (P < 0.001).
FIG. 3.
Cytopathogenicity of F. tularensis strains. J774 cells were infected for 1 h with the indicated F. tularensis strains at an MOI of 5 in the absence or presence of IFN-γ. Cytopathogenicity was estimated by assaying LDH activity in culture supernatants and was expressed as a percentage of the level for noninfected lysed cells. The bars indicate the means and the error bars indicate the standard errors of the means for triplicate wells from one of three experiments. Asterisks indicate that the cytopathogenicity levels were significantly higher than those of uninfected cells, as determined by one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Comparisons were performed separately for IFN-γ-treated cultures.
IFN-γ-treated J774 cells resisted the cytopathic effects of F. tularensis LVS infection. At 72 h, similar levels of LDH release (∼55%) occurred in LVS-infected cultures and cultures of noninfected cells. The same was true for cells infected with any of the mutants. Thus, in the absence of IFN-γ, expression of the Igl proteins and MglA was required for induction of cytopathogenicity. In IFN-γ-activated J774 cells, no cytopathic effects of LVS or any of the mutants were observed.
Importance of MglA and Igl proteins for the escape of F. tularensis from phagosomes.
Similar to fully virulent F. tularensis strains, LVS is capable of escaping into the host cell cytoplasm and replicating in this permissive location (7, 9, 14). By contrast, the ΔiglC mutant of LVS is confined to the phagosome (22). In the present study of igl and mgl mutants, we used confocal microscopy to estimate the percentage of bacteria colocalizing with markers of intracellular compartments.
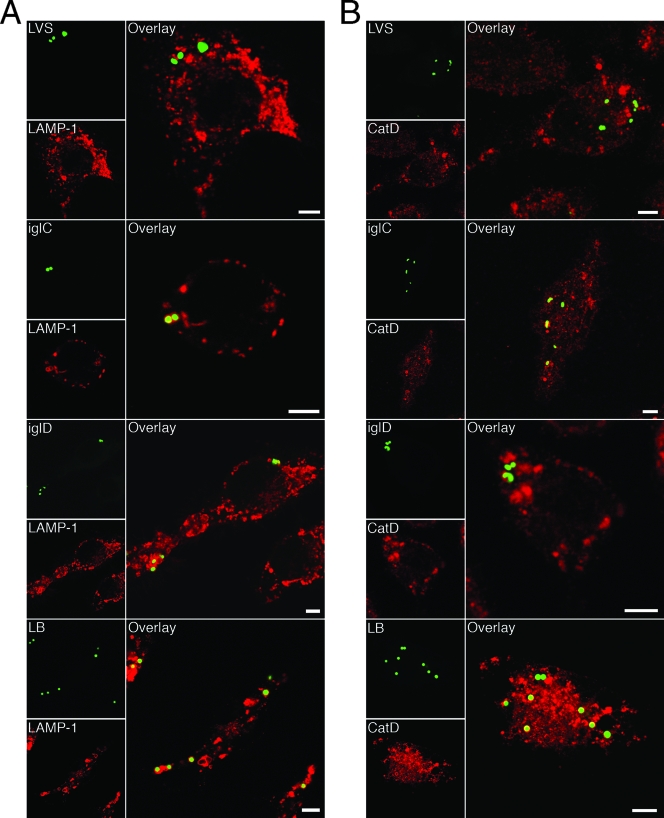
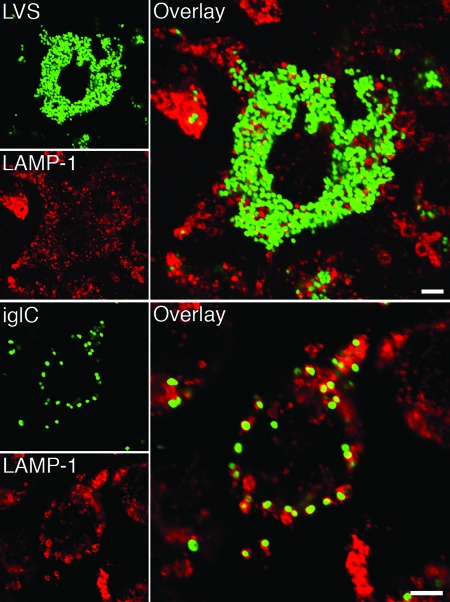
Infection with any of the three mutants (ΔiglC, ΔiglD, or ΔmglA) resulted in a much higher percentage of colocalization with the late endosomal and lysosomal marker LAMP-1 than infection with F. tularensis LVS. At 1 and 3 h after infection, only 40 and 19%, respectively, of the LVS bacteria colocalized with LAMP-1, compared to 65 to 76% (P < 0.05) and 75 to 81% (P < 0.001) of the mutant bacteria (Table 1 and Fig. 4A). The percentage of colocalization with LAMP-1 for each of the mutants was similar to that of FK-LVS bacteria or latex beads (Table 1 and Fig. 4A).
