Abstract
Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans is a pathogen that causes localized aggressive periodontitis and extraoral infections including infective endocarditis. Recently, we reported that A. actinomycetemcomitans is beta-hemolytic on certain growth media due to the production of leukotoxin (LtxA). Based on this observation and our ability to generate random transposon insertions in A. actinomycetemcomitans, we developed and carried out a rapid screen for LtxA mutants. Using PCR, we mapped several of the mutations to genes that are known or predicted to be required for LtxA production, including ltxA, ltxB, ltxD, and tdeA. In addition, we identified an insertion in a gene previously not recognized to be involved in LtxA biosynthesis, ptsH. ptsH encodes the protein HPr, a phosphocarrier protein that is part of the sugar phosphotransferase system. HPr results in the phosphorylation of other proteins and ultimately in the activation of adenylate cyclase and cyclic AMP (cAMP) production. The ptsH mutant showed only partial hemolysis on blood agar and did not produce LtxA. The phenotype was complemented by supplying wild-type ptsH in trans, and real-time PCR analysis showed that the ptsH mutant produced approximately 10-fold less ltxA mRNA than the wild-type strain. The levels of cAMP in the ptsH mutant were significantly lower than in the wild-type strain, and LtxA production could be restored by adding exogenous cAMP to the culture.
Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans is a gram-negative capnophilic bacterium that colonizes the oral cavities of humans and Old World primates (13, 45, 53). A. actinomycetemcomitans has been associated with various diseases including localized aggressive periodontitis (45, 52) and subacute infective endocarditis (4, 6, 42). Localized aggressive periodontitis is an aggressive disease of the mouth that presents in adolescents and results in the loss of periodontal bone and ligament. When untreated, loss of teeth occurs. The bacterium expresses several putative virulence factors including a leukotoxin (LtxA) (30, 33). LtxA is a 113-kDa protein that has been reported to kill specifically white blood cells of humans and Old World primates (46-48). Production of LtxA by A. actinomycetemcomitans is believed to be one mechanism used by the bacterium to evade host immune responses (1). Clinical evidence suggests that LtxA is an important virulence factor for A. actinomycetemcomitans (21-23). In addition, we have recently shown that LtxA can act as a hemolysin that may be used during systemic infection for acquiring iron from the host (2). Indeed, iron, but not other metals, regulates LtxA secretion in A. actinomycetemcomitans (3).
LtxA belongs to the RTX (repeats in the toxin) family of toxins, which includes Escherichia coli α-hemolysin, Bordetella pertussis adenylate cyclase, Mannheimia haemolytica leukotoxin, Vibrio cholerae RTX toxin, and Actinobacillus pleuropneumoniae Apx toxins (16, 37, 51). The current model for the production, activation, and secretion of LtxA in A. actinomycetemcomitans is based primarily on work done with other RTX toxins, especially E. coli α-hemolysin (7, 17). Biosynthesis of LtxA is apparently dependent on an operon that consists of four genes, ltxCABD. ltxC encodes a protein presumed to be responsible for the acylation of the toxin, ltxA encodes the toxin, and ltxB and ltxD code for predicted components of the membrane transport system. In addition, we recently discovered a TolC-like protein, TdeA, which is required for both LtxA secretion and drug efflux (5). There is also evidence that LtxA biosynthesis is regulated by other genes outside the LtxA operon. Indeed, Fong et al. (15) showed that LtxA expression is at least partly regulated by LuxS (autoinducer II), while Inoue et al. (25) found that the levels of LtxA are affected by cyclic AMP (cAMP) concentration within the cell. However, the genes and mechanisms involved in regulation are largely unknown.
An effective way to identify novel genes involved in the regulation of ltxA in A. actinomycetemcomitans is to isolate nonleukotoxic mutants. To date, there is no practical way to screen for mutants of A. actinomycetemcomitans that are defective in LtxA production. We recently found that A. actinomycetemcomitans is beta-hemolytic on certain growth media and that this phenotype is due to the production of LtxA (2). Based on this novel finding, we have developed a rapid assay to screen for LtxA mutants of A. actinomycetemcomitans. Herein, we describe the genetic screen and present data that support its use as an effective method for identifying novel LtxA genes in A. actinomycetemcomitans and potentially related bacteria. In addition, we have used this screen to characterize a gene that is involved in the cAMP-dependent regulation of ltxA.
MATERIALS AND METHODS
Bacterial strains and media.
A. actinomycetemcomitans strain JP2N (a nalidixic acid-resistant variant of JP2) (50) was used for all experiments except where noted. AA1704 is an ltxA mutant of JP2N and has been previously described (2). Strain AA1698 is a derivative of JP2N that harbors the transposon mutagenesis plasmid, pVJT128 (27, 49) (Table 1). Nalidixic acid was used for counter-selection during conjugation experiments with E. coli to construct the A. actinomycetemcomitans strains used here.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| AA1698 | JP2N containing pVJT128 | 2 |
| AA1704 | JP2N ltxA::IS903φkan | 2 |
| CU1000 | Rough clinical isolate; serotype f | 27 |
| CU1000N | Spontaneous Nalr variant of CU1000 | 11 |
| JK1025 | CU1000N ptsI::IS903φkan | This study |
| JP2 | Highly leukotoxic; serotype b | 50 |
| JP2N | Spontaneous Nalr variant of JP2 | 2 |
| MP046 | JP2N ptsH::IS903φkan | This study |
| Plasmids | ||
| pJAK16 | IncQ broad-host-range vector; tacp Cmr | J. A. Kornacki |
| pJK617 | pJAK16 containing ptsI | This study |
| pMP021 | pJAK16 containing ptsH | This study |
| pVJT128 | Plasmid containing inducible IS903φkan | 27, 49 |
A. actinomycetemcomitans was grown on AAGM plates (40 g of trypticase soy agar and 6 g of yeast extract per liter, 0.75% glucose, and 0.4% NaHCO3; glucose and NaHCO3 were added after the medium was autoclaved) (12). For liquid cultures, cells were grown in AAGM broth prepared by replacing trypticase soy agar with trypticase soy broth (30 g). Where indicated, medium for A. actinomycetemcomitans was supplemented with 2 μg/ml of chloramphenicol and 20 μg/ml of kanamycin. Plates were incubated at 37°C in a 10% CO2 environment for 72 h, and liquid cultures were incubated for 24 to 48 h.
