Abstract
The mycobacterial arabinan is an elaborate component of the cell wall with multiple glycosyl linkages and no repeating units. In Mycobacterium spp., the Emb proteins (EmbA, EmbB, and EmbC) have been identified as putative mycobacterial arabinosyltransferases implicated in the biogenesis of the cell wall arabinan. Furthermore, it is now evident that the EmbA and EmbB proteins are involved in the assembly of the nonreducing terminal motif of arabinogalactan and EmbC is involved in transferring arabinose, perhaps in the early stage of arabinan synthesis in lipoarabinomannan. It has also been shown that the Emb proteins are a target of the antimycobacterial drug ethambutol (EMB). In the search for additional mycobacterial arabinosyltransferases in addition to the Emb proteins, we disrupted MSMEG_6386 (an orthologue of Rv3792 and a gene upstream of embC) in Mycobacterium smegmatis. Allelic exchange at the chromosomal MSMEG_6386 locus of M. smegmatis could only be achieved in the presence of a rescue plasmid carrying a functional copy of MSMEG_6386 or Rv3792, strongly suggesting that MSMEG_6386 is essential. An in vitro arabinosyltransferase assay using a membrane preparation from M. smegmatis expressing Rv3792 and synthetic β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl and β-d-Galf-(1→6)-β-d-Galf-(1→5)-β-d-Galf-octyl showed that Rv3792 gene product can transfer an arabinose residue to the C-5 position of the internal 6-linked galactose. The reactions were insensitive to EMB, and when α-d-Manp-(1→6)-α-d-Manp-(1→6)-α-d-Manp-octylthiomethyl was used as an acceptor, no product was formed. These observations indicate that transfer of the first arabinofuranose residue to galactan is essential for M. smegmatis viability.
Mycobacterium tuberculosis, the causative agent of tuberculosis, is a major cause of mortality and morbidity worldwide (21, 34). Common to all pathogenic Mycobacterium spp. is a complex cell wall which is essential for growth, functional integrity, and survival inside the host. The biogenesis of the mycobacterial cell envelope is the target of many of the first-line drugs currently in use to combat tuberculosis.
A significant portion of the mycobacterial cell wall is made up of d-arabinan, a common constituent of both arabinogalactan (AG) and lipoarabinomannan (LAM) (8, 20). In AG, the arabinan maintains the structural integrity by tethering the mycolic acids to form the mycolylarabinogalactan-peptidoglycan (mAGP) complex. Its most characteristic structural feature is a terminal hexa-arabinosyl motif, [β-d-Araf-(1→2)-α-d-Araf-(1→)]2-(3,5)-α-d-Araf-(1→5)-α-d-Araf (Ara6). Both terminal β-d-Araf and penultimate 2-α-Araf can be covalently linked to mycolic acids (19). LAM arabinan on the other hand is more elaborate, less structured, and has extended linear β-d-Araf-(1→2)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf (Ara4) arabinan chains not found in AG, along with Ara6 as in AG (Fig. 1). With various degrees of mannose capping at the nonreducing Ara4 and Ara6 termini, the arabinan in LAM plays a pivotal role in the pathogenesis of the disease (7).
FIG. 1.
Only the nonreducing end arabinan domains in LAM and AG are shown here. Arabinan consists of the Ara2 internal motifs and Ara4 and Ara6 terminal motifs. The Ara2 motif consists of α-d-Araf-(1→5)-α-d-Araf and forms the linear backbone of arabinan, the Ara6 motif is [β-d-Araf-(1→2)-α-d-Araf-(1→)]2-(3,5)-α-d-Araf-(1→5)-α-d-Araf (A), and the Ara4 motif is β-d-Araf-(1→2)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf (B). The Ara6 motif is the most characteristic structural feature in AG: both terminal β-d-Araf and penultimate 2-α-Araf can be covalently linked to mycolic acids, while Ara4 motifs are not found in AG. On the other hand, both Ara4 and Ara6 motifs can be found in LAM, and with various degrees of mannose capping at the nonreducing Ara4 and Ara6 termini, the arabinan in LAM plays a pivotal role in the pathogenesis of the disease.
The assembly of the polymeric arabinan on either the lipomannan core or the galactan is a key process in the biogenesis of the cell wall. Ethambutol (EMB) is known to inhibit arabinosylation by acting on the putative arabinosyltransferases encoded by the embCAB gene cluster (2, 31). Although the Emb proteins have never been isolated, they have been shown to play pivotal role in cell wall arabinan synthesis (5, 12, 27, 33). Biochemical analysis of the viable mutant with individual disruption of embA, embB, and embC in Mycobacterium smegmatis revealed that the embA and embB gene products are responsible for the formation of the 3-linked branch of the Ara6 motif, whereas the embC is involved in the arabinan formation of LAM. The emerging biochemical data also indicated that despite overall similarity, the arabinans of LAM and AG could be distinguished by virtue of the additional presence of extended linear termini in LAM, which entails some as yet unknown feature of the EmbC protein for the proper synthesis (27). For the assembly of the arabinan domain of LAM and AG, logically, several different arabinosyltransferases should be present to conform to different linkages involved. In total, five major arabinosyltransferases have been identified, and there are clear indications that two more could exist (J. Zhang and D. Chatterjee, unpublished observations) which could be involved in the internal α3,5-Araf branching and α1→5-Araf chain elongation.