TABLE 1.
Quantitation of endocytic marker acquisition by phagosomes during infection with F. tularensis strains or after uptake of formalin-killed F. tularensis or latex beads after 60 or 180 mina
| Incubation time (min) | Group | Marker colocalization (%)b
|
|||||
|---|---|---|---|---|---|---|---|
| LAMP-1
|
CatD
|
Lysotracker Red
|
|||||
| Without IFN-γ | With IFN-γ | Without IFN-γ | With IFN-γ | Without IFN-γ | With IFN-γ | ||
| 60 | LVS | 40 ± 8.0 | 62 ± 7.6 | 40 ± 9.3 | 37 ± 24.6 | 16 ± 6.4 | 16 ± 3.1 |
| ΔiglC | 72 ± 6.6* | 78 ± 0.8 | 36 ± 5.9 | 47 ± 2.7 | 33 ± 10.7 | 88 ± 6.7*** | |
| ΔiglD | 65 ± 3.9 | 79 ± 1.2* | 36 ± 5.7 | 54 ± 2.3 | 16 ± 5.6 | 75 ± 1.5*** | |
| ΔmglA | 76 ± 2.2* | 79 ± 1.3* | 35 ± 10.9 | 47 ± 3.4 | 30 ± 4.9 | 74 ± 1.7*** | |
| FK-LVS | 83 ± 3.2** | 80 ± 2.9* | 47 ± 5.8 | 41 ± 5.8 | 63 ± 8.1** | 85 ± 2.4*** | |
| Latex beads | 76 ± 5.4* | 80 ± 2.0* | 68 ± 9.5 | 75 ± 5.0 | 76 ± 4.5** | 71 ± 4.0*** | |
| 180 | LVS | 19 ± 0.1 | 25 ± 1.9 | 22 ± 3.4 | 43 ± 12.1 | 2 ± 1.1 | 10 ± 1.0 |
| ΔiglC | 75 ± 3.4*** | 89 ± 3.1*** | 22 ± 5.5 | 61 ± 4.1 | 42 ± 4.8** | 86 ± 2.0*** | |
| ΔiglD | 81 ± 0.6*** | 90 ± 0.9*** | 28 ± 11.3 | 57 ± 3.5 | 44 ± 9.5** | 91 ± 2.7*** | |
| ΔmglA | 80 ± 6.6*** | 81 ± 1.3*** | 29 ± 6.1 | 46 ± 5.2 | 41 ± 2.2** | 82 ± 1.5*** | |
| FK-LVS | 86 ± 0.6*** | 89 ± 1.5*** | 44 ± 13.0 | 41 ± 1.0 | 69 ± 6.3*** | 79 ± 2.4*** | |
| Latex beads | 88 ± 1.9*** | 97 ± 0.9*** | 77 ± 5.6* | 73 ± 7.4 | 87 ± 4.6*** | 88 ± 0.9*** | |
J774 cells were infected for 1 h with the indicated live or formalin-killed F. tularensis strains at an MOI of 30 or with latex beads at an MOI of 10 and then incubated for 60 or 180 min.
The percentages indicate the fraction of F. tularensis-containing phagosomes stained for each of the following late endosomal/lysosomal markers: LAMP-1, CatD, and the lysomotropic substance Lysotracker Red. The results are expressed as means ± standard errors of the means from three separate experiments, and 100 bacteria on triplicate coverslips were counted in each experiment. Asterisks indicate that the colocalization level was significantly different (as determined by a one-way ANOVA) from that of LVS within each column for each incubation time, as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
FIG. 4.