For examination of the hemolytic phenotype, Columbia agar (Accumedia, Lansing, MI) was supplemented with 5% sheep blood (PML Microbiologicals, Inc., Wilsonville, OR) after the medium cooled to 48°C. Plates were incubated as described above.
Mutagenesis of A. actinomycetemcomitans.
Strain AA1698 was grown on chloramphenicol plates, and individual colonies were selected and grown overnight in AAGM broth containing chloramphenicol and 1.0 mM isopropyl-beta-d-thiogalactopyranoside (IPTG). IPTG was used to induce the transposon system as previously described (27, 49). After incubation, dilutions were made from the suspensions and plated onto kanamycin plates. Individual colonies, representing potentially independent mutants, were picked and patched onto blood plates. Nonhemolytic mutants were characterized by the lack of a zone of clearing around the patch.
Strain JK1025 was constructed by mutagenizing strain CU1000N with pVJT128 and selecting mutants that produced rough-textured colonies on agar (29).
PCR.
DNA was PCR amplified using Taq DNA polymerase (Qiagen, Valencia, CA). Colony PCR was performed by adding a small amount of a colony into the PCR tube with a sterile pipette tip. For all reactions, the denaturation step was carried out at 94°C for 1 min, and the primer extension step was carried out at 72°C for 1 min. For amplification of ltxA, an annealing temperature of 60°C was used for 3 min. For ltxC, ltxB, and ltxD, annealing temperatures of 55°C, 60°C, and 55°C, respectively, were used for 1 min. All PCR products were confirmed by DNA agarose gel electrophoresis. The DNA sequences of primers used are noted in Table 2.
TABLE 2.
Primers used in this study
| Gene | Primer | Sequence (5′→3′) |
|---|---|---|
| PCR target genes | ||
| ltxA | ltxAup | GCCCGGGATGGCAACTACTACACTGCTAAATAC |
| ltxAdown | GCCCGGGCAGTAGTTGCTAACGAATTTGC | |
| ltxB | ltxBup | CGCAAATTCGTTAGCAACTACTGC |
| ltxBdown | CAAGTTTTCATTATCGTTCGTTCC | |
| ltxC | ltxCup | AAAAACTATTGGAATACCAAGTAC |
| ltxCdown | GTAGTAGTTGCCATAATCTATTCTC | |
| ltxD | ltxDup | ATTACAAGTAAATTAAAGGAACGAAC |
| ltxDdown | GAAAACCGGAATGTTATATTCTATC | |
| tdeA | tdeAup | CGCCATGGGGCGTGCCGCATGCG |
| tdeAdown | CCGATTACAGCGTTGGCGCAAG | |
| ptsH | ptsHup | TCCTGAAATCATAGTAACCTTC |
| ptsHdown | GGTTTGTGGCATCTGGAAGATG | |
| ptsI | ptsIup | GCGGATCCCAAGGAGATTTAATATGATTTCAGGAATTCCGGC |
| ptsIdown | CGCAAGCTTCTAATTTAATGCTTTTTCATAC | |
| RT-PCR target genesa | ||
| ltxA | ltxA-s | GTGCTAGGTAAACATCGGTAAAG |
| ltxA-as | GACCACAGAGGCAATTAACC | |
| glyA | glyA-s | CCCAATTCACCAACAAATATGC |
| glyA-as | ATTCTTTCGCACGCTCAATAG |
RT-PCR, real-time PCR.
Examination of LtxA.
To confirm that mutants were defective in LtxA production, we isolated total protein from supernatants of cultures as previously described (26). Briefly, after cells were grown overnight in AAGM broth, cultures were centrifuged, and 500 μl of supernatant was precipitated with 1 ml of ice-cold ethanol. The pelleted protein was resuspended in sodium dodecyl sulfate (SDS) loading dye and run on an SDS-polyacrylamide gel electrophoresis (PAGE) gel. The gel was stained with Coomassie blue and visualized. For examination of cell-associated protein, the bacterial pellet was resuspended directly in SDS loading dye and run on an SDS-PAGE gel. Proteins were transferred to a nitrocellulose membrane and processed for Western blot analysis probing with anti-LtxA antibody (9).
Inverse PCR.
Inverse PCR on genomic DNA was performed as described previously (27). Briefly, 10 to 20 μg of genomic DNA was digested with EcoRI (which does not cleave IS903φkan). Precipitation of the fragments was done by ethanol precipitation, followed by dilution and ligation to circularize the fragments. Resulting fragments were amplified for 30 cycles using primers directed outward from the ends or the transposon. Amplified fragments were cloned into pCR2.1 (Invitrogen, Carlsbad, CA) for sequencing. Nucleotide sequence was determined by the University of Medicine and Dentistry of New Jersey Molecular Research Facility.
Genetic complementation.
ptsH was amplified by PCR, cloned into pCR2.1, sequenced, and subcloned into pJAK16 (49). The derivative plasmid was mobilized by conjugation from an E. coli donor to strain MP046 as previously described (27, 49). IPTG at a final concentration of 1.0 mM was added to plates and liquid cultures for complementation assays.
Real-time PCR.
Total bacterial RNA was isolated using TRIzol according to the manufacturer's protocol (Invitrogen Corporation, Carlsbad, CA) and further purified by DNase I treatment, followed by passage through an RNeasy spin column (Qiagen, Valencia, CA). Reverse transcription-PCR was subsequently carried out using TaqMan Reverse Transcription Reagents (Applied Biosystems, Branchburg, NJ). Quantitative real-time PCR was performed using the Sybr Green PCR Master Mix (Applied Biosystems, Warrington, United Kingdom). The DNA sequences of primers used are noted in Table 2. The threshold cycle (CT) of the internal control gene glyA was subtracted from the CT of the target gene to obtain the ΔCT. The normalized relative changes of the mRNA expression level of each target gene was expressed as the 2−ΔΔCT, where ΔΔCT is equal to the ΔCT of the ptsH mutant minus ΔCT of JP2. These experiments were performed in triplicate.