Colocalization of GFP-expressing F. tularensis strains and late endosomal markers. J774 cells were infected for 1 h with live or formalin-killed F. tularensis strains at an MOI of 30, or latex beads were added at an MOI of 10 and, after washing, incubated for 3 h. Fixed specimens were labeled for either (A) the late endosomal and lysosomal marker LAMP-1 or (B) the lysosomal marker CatD. In the representative confocal images, green indicates bacteria or latex particles and red indicates the endocytic marker. Confocal images were acquired with the Leica SP2 confocal microscope (Leica Microsystems, Bensheim, Germany) and were assembled using Adobe Photoshop CS2 (Adobe Systems, San Jose, CA). LB, latex beads. Bars = 4 μm.
The extents of colocalization of the lysosomal hydrolase CatD were similar irrespective of whether the macrophages were infected with F. tularensis LVS or were infected with any of the three mutants. At 1 h, 40% of the LVS bacteria and 35 to 36% of the mglA or igl mutant bacteria colocalized with CatD (Table 1). At 3 h, 22% of the LVS bacteria colocalized with CatD, and the levels of colocalization of the mutants with the lysosomal marker were similar, 22 to 29% (Table 1 and Fig. 4B). Colocalization of formalin-killed F. tularensis LVS with CatD occurred for 47% of the bacteria at 1 h and for 44% of the bacteria at 3 h. These percentages were lower than those recorded for latex beads (68 and 77%, respectively) (Table 1 and Fig. 4B).
In essence, the investigated mutants all showed similar colocalization patterns and patterns similar to the LVS pattern for CatD, but they differed from LVS by having an increased propensity for colocalization with LAMP-1. The results obtained for the mutant strains are in agreement with previously published data for the LVS iglC mutant showing impaired escape into the cytosol (22). The high degree of colocalization of the mutants with LAMP-1 indicated that the phagosomal membranes were intact, while the intermediate level of colocalization with CatD indicated that F. tularensis possesses a mechanism(s) independent of the Igl/Mgl proteins for evading the fusion of phagosomes with lysosomes and that this mechanism(s) is active even after uptake of killed bacteria.
Colocalization was also studied using IFN-γ-activated J774 cells. The colocalization patterns with the endosomal markers were remarkably similar to those observed with nonactivated cells. The only notable differences were the somewhat higher levels of colocalization of CatD and the three mutants in IFN-γ-activated cells (Table 1).
To study the acidification of Francisella-containing vacuoles, we used Lysotracker Red, a substance which diffuses through the cells and accumulates in lysosomes at low pH. At 1 h, only 16% of the LVS bacteria colocalized with this dye, and at 3 h 2% of the LVS bacteria colocalized with the dye (Table 1). The average percentage of colocalization of the mutants and Lysotracker Red at 1 h was 26%, and at 3 h the level was significantly higher (P < 0.01) (42%) compared to LVS (Table 1). A total of 63% of the vacuoles containing FK-LVS were acidified at 1 h, and 69% were acidified at 3 h, levels that were distinct from (although in most cases not significantly higher than) those for either of the mutants, whereas the corresponding percentages for latex beads were significantly higher (P < 0.01) (76% and 87%, respectively) (Table 1). The data suggest that each of the proteins missing in the mutants is needed for complete prevention of acidification but that there are also Igl/Mgl-independent mechanisms that contribute to the prevention of acidification.
IFN-γ activation of the J774 cells resulted in significantly increased acidification of the vacuoles containing mutant bacteria; on average, 79 and 86% of bacteria colocalized with the Lysotracker Red at 1 and 3 h, respectively (Table 1). Despite IFN-γ treatment, LVS colocalization with the Lysotracker Red did not increase markedly at any time point (Table 1).
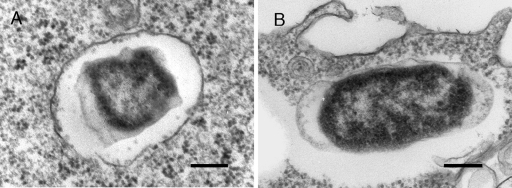
The colocalization patterns of intracellular LVS, ΔiglC, ΔiglD, and ΔmglA bacteria were determined up to 72 h. At this time, 93 to 100% of the ΔiglC, ΔiglD, and ΔmglA bacteria were colocalized with LAMP-1, whereas <10% of the LVS bacteria were colocalized. Moreover, the mutant bacteria were enclosed by individual LAMP-1-positive membranes throughout the infection (Fig. 5), while at the same time the igl mutants were marginally replicating (Fig. 2). To corroborate the data obtained by confocal microscopy, an electron microscopy analysis was performed with J774 cells infected with LVS or the ΔiglC mutant after 20 h. A total of 84% of the ΔiglC bacteria were enclosed by intact or slightly damaged vacuolar membranes, whereas the corresponding value for LVS was only 3% (Fig. 6). Thus, collectively, all the data support the notion that the investigated mutant bacteria proliferated in a LAMP-1-positive vacuole. It should be noted that in some cells, clusters of LVS bacteria located in LAMP-1-positive organelles were observed between 20 and 72 h (data not shown). These organelles were similar to the autophagic structures described by Checroun et al. (7).