Determination of cAMP concentration.
Intracellular cAMP levels were determined using a cAMP enzyme immunoassay system (GE Healthcare, Piscataway, NJ). Cells were collected at 24 h by centrifugation and resuspended in the lysis buffer provided by the manufacturer. Samples were boiled at 100°C for 15 min and centrifuged at 2,000 × g for 5 min at 4°C. The cAMP concentrations in supernatants were determined according to the manufacturer's instructions using standards provided in the kit. The average intracellular cAMP concentration (determined in duplicate) was expressed in femtomoles per milligram of total protein.
RESULTS
A screen for LtxA mutants.
Knowledge of LtxA genetics in A. actinomycetemcomitans has been hampered by the lack of a rapid and efficient screen for mutants. We have previously used an assay based on killing of HL-60 cells by A. actinomycetemcomitans culture supernatants (M. P. Isaza and S. C. Kachlany, unpublished results). While effective, this bioassay is time-consuming and not easily amenable to high-throughput screening. Therefore, we sought to take advantage of our recent observation that LtxA is required for β-hemolysis on Columbia blood agar (2).
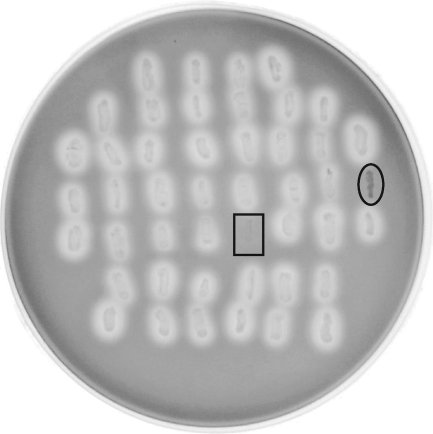
We began by generating random transposon mutants in A. actinomycetemcomitans as previously described (27, 28, 43). The transposon IS903φkan creates nonpolar insertions with no apparent hot spots (27, 49). After transposon mutants were selected on kanamycin, the colonies were patched onto blood agar in a grid format. Figure 1 shows a plate from one typical experiment. The boxed patch represents AA1704, a known ltxA mutant strain (2), and the circled patch is a transposon mutant isolated with our screen.
FIG. 1.
Blood agar screen for non-beta-hemolytic mutants of A. actinomycetemcomitans. Random transposon mutants were patched onto blood agar and grown for 3 days. The squared patch represents a known ltxA mutant, and the circled patch is a non-beta-hemolytic mutant isolated with this screen.
Examination of LtxA mutants.
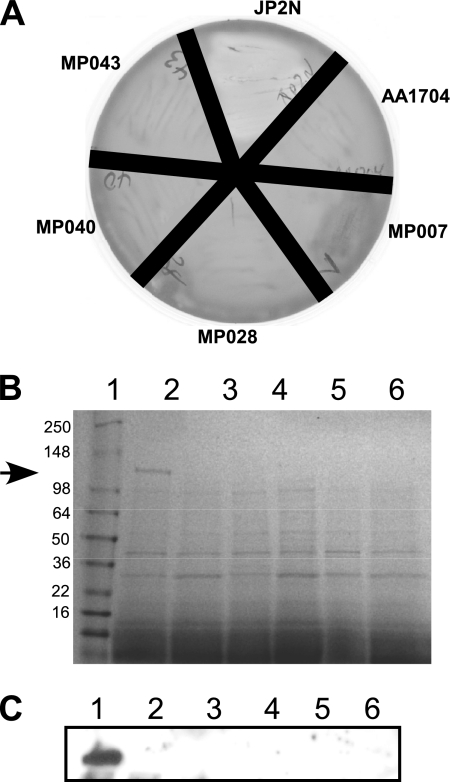
After screening approximately 2,500 random transposon mutants on blood agar, we identified 15 that were partially or non-beta-hemolytic. We wished to further confirm the non-beta-hemolytic phenotype of several of these mutants (Fig. 2A). As expected, when streaked onto blood agar, wild-type JP2N was hemolytic. In contrast, the control ltxA mutant strain, AA1704, and others isolated here (MP007, MP028, MP040, and MP043) were all non-beta-hemolytic.
FIG. 2.
Examination of non-beta-hemolytic mutants. (A) Several mutants were restreaked on blood agar. JP2N is the wild-type strain; AA1704 is an isogenic ltxA mutant of JP2N; and MP007, MP028, MP040, and MP043 are mutants isolated here. (B) Supernatants were isolated from bacterial cultures as described in Materials and Methods and subjected to SDS-PAGE analysis. The gel was stained with Coomassie blue. The arrow indicates the position of LtxA. Lane 1, JP2N; lane 2, AA1704; lane 3, MP007; lane 4, MP028; lane 5, MP040; lane 6, MP043. (C) Western blot analysis of cell-associated protein from the strains described above. Lanes are identical to those in panel B. The blot was probed with anti-LtxA antibody to detect cell-associated LtxA.
To determine if LtxA was being produced in these strains, we examined secreted and cell-associated protein from the wild-type and mutant strains (Fig. 2B and C). SDS-PAGE analysis of supernatant (secreted) protein from JP2N revealed a band at the expected size representing LtxA (Fig. 2B, arrow). In contrast, the supernatants from all other mutant strains lacked the LtxA band, indicating that any LtxA produced by these strains was below the limit of detection. We then examined cell-associated protein using Western blot analysis with anti-LtxA antibody (Fig. 2C). Similar to our results with supernatants, only JP2N produced LtxA. These results indicate that mutants unable to perform β-hemolysis on blood agar are also defective for LtxA production or secretion. This fact further confirms the correlation between β-hemolysis and LtxA production in A. actinomycetemcomitans.
PCR mapping of transposon insertions.
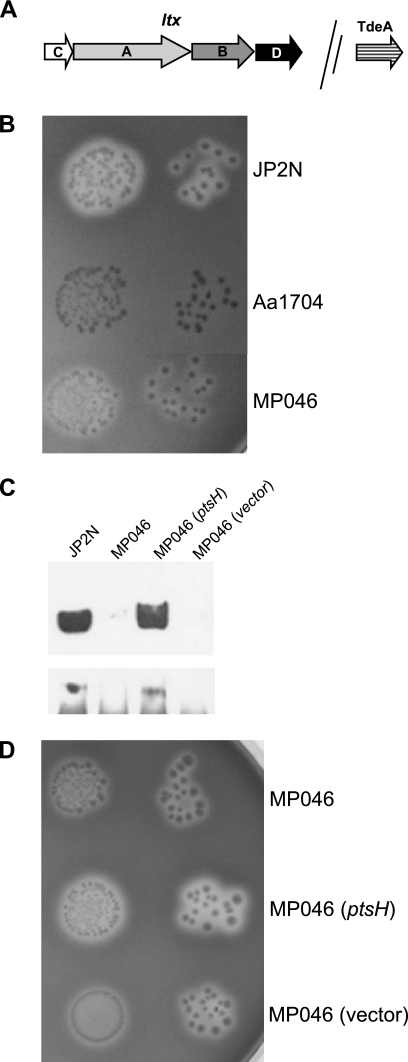
The LtxA operon contains four genes (Fig. 3A), all putatively required for LtxA production and activity, and an unlinked tolC-like gene, tdeA, which encodes a protein required for the export of LtxA (5). To identify the insertion sites of IS903φkan in several of our mutants, we designed PCR primers to amplify each of four ltx genes and tdeA and performed PCR. Genes with a transposon insertion show a PCR product that is approximately 1 kb larger than the wild-type size (data not shown). Table 3 shows that we isolated ltxA, ltxB, ltxD, and tdeA mutants but not an ltxC mutant. However, we have recently isolated an ltxC mutant using a direct screening method, indicating that mutations in ltxC are not lethal (unpublished data). Furthermore, one of the mutants (MP046) apparently contained the transposon insertion outside of the known ltx genes.
FIG. 3.
Characterization of the ptsH mutant. (A) Map of the genes that constitute the LtxA operon in A. actinomycetemcomitans. (B) Partial hemolysis by MP046 on blood agar. JP2N is the wild-type strain, and AA1704 is an ltxA mutant. (C) Western blot analysis of MP046. The top panel represents supernatant (secreted) protein, and the bottom panel shows cell-associated protein. The last two lanes represent complementation experiments with MP046. The blot was probed with anti-LtxA antibody. (D) Complementation of hemolysis on blood agar.
TABLE 3.
PCR mapping of LtxA mutants
| Strain | Hemolysis | Relative product size after PCR amplification of the indicated genea
|
||||
|---|---|---|---|---|---|---|
| ltxC | ltxA | ltxB | ltxD | tdeA | ||
| MP07 | +/− | WT | WT | WT | WT | Mut |
| MP028 | − | WT | WT | Mut | WT | WT |
| MP035 | − | WT | Mut | WT | WT | WT |
| MP038 | − | WT | Mut | WT | WT | WT |
| MP040 | − | WT | WT | Mut | Mut | WT |
| MP041 | +/− | WT | WT | WT | WT | Mut |
| MP042 | − | WT | Mut | WT | WT | WT |
| MP043 | − | WT | Mut | WT | WT | WT |
| MP046 | +/− | WT | WT | WT | WT | WT |
| MP047 | − | WT | WT | WT | WT | Mut |
WT, product size equal to that of the wild type; Mut, increase in the product size of approximately 1 kbp.
Identification of a ptsH mutant.
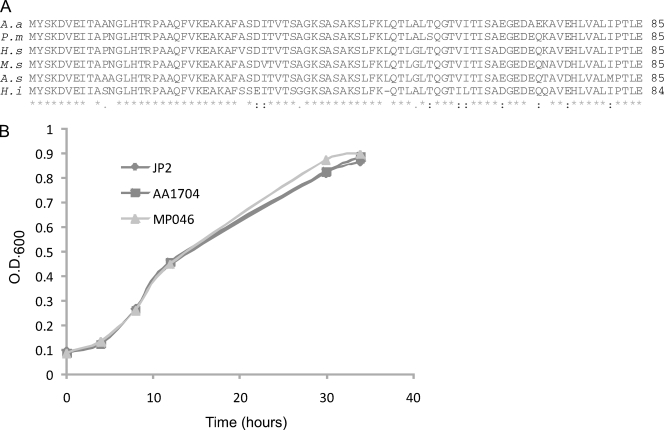
One of the mutants (MP046) that contained a transposon insertion outside of the known ltx genes displayed a partial hemolytic phenotype on blood agar compared to an ltxA mutant, AA1704 (Fig. 3B). LtxA was not detected in the supernatants or cell pellets of MP046 (Fig. 3C). Because of the unique phenotype of this mutant, we wished to further study the genetic basis of this observation. To map the sites where IS903φkan inserted outside of the ltx operon, we performed inverse PCR on genomic DNA as described previously (27). The products were cloned into plasmid pCR-TOPO2.1 and sequenced using universal primers. The sequence indicated an insertion of the IS903φkan transposon within the first codon of the open reading frame. This putative open reading frame, designated ptsH, would encode a protein with significant sequence similarity to histidine-containing protein, HPr (Fig. 4A). To further confirm the location of this mutation, we designed primers that amplify the region flanking the IS903φkan insertion. Strain MP046 showed a PCR product that is approximately 1 kb larger than the wild-type size (data not shown), confirming that the site of insertion was within the ptsH gene.
FIG. 4.