FIG. 5.
Colocalization of GFP-expressing F. tularensis strains and LAMP-1. J774 cells were infected for 1 h with F. tularensis strains at an MOI of 5 and then incubated for 36 h. Fixed slides were labeled for the late endosomal and lysosomal marker LAMP-1. In the representative confocal images of cells infected with either LVS-GFP or ΔiglC-GFP, bacteria are indicated by green and LAMP-1 is indicated by red. Confocal images were acquired with the Leica SP2 confocal microscope and were assembled using Adobe Photoshop CS2. Bars = 4 μm.
FIG. 6.
Electron micrographs of J774 cells infected for 1 h with either F. tularensis LVS (B) or the ΔiglC mutant (A) and then incubated for 20 h. Electron micrographs were acquired with the JEOL 1200 EX-II electron microscope (JEOL Ltd., Tokyo, Japan) and were assembled using Adobe Photoshop CS2. Bars = 0.2 μm.
Complementation in trans of the iglC and iglD mutants restores intracellular growth and the ability to escape from the phagosome.
The effects of complementation in trans with the iglC or iglD gene on the ability to proliferate intracellularly and colocalize with LAMP-1 were determined. Intracellular growth of the complemented strains was restored and the intracellular numbers of these bacteria increased by 4.0 log10 during 72 h; this growth even exceeded the growth of LVS bacteria, whose number increased by 3.0 log10. Complementation also restored the ability of the mutant strains to induce host cell cytopathogenicity (data not shown). A colocalization analysis for the LAMP-1 marker at 3 h showed that whereas 79% of the ΔiglC bacteria and 85% of the ΔiglD bacteria colocalized with LAMP-1, the corresponding values for the complemented strains were 22 and 28% (Table 2). The level of colocalization of LVS with this endosomal membrane marker was 18%, indicating that the complemented strains escaped essentially as effectively as the wild type. Similar colocalization patterns were observed for the strains at 24 h (Table 2). Altogether, the complementation in trans successfully restored the intracellular virulence of the iglC and iglD mutants, indicating that the phenotypes of these mutants were dependent on the lack of IglC and IglD, respectively.
TABLE 2.
Quantitation of endocytic marker LAMP-1 acquisition by phagosomes during infection with F. tularensis strains or after uptake of formalin-killed F. tularensis or latex beads after 3 or 24 ha
| Group | LAMP-1 colocalization (%)b
|
|
|---|---|---|
| 3 h | 24 h | |
| LVS | 18 ± 1.2 | 5 ± 3.7 |
| ΔiglC | 79 ± 2.8*** | 95 ± 1.8*** |
| ΔiglD | 85 ± 2.0*** | 94 ± 0.9*** |
| ΔiglC and iglC | 22 ± 3.0 | 5 ± 2.9 |
| ΔiglD and iglD | 28 ± 1.5 | 3 ± 3.0 |
| FK-LVS | 75 ± 5.7*** | 74 ± 3.0*** |
| Latex beads | 99 ± 0.0*** | 99 ± 0.3*** |
J774 cells were infected for 1 h with the indicated live or formalin-killed F. tularensis strains at an MOI of 30 or with latex beads at an MOI of 10 and then incubated for 3 or 24 h.
The percentages indicate the fraction of F. tularensis-containing phagosomes stained for the late endosomal and lysosomal marker LAMP-1. One hundred bacteria on triplicate coverslips were counted, and the results are expressed as means ± standard errors of the means from one of the two experiments performed. Three asterisks indicate that the colocalization level was significantly different from that of LVS within each column (P < 0.001, as determined by one-way ANOVA).