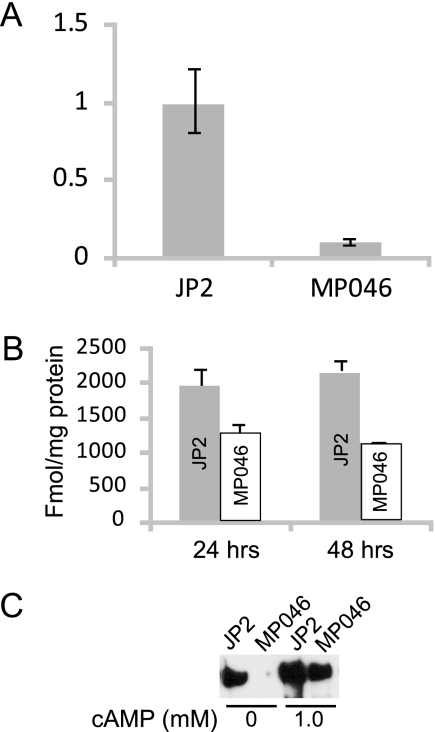
(A) Amino acid sequence alignment HPr from related bacteria: A. actinomycetemcomitans (A.a), Pasteurella multocida (P.m; accession no. AE006128.1), Haemophilus somnus (H.s; accession no. NZ AAB002000004.1), Mannheimia succiniciproducens (M.s; accession no. AE016827.1), Actinobacillus succinogenes (A.s; accession no. NZ AAKC01000013.1), and Haemophilus influenzae (H.i; accession no. NZ AADP01000001.1). (B) Growth curve comparing MP046 to the wild-type strain (JP2) and the ltxA mutant, AA1704. The line for JP2 partially overlaps the strain AA1704 curve.
HPr is part of phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS), which is responsible for the transport and phosphorylation of sugars into the cell (8). Because the transposon inserted into one of the genes that controls sugar transport, we wished to determine if this mutation was affecting the overall growth of the strain. We performed growth experiments with glucose and found that strain MP046 grew at the same rate as the parent strain in the complex medium used here (Fig. 4B).
To confirm that our insertion was not polar on downstream genes, we cloned and expressed wild-type ptsH in trans in strain MP046. The complemented mutant strain regained the hemolytic phenotype (Fig. 3D), and LtxA was detected in the cell supernatant and pellet (Fig. 3C). Thus, the insertion in ptsH is responsible for the observed defect in LtxA production.
Characterization of the ptsH mutant.
We performed real-time PCR to determine if the phenotype observed in MP046 was the result of transcriptional regulation of ltxA. Our results indicate that, relative to the expression of the housekeeping gene glyA (39), the level of ltxA transcription in strain MP046 was approximately 1/10 of the wild-type level (Fig. 5A). Thus, ptsH apparently plays a role in the transcription of ltxA. While a decrease in mRNA stability could also explain these results, MP046 expressed glyA at the same level as the wild-type strain, indicating that a general effect on mRNA stability did not occur.
FIG. 5.
(A) Real-time PCR analysis of ltxA transcription in JP2 and MP046. The y axis represents relative levels of mRNA. The glyA housekeeping gene was used to standardize values as described in Materials and Methods. (B) Levels of cAMP in fmol/mg protein from JP2 and MP046 at 24 and 48 h. (C) Western blot analysis of supernatant protein of MP046 grown in the presence of 1.0 mM cAMP for 24 h. The blot was probed with anti-LtxA antibody.
Inoue et al. (25) have shown that cAMP was involved in the regulation of ltxA since the levels of LtxA and cAMP decreased as fermentable sugars were added to the culture. They proposed a catabolite repression mechanism to explain their results. To determine if cAMP levels were altered in the ptsH mutant, we examined intracellular levels of cAMP in the MP046 and the wild-type strains. Our results indicate that the cAMP levels in the ptsH mutant are decreased by approximately half of the wild-type levels (Fig. 5B). Furthermore, when exogenous cAMP (1.0 mM) was added to the culture, LtxA was detected in the supernatant of the ptsH mutant (Fig. 5C). Thus, the partial hemolysis and absence of LtxA may be attributed to the diminished levels of intracellular cAMP due to a defect in the PTS machinery.
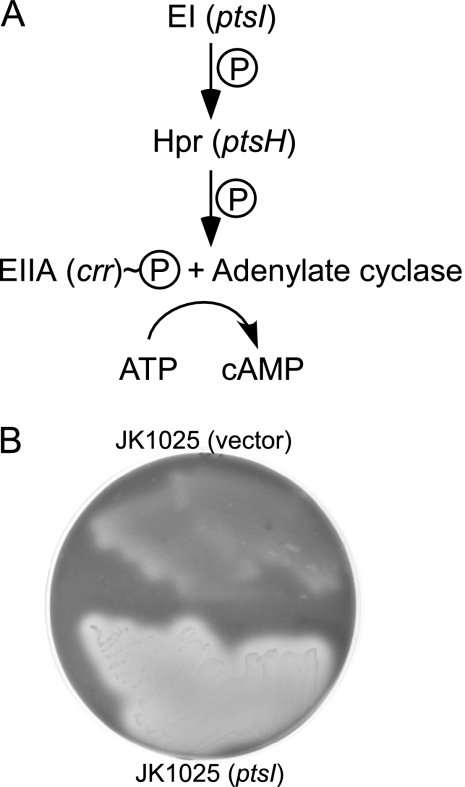
Another PTS gene, ptsI, is required for LtxA production.
We wished to determine if the observed effects on LtxA production are specific for HPr or whether other PTS genes are involved. Enzyme I (EI; encoded by ptsI) is the protein that acts just upstream of HPr in the PTS pathway and is responsible for transferring a phosphate group to HPr (Fig. 6A) (8). We tested a ptsI mutant (JK1025) for hemolysis and found that it was also a nonhemolytic mutant whose phenotype could be complemented by supplying wild-type ptsI in trans but not with the empty vector (Fig. 6B). Thus, EI is also involved in LtxA biosynthesis.
FIG. 6.
(A) The pathway for activation of adenylate cyclase by EI and HPr. The model is based on the E. coli PTS system. (B) Hemolysis of a ptsI mutant on blood agar. JK1025 is a ptsI transposon mutant derived from strain CU1000 (11, 27).
DISCUSSION
We report here an efficient genetic screen for the identification of genes that play a role in LtxA production in A. actinomycetemcomitans. This is the first report of an approach to screen random mutants of A. actinomycetemcomitans for loss of the leukotoxic phenotype. Using this screen, we have confirmed the importance of several genes known or expected to be involved in LtxA biosynthesis, and we identified and characterized a gene that may play a novel role in ltxA expression. Importantly, we have also demonstrated the strong correlation between LtxA production and hemolysis. All of our mutants affected LtxA biosynthesis, confirming that this toxin is apparently the only hemolysin that A. actinomycetemcomitans produces. Thus, hemolysis proves to be a useful assay in which to study LtxA biology.
The ltx operon consists of four genes, ltxCABD. In addition, tdeA has been shown to be required for LtxA secretion (5) and serves as the TolC-like outer membrane channel (31). We found that the phenotypes of the ltxB and ltxD mutants (MP028 and MP040, respectively) on blood agar were essentially indistinguishable from ltxA mutants. As suggested by sequence similarities to other RTX secretion genes, it is likely that LtxB and LtxD are involved in LtxA secretion (18, 20, 34). Thus, one prediction for ltxB and ltxD mutants is that they would produce LtxA but not secrete it. However, we found that these mutants neither secreted LtxA nor retained it in the cell pellet (Fig. 2B and C). The fact that LtxA was not produced in these mutants is consistent with the findings of Guthmiller et al. (19), who reported that, while expression of ltxA was unaffected in ltxB and ltxD mutants, the amount of intracellular LtxA decreased significantly. A likely explanation for this observation is that nonsecreted LtxA may be proteolyzed inside the cell. Indeed, studies on other RTX toxins suggest that the low or absent intracellular levels of RTX toxins in certain mutants can be attributed to the instability of the protein retained in the cytoplasm due to the lack of a complete transport system (41).
Using the hemolysis screen, we identified a gene that was previously not implicated in LtxA regulation. HPr is a small protein (∼90 residues) encoded by ptsH and is part of the PTS. In both gram-negative and gram-positive bacteria, HPr participates in the regulation of various cellular processes, including transport and phosphorylation of sugars and amino acids and catabolite repression. In addition, in some bacteria, HPr can regulate non-PTS proteins, including transcriptional activators (14, 24, 36). The PTS has two general cytoplasmic proteins, EI and HPr, that can be paired with a number of enzymes with sugar specificity (EII). The overall reaction of sugar transport can be described as a cascade that starts with the removal of a phosphoryl group from phosphoenolpyruvate by EI and then phosphorylation of HPr at His-15 with HPr transferring its phosphoryl group to EII (8). EIIAGlc, the glucose-specific enzyme, has been shown to regulate levels of cAMP by activating adenylate cyclase (Fig. 6A) (32). Intracellular levels of cAMP are highest when EIIAGlc is in the phosphorylated state. Therefore, PTS mutants express low levels of cAMP (10, 35, 38). Interestingly, we found that the ptsH mutant grew at the same rate as the wild-type strain, even though this mutant presumably has a defect in sugar transport. However, this result is consistent with reports for other bacterial ptsH mutants (40). One explanation for this observation is that A. actinomycetemcomitans possesses HPr-like proteins, such as diphosphoryltransfer protein, that can partially substitute for HPr when grown in certain media (44). In addition, the bacteria may be utilizing other carbon sources in the complex medium. To date, no minimal medium exists for A. actinomycetemcomitans, and so we are unable to determine if there is a defect in sugar transport. Nonetheless, under our experimental conditions, the mutant strain grows as well as the wild-type strain.
We have shown here that a defect in HPr in A. actinomycetemcomitans results in low levels of ltxA expression, and we correlate this observation to decreased production of cAMP. This finding is consistent with previous studies in E. coli that demonstrated low adenylate cyclase activity in ptsH mutants (10, 35, 38) and with the study of Inoue et al. (25), who reported that low intracellular levels of cAMP correlate with low levels of LtxA. The low, albeit detectable, levels of cAMP may explain why the mutant is partially hemolytic. Production of cAMP over time may allow for some ltxA to be transcribed and eventual accumulation of LtxA. cAMP can regulate several operons through binding of cyclic AMP receptor protein (CRP) to specific sites on the DNA. Inoue et al. (25) noted that the CRP binding site consensus sequence is not present within the ltx operon; however, A. actinomycetemcomitans CRP is 84% identical to E. coli CRP and therefore could recognize a slightly different sequence motif.
We found that another PTS gene, ptsI, is also required for LtxA-mediated hemolysis. This observation is not surprising, given that EI phosphorylates HPr and would result in a nonfunctional HPr, similar to the result when ptsH is inactivated. Thus, our current model proposes that HPr regulates levels of ltxA through its interaction with other PTS proteins, which affects the levels of cAMP. In turn, cAMP regulates levels of ltxA mRNA through an unknown mechanism. While HPr plays a significant role in transcriptional control in many gram-positive bacteria (14, 24, 36), a similar bifunctional mechanism has not yet been established for HPr in gram-negative organisms. Examining the effects of other PTS mutants and protein interactions with HPr will shed more light on our model.
In summary, we have developed and utilized a method to isolate A. actinomycetemcomitans mutants that are defective in LtxA production based on β-hemolysis on blood agar. This rapid screen may allow for the identification of new target proteins for the development of therapeutics that interfere with LtxA activity and A. actinomycetemcomitans pathogenesis.
Acknowledgments
We thank Nataliya Balashova and Juan Crosby for thoughtful comments and suggestions throughout the project.
This work was generously supported by grants from the National Institute of Dental and Craniofacial Research (R01DE16133 to S.C.K. and F31DE018099 to M.P.I.) and the Sloan Foundation (to M.P.I.).
Editor: V. J. DiRita
Footnotes
Published ahead of print on 9 June 2008.