DISCUSSION
F. tularensis possesses a number of mechanisms for surviving in the hostile environment of macrophages. It has been demonstrated that F. tularensis-containing phagosomes are not efficiently acidified and that this bacterium can escape from the phagosomes (7, 9, 14). The escape of F. tularensis LVS is dependent on expression of the IglC protein (22), but the roles of the other Igl proteins in the intracellular evasion mechanisms have not been characterized. All information obtained so far has indicated that the marked attenuation resulting from the absence of Igl proteins is related to the critical roles that these proteins play during the various phases of intracellular infection. In the present study, we observed very similar phenotypes for the IglC and IglD mutants, as well as the MglA mutant, inside macrophages, which confirmed and extended previous reports on the iglC mutant (15, 22). The findings demonstrate that the F. tularensis IglC and IglD proteins, as well as the regulator of the igl operon, MglA, directly or indirectly contribute to prevention of the acidification of the phagosomes and are required for the escape of F. tularensis from phagosomes. Among the investigated parameters, only the relatively low colocalization with CatD appeared to be independent of Igl expression.
We observed that the mglA mutant was more defective for intracellular replication than any of the igl mutants. The reason for the pronounced attenuation of the mglA mutant is unclear, but it is noteworthy that the regulation of some 100 genes was affected by the absence of MglA in F. novicida (4). Despite the more impaired survival of the mglA mutant intracellularly, all mutants showed similar levels of colocalization with LAMP-1, CatD, and Lysotracker Red. Therefore, the more impaired survival of the mglA mutant may depend on dysregulation of proteins other than IglC or IglD that in LVS contribute to its ability to survive and replicate intracellularly but do not modulate the biogenesis of the phagosome.
The igl and mglA mutants, in contrast to LVS, colocalized with LAMP-1, indicating that there was phagosomal localization of the mutants. Since no previous data have demonstrated that F. tularensis is capable of replicating in the phagosome, the finding that there was a 20-fold increase in the numbers of iglC and iglD mutant bacteria was intriguing. The finding of Checroun et al. that F. tularensis can reenter the endocytic pathway from the cytoplasm could indicate one mechanism that explains our finding (7). However, despite careful examination, there was no indication that the mutant bacteria were free in the cytoplasm up to 72 h. This finding resembles findings obtained in studies showing that Salmonella replicates in a vacuole that after transient interaction with early endosomes expresses LAMP-1, but avoids fusion with lysosomes (5). Brucella-containing vacuoles initially interacted with early endosomes and acquired LAMP-1, but after this the bacteria multiplied enclosed by LAMP-1-negative endoplasmic reticulum-derived membranes (6). Interestingly, the Brucella-containing vacuoles contained only individual bacteria, as observed for the ΔiglC and ΔiglD mutants in the present study. Thus, in J774 cells the ΔiglC and ΔiglD strains appear to behave like an intracellular bacterium that successfully subverts the maturation of the phagolysosome and thereby survives without escaping into the cytoplasm. Our observation that there was distinct colocalization of the ΔiglC and ΔiglD mutants compared with latex beads supports this hypothesis. Nevertheless, we cannot completely exclude the possibility that the small number of mutant bacteria that were not enclosed by membranes were responsible for the replication up to 72 h.
It is noteworthy that in the absence of IFN-γ activation, less than 50% of the Δigl and ΔmglA bacteria colocalized with Lysotracker Red, while a majority of both FK-LVS and latex bead-containing vacuoles colocalized. Therefore, the relatively inefficient killing may depend on the localization of most bacteria in nonacidified organelles. This suggests that F. tularensis possesses an Igl/Mgl-independent mechanism to prevent effective acidification of the phagosome. Our observation of a relatively high percentage of nonacidified wild-type F. tularensis-containing phagosomes is in agreement with findings described in a previous report (9). Since CatD is a marker of lysosome fusion, a process strictly dependent upon phagosomal acidification, one unexpected finding was the partial accumulation of CatD in the F. tularensis-containing phagosomes. One possible explanation for this finding is that the phagosomes initially undergo rapid, transient acidification but that F. tularensis-specific mechanisms subsequently counteract this acidification.
We observed that IFN-γ activation contributed significantly to control intracellular replication of LVS, but at the same time, the lack of colocalization of LVS with endosomal markers indicated that only a minority of the bacteria were in phagolysosomes after IFN-γ treatment. This is in agreement with our previous demonstration of phagosomal escape of LVS in IFN-γ-activated murine macrophages (22). It is possible that after IFN-γ activation, LVS is exposed to cidal mechanisms while it is localized in the phagosome but this does not prevent the escape.