REFERENCES
- 1.Baehni, P. C., C. C. Tsai, W. P. McArthur, B. F. Hammond, B. J. Shenker, and N. S. Taichman. 1981. Leukotoxic activity in different strains of the bacterium Actinobacillus actinomycetemcomitans isolated from juvenile periodontitis in man. Arch. Oral Biol. 26671-676. [DOI] [PubMed] [Google Scholar]
- 2.Balashova, N. V., J. A. Crosby, L. Al Ghofaily, and S. C. Kachlany. 2006. Leukotoxin confers beta-hemolytic activity to Actinobacillus actinomycetemcomitans. Infect. Immun. 742015-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balashova, N. V., R. Diaz, S. V. Balashov, J. A. Crosby, and S. C. Kachlany. 2006. Regulation of Aggregatibacter (Actinobacillus) actinomycetemcomitans leukotoxin secretion by iron. J. Bacteriol. 1888658-8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berbari, E. F., F. R. Cockerill III, and J. M. Steckelberg. 1997. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin. Proc. 72532-542. [DOI] [PubMed] [Google Scholar]
- 5.Crosby, J. A., and S. C. Kachlany. 2007. TdeA, a TolC-like protein required for toxin and drug export in Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene 38883-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, M., A. D. Badley, F. R. Cockerill, J. M. Steckelberg, and W. R. Wilson. 1997. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 4825-33. [DOI] [PubMed] [Google Scholar]
- 7.Delepelaire, P. 2004. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694149-161. [DOI] [PubMed] [Google Scholar]
- 8.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz, R., L. A. Ghofaily, J. Patel, N. V. Balashova, A. C. Freitas, I. Labib, and S. C. Kachlany. 2006. Characterization of leukotoxin from a clinical strain of Actinobacillus actinomycetemcomitans. Microb. Pathog. 4048-55. [DOI] [PubMed] [Google Scholar]
- 10.Feucht, B. U., and M. H. Saier, Jr. 1980. Fine control of adenylate cyclase by the phosphoenolpyruvate:sugar phosphotransferase systems in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 141603-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fine, D. H., D. Furgang, J. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 441063-1076. [DOI] [PubMed] [Google Scholar]
- 12.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 1451335-1347. [DOI] [PubMed] [Google Scholar]
- 13.Fine, D. H., J. B. Kaplan, S. C. Kachlany, and H. C. Schreiner. 2006. How we got attached to Actinobacillus actinomycetemcomitans: a model for infectious diseases. Periodontol. 2000 42114-157. [DOI] [PubMed] [Google Scholar]
- 14.Flores, N., R. de Anda, S. Flores, A. Escalante, G. Hernandez, A. Martinez, O. T. Ramirez, G. Gosset, and F. Bolivar. 2004. Role of pyruvate oxidase in Escherichia coli strains lacking the phosphoenolpyruvate:carbohydrate phosphotransferase system. J. Mol. Microbiol. Biotechnol. 8209-221. [DOI] [PubMed] [Google Scholar]
- 15.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 697625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey, J., and P. Kuhnert. 2002. RTX toxins in Pasteurellaceae. Int. J. Med. Microbiol. 292149-158. [DOI] [PubMed] [Google Scholar]
- 17.Gentschev, I., G. Dietrich, and W. Goebel. 2002. The E. coli alpha-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 1039-45. [DOI] [PubMed] [Google Scholar]
- 18.Guthmiller, J. M., D. Kolodrubetz, M. P. Cagle, and E. Kraig. 1990. Sequence of the lktB gene from Actinobacillus actinomycetemcomitans. Nucleic Acids Res. 185291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guthmiller, J. M., D. Kolodrubetz, and E. Kraig. 1995. Mutational analysis of the putative leukotoxin transport genes in Actinobacillus actinomycetemcomitans. Microb. Pathog 18307-321. [DOI] [PubMed] [Google Scholar]
- 20.Guthmiller, J. M., E. Kraig, M. P. Cagle, and D. Kolodrubetz. 1990. Sequence of the lktD gene from Actinobacillus actinomycetemcomitans. Nucleic Acids Res. 185292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haraszthy, V. I., G. Hariharan, E. M. Tinoco, J. R. Cortelli, E. T. Lally, E. Davis, and J. J. Zambon. 2000. Evidence for the role of highly leukotoxic Actinobacillus actinomycetemcomitans in the pathogenesis of localized juvenile and other forms of early-onset periodontitis. J. Periodontol. 71912-922. [DOI] [PubMed] [Google Scholar]
- 22.Haubek, D., J. M. Dirienzo, E. M. Tinoco, J. Westergaard, N. J. Lopez, C. P. Chung, K. Poulsen, and M. Kilian. 1997. Racial tropism of a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J. Clin. Microbiol. 353037-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haubek, D., O. K. Ennibi, K. Poulsen, S. Poulsen, N. Benzarti, and M. Kilian. 2001. Early-onset periodontitis in Morocco is associated with the highly leukotoxic clone of Actinobacillus actinomycetemcomitans. J. Dent. Res. 801580-1583. [DOI] [PubMed] [Google Scholar]
- 24.Horstmann, N., G. Seidel, L. M. Aung-Hilbrich, and W. Hillen. 2007. Residues His-15 and Arg-17 of HPr participate differently in catabolite signal processing via CcpA. J. Biol. Chem. 2821175-1182. [DOI] [PubMed] [Google Scholar]
- 25.Inoue, T., I. Tanimoto, T. Tada, T. Ohashi, K. Fukui, and H. Ohta. 2001. Fermentable-sugar-level-dependent regulation of leukotoxin synthesis in a variably toxic strain of Actinobacillus actinomycetemcomitans. Microbiology 1472749-2756. [DOI] [PubMed] [Google Scholar]