The colocalization pattern of the LVS mutants with endocytic markers was relatively similar to that of FK-LVS but distinct from that of the latex bead-containing phagosomes, which displayed all markers typical of mature phagolysosomes (12). Remarkably, killed bacteria also possess mechanisms to evade the fusion of phagosomes with lysosomes, as shown by their relatively low CatD colocalization. Thus, there appears to be a propensity for F. tularensis bacteria to modulate their intracellular fate even in the absence of active protein synthesis or secretion. It is possible that the primary association with and uptake of bacteria by the host monocytes are decisive for the intracellular localization regardless of bacterial viability.
The intracellular localization of type strain U112 of F. novicida and mglA, iglC, and iglD mutants of this strain has been characterized. It was observed that the wild-type bacterium and the iglD mutant colocalized with LAMP-1 but not with CatD or lysosomes before escape into the cytoplasm, whereas the mglA and iglC mutants localized in phagosomes displaying LAMP-1, CatD, and a lysosomal tracer (28, 29). Thus, LVS appears to prevent maturation of phagosomes at an earlier stage than the U112 strain since it is characterized by only a transient interaction with late endosomes (LAMP-1) but not with lysosomes (CatD or Lysotracker Red). Also, a majority of the LVS ΔmglA, ΔiglC, and ΔiglD bacteria containing vacuoles showed no accumulation of CatD and fusion with acidic organelles, in marked contrast to the corresponding F. novicida mutants. The most striking difference is the recent report describing phagosomal escape of the iglD mutant of F. novicida (28). Our findings do not indicate that there are any fundamental differences between the phenotypes of the iglC and iglD mutants of LVS, and therefore the distinct phenotype of the iglD mutant of F. novicida may be limited to this species. Also, differences in types of host cells and methodologies could explain the discrepancy.
Altogether, our results contribute to the emerging picture of a unique survival strategy of F. tularensis and specifically to our understanding of the role of the Igl proteins and MglA. The strategy is dependent on prevention of the normal maturation and acidification of phagosomes, which allows bacteria to evade the generation of an otherwise hostile intracellular environment in monocytic cells. Each of the evasion mechanisms appears to directly or indirectly require the expression of several Igl proteins and MglA. Moreover, we also demonstrated that under certain conditions, even igl mutants can proliferate intracellularly, but they do so by means distinct from those of the LVS strain. Our findings identify several avenues for future exploration in order to fully understand the intracellular survival mechanisms of this enigmatic pathogen.
Acknowledgments
We thank Lena Carlsson for assistance with electron microscopy experiments and Konstantin Kadzhaev for construction of the pKK289Km-gfp plasmid.
Grant support was provided by the Swedish Medical Research Council, Samverkansnämnden, Norra Sjukvårdsregionen, Umeå, the J. C. Kempes Minnesfond Foundation, and the Medical Faculty, Umeå University, Umeå, Sweden.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 12 May 2008.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandström, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ Microbiol. 69600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, L. S., P. J. Morrissey, and F. E. Nano. 1992. Growth inhibition of Francisella tularensis live vaccine strain by IFN-gamma-activated macrophages is mediated by reactive nitrogen intermediates derived from l-arginine metabolism. J. Immunol. 1481829-1834. [PubMed] [Google Scholar]
- 3.Balagopal, A., A. S. MacFarlane, N. Mohapatra, S. Soni, J. S. Gunn, and L. S. Schlesinger. 2006. Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect. Immun. 745114-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brotcke, A., D. S. Weiss, C. C. Kim, P. Chain, S. Malfatti, E. Garcia, and D. M. Monack. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 746642-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumell, J. H., and S. Grinstein. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 778-84. [DOI] [PubMed] [Google Scholar]
- 6.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizarro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Checroun, C., T. D. Wehrly, E. R. Fischer, S. F. Hayes, and J. Celli. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA 10314578-14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2005. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 735892-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 723204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlan, J. W. 2004. Vaccines against Francisella tularensis--past, present and future. Expert Rev. Vaccines 3307-314. [DOI] [PubMed] [Google Scholar]
- 11.de Bruin, O. M., J. S. Ludu, and F. E. Nano. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desjardins, M., and G. Griffiths. 2003. Phagocytosis: latex leads the way. Curr. Opin. Cell Biol. 15498-503. [DOI] [PubMed] [Google Scholar]
- 13.Duclos, S., and M. Desjardins. 2000. Subversion of a young phagosome: the survival strategies of intracellular pathogens. Cell. Microbiol. 2365-377. [DOI] [PubMed] [Google Scholar]
- 14.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjöstedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 715940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovliov, I., A. Sjöstedt, A. Mokrievich, and V. Pavlov. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222273-280. [DOI] [PubMed] [Google Scholar]
- 16.Gray, C. G., S. C. Cowley, K. K. M. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Immunol. 21553-56. [DOI] [PubMed] [Google Scholar]
- 17.Haas, A. 2007. The phagosome: compartment with a license to kill. Traffic 8311-330. [DOI] [PubMed] [Google Scholar]
- 18.Keim, P., A. Johansson, and D. M. Wagner. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 110530-66. [DOI] [PubMed] [Google Scholar]
- 19.Lai, X. H., I. Golovliov, and A. Sjöstedt. 2004. Expression of IglC is necessary for intracellular growth and induction of apoptosis in murine macrophages by Francisella tularensis. Microb. Pathog. 37225-230. [DOI] [PubMed] [Google Scholar]
- 20.Larsson, P., P. C. F. Oyston, P. Chain, M. C. Chu, M. Duffield, H.-H. Fuxelius, E. Garcia, G. Hälltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. G. Prior, S. Malfatti, A. Sjöstedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. E. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37153-159. [DOI] [PubMed] [Google Scholar]
- 21.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA 1014246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren, H., I. Golovliov, V. Baranov, R. K. Ernst, M. Telepnev, and A. Sjöstedt. 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53953-958. [DOI] [PubMed] [Google Scholar]
- 23.Oyston, P. C., A. Sjöstedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2967-978. [DOI] [PubMed] [Google Scholar]
- 24.Petrosino, J. F., Q. Xiang, S. E. Karpathy, H. Jiang, S. Yerrapragada, Y. Liu, J. Gioia, L. Hemphill, A. Gonzalez, T. M. Raghavan, A. Uzman, G. E. Fox, S. Highlander, M. Reichard, R. J. Morton, K. D. Clinkenbeard, and G. M. Weinstock. 2006. Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J. Bacteriol. 1886977-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierini, L. M. 2006. Uptake of serum-opsonized Francisella tularensis by macrophages can be mediated by class A scavenger receptors. Cell. Microbiol. 81361-1370. [DOI] [PubMed] [Google Scholar]
- 26.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 1031528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohmer, L., C. Fong, S. Abmayr, M. Wasnick, T. J. Larson Freeman, M. Radey, T. Guina, K. Svensson, H. S. Hayden, M. Jacobs, L. A. Gallagher, C. Manoil, R. K. Ernst, B. Drees, D. Buckley, E. Haugen, D. Bovee, Y. Zhou, J. Chang, R. Levy, R. Lim, W. Gillett, D. Guenthener, A. Kang, S. A. Shaffer, G. Taylor, J. Chen, B. Gallis, D. A. D'Argenio, M. Forsman, M. V. Olson, D. R. Goodlett, R. Kaul, S. I. Miller, and M. J. Brittnacher. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santic, M., M. Molmeret, J. R. Barker, K. E. Klose, A. Dekanic, M. Doric, and Y. Abu Kwaik. 2007. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell. Microbiol. 92391-2403. [DOI] [PubMed] [Google Scholar]
- 29.Santic, M., M. Molmeret, K. E. Klose, S. Jones, and Y. A. Kwaik. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7969-979. [DOI] [PubMed] [Google Scholar]
- 30.Schulert, G. S., and L. A. Allen. 2006. Differential infection of mononuclear phagocytes by Francisella tularensis: role of the macrophage mannose receptor. J. Leukoc. Biol. 80563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Twine, S., M. Byström, W. Chen, M. Forsman, I. Golovliov, A. Johansson, J. Kelly, H. Lindgren, K. Svensson, C. Zingmark, W. Conlan, and A. Sjöstedt. 2005. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 738345-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieira, O. V., R. J. Botelho, and S. Grinstein. 2002. Phagosome maturation: aging gracefully. Biochem. J. 366689-704. [DOI] [PMC free article] [PubMed] [Google Scholar]