- 26.Kachlany, S. C., D. H. Fine, and D. H. Figurski. 2000. Secretion of RTX leukotoxin by Actinobacillus actinomycetemcomitans. Infect. Immun. 686094-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 1826169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kachlany, S. C., P. J. Planet, R. Desalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40542-554. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan, J. B., M. B. Perry, L. L. MacLean, D. Furgang, M. E. Wilson, and D. H. Fine. 2001. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 695375-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolodrubetz, D., T. Dailey, J. Ebersole, and E. Kraig. 1989. Cloning and expression of the leukotoxin gene from Actinobacillus actinomycetemcomitans. Infect. Immun. 571465-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koronakis, V. 2003. TolC—the bacterial exit duct for proteins and drugs. FEBS Lett. 55566-71. [DOI] [PubMed] [Google Scholar]
- 32.Krin, E., O. Sismeiro, A. Danchin, and P. N. Bertin. 2002. The regulation of Enzyme IIA(Glc) expression controls adenylate cyclase activity in Escherichia coli. Microbiology 1481553-1559. [DOI] [PubMed] [Google Scholar]
- 33.Lally, E. T., E. E. Golub, I. R. Kieba, N. S. Taichman, J. Rosenbloom, J. C. Rosenbloom, C. W. Gibson, and D. R. Demuth. 1989. Analysis of the Actinobacillus actinomycetemcomitans leukotoxin gene. Delineation of unique features and comparison to homologous toxins. J. Biol. Chem. 26415451-15456. [PubMed] [Google Scholar]
- 34.Lally, E. T., I. R. Kieba, N. S. Taichman, J. Rosenbloom, C. W. Gibson, D. R. Demuth, G. Harrison, and E. E. Golub. 1991. Actinobacillus actinomycetemcomitans leukotoxin is a calcium-binding protein. J. Periodontal Res. 26268-271. [DOI] [PubMed] [Google Scholar]
- 35.Levy, S., G. Q. Zeng, and A. Danchin. 1990. Cyclic AMP synthesis in Escherichia coli strains bearing known deletions in the pts phosphotransferase operon. Gene 8627-33. [DOI] [PubMed] [Google Scholar]
- 36.Muller, W., N. Horstmann, W. Hillen, and H. Sticht. 2006. The transcription regulator RbsR represents a novel interaction partner of the phosphoprotein HPr-Ser46-P in Bacillus subtilis. FEBS J. 2731251-1261. [DOI] [PubMed] [Google Scholar]
- 37.Narayanan, S. K., T. G. Nagaraja, M. M. Chengappa, and G. C. Stewart. 2002. Leukotoxins of gram-negative bacteria. Vet. Microbiol. 84337-356. [DOI] [PubMed] [Google Scholar]
- 38.Nelson, S. O., B. J. Scholte, and P. W. Postma. 1982. Phosphoenolpyruvate:sugar phosphotransferase system-mediated regulation of carbohydrate metabolism in Salmonella typhimurium. J. Bacteriol. 150604-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen, K. K., and M. Boye. 2005. Real-time quantitative reverse transcription-PCR analysis of expression stability of Actinobacillus pleuropneumoniae housekeeping genes during in vitro growth under iron-depleted conditions. Appl. Environ. Microbiol. 712949-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nothaft, H., S. Parche, A. Kamionka, and F. Titgemeyer. 2003. In vivo analysis of HPr reveals a fructose-specific phosphotransferase system that confers high-affinity uptake in Streptomyces coelicolor. J. Bacteriol. 185929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oropeza-Wekerle, R. L., E. Muller, P. Kern, R. Meyermann, and W. Goebel. 1989. Synthesis, inactivation, and localization of extracellular and intracellular Escherichia coli hemolysins. J. Bacteriol. 1712783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paturel, L., J. P. Casalta, G. Habib, M. Nezri, and D. Raoult. 2004. Actinobacillus actinomycetemcomitans endocarditis. Clin. Microbiol. Infect. 1098-118. [DOI] [PubMed] [Google Scholar]
- 43.Planet, P. J., S. C. Kachlany, D. H. Fine, R. DeSalle, and D. H. Figurski. 2003. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34193-198. [DOI] [PubMed] [Google Scholar]
- 44.Reichenbach, B., D. A. Breustedt, J. Stulke, B. Rak, and B. Gorke. 2007. Genetic dissection of specificity determinants in the interaction of HPr with enzymes II of the bacterial phosphoenolpyruvate:sugar phosphotransferase system in Escherichia coli. J. Bacteriol. 1894603-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slots, J., and M. Ting. 1999. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol. 2000 2082-121. [DOI] [PubMed] [Google Scholar]
- 46.Taichman, N. S., R. T. Dean, and C. J. Sanderson. 1980. Biochemical and morphological characterization of the killing of human monocytes by a leukotoxin derived from Actinobacillus actinomycetemcomitans. Infect. Immun. 28258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taichman, N. S., D. L. Simpson, S. Sakurada, M. Cranfield, J. DiRienzo, and J. Slots. 1987. Comparative studies on the biology of Actinobacillus actinomycetemcomitans leukotoxin in primates. Oral Microbiol. Immunol. 297-104. [DOI] [PubMed] [Google Scholar]
- 48.Taichman, N. S., and J. M. Wilton. 1981. Leukotoxicity of an extract from Actinobacillus actinomycetemcomitans for human gingival polymorphonuclear leukocytes. Inflammation 51-12. [DOI] [PubMed] [Google Scholar]
- 49.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 1817298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai, C. C., B. J. Shenker, J. M. DiRienzo, D. Malamud, and N. S. Taichman. 1984. Extraction and isolation of a leukotoxin from Actinobacillus actinomycetemcomitans with polymyxin B. Infect. Immun. 43700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welch, R. A. 2001. RTX toxin structure and function: a story of numerous anomalies and few analogies in toxin biology. Curr. Top. Microbiol. Immunol. 25785-111. [DOI] [PubMed] [Google Scholar]
- 52.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 121-20. [DOI] [PubMed] [Google Scholar]
- 53.Zambon, J. J. 1996. Periodontal diseases: microbial factors. Ann. Periodontol. 1879-925. [DOI] [PubMed] [Google Scholar]