In search of mycobacterial arabinosyltransferases in addition to the ones discussed above, we pursued the MSMEG_6386 gene (an orthologue of Rv3792 in Mycobacterium smegmatis), organized immediate upstream of the emb gene cluster with a similar arrangement in Mycobacterium leprae and M. tuberculosis H37Rv. It belonged to the GT-C superfamily of integral membrane glycosyltransferases (4), and topological analysis suggested the existence of conserved D and R residues located in the second loop outside of the periplasm in the N terminus, which are predicted to be involved in the transfer of Araf from decaprenylphosphoryl arabinose (DPA). Herein, we show that the MSMEG_6386 gene is essential for the growth of M. smegmatis. In addition, we establish that in an in vitro arabinosyltransferase assay using synthetic oligosaccharide acceptors such as β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl and β-d-Galf-(1→6)-β-d-Galf-(1→5)-β-d-Galf-octyl, one arabinose residue was added by Rv3792 in each case.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacteria and plasmids used in this study are listed in Table 1. M. smegmatis strain mc2155 (28) was grown at 30°C or 42°C in Luria-Bertani (LB) broth (Difco) supplemented with 0.05% Tween 80. LB medium was used as the solid medium for all bacteria. Antibiotics were added, where appropriate, at the following concentrations: ampicillin (Amp; Sigma), 100 μg/ml; kanamycin (Kan; Sigma); hygromycin B (Hyg; Calbiochem), 50 μg/ml; gentamicin (Gen; Sigma), 5 μg/ml; and streptomycin (Str; Sigma), 20 μg/ml. When required, 10% sucrose (Suc; Fisher) was added to the solid medium.
TABLE 1.
Key M. smegmatis strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| mc2155 | Fast-growing mycobacterium; strain harboring all plasmids used herein | 28 |
| LL1 | M. smegmatis mc2155 with pLL1 integrated into MSMEG_6386 locus | This study |
| LL2 | Mutant with intrachromosomal allelic exchange at MSMEG_6386 locus in presence of pCG76:MSMEG_6386 | This study |
| LL3 | Mutant with intrachromosomal allelic exchange at MSMEG_6386 locus in presence of pCG76:Rv3792 | This study |
| Plasmids | ||
| pPR27 | Temperature-sensitive mycobacterial origin of replication; carries sacB gene and xylE reporter gene, Genr cassette, and E. coli origin of replication | 23 |
| pLL1 | pPR27 derivative carrying MSMEG_6386::Hygr | This study |
| pCG76 | E. coli and Mycobacterium shuttle vector carrying temperature-sensitive mycobacterial origin of replication and Strr cassette | 14 |
| pCG76:MSMEG_6386 | pCG76 plasmid carrying MSMEG_6386 (under control of Phsp60) | This study |
| pVV16 | pMV261 with Hygr cassette, Kanr cassette, and Phsp60 | 17 |
| pVV16:Rv3792 | pVV16 plasmid carrying Rv3792 | This study |
Construction of conditional replication plasmid carrying MSMEG_6386::Hygr.
Two DNA fragments were amplified from M. smegmatis mc2155 genomic DNA by PCR with rTth DNA polymerase (Roche). One DNA fragment named PCRI included 896-bp sequences upstream of MSMEG_6386 and the first 128-bp sequences from MSMEG_6386 and was amplified using the following primers: AAGCTTGACACACAACCACCTGGAC (HindIII) and CATATGAGCTGGTTGGACGTGTTGTA (NdeI). The other fragment named PCRII included the last 107-bp sequences from MSMEG_6386 and 968-bp sequences downstream of MSMEG_6386 and was amplified using the following primers: CATATGTGGACTCTGCGGTGTTCGAC (NdeI) and GCGGCCGCGGTCGTAGTACCAGCCGAAC (NotI). The M. smegmatis mc2155 genomic DNA sequences used in this study were obtained from the TIGR Center (www.tigr.org). PCRI and PCRII were cloned into pCR4Blunt-TOPO blunt vector (Invitrogen). The Hyg resistance cassette (Hygr) amplified from vector pVV16 was inserted into the blunt-ended NdeI site between PCRI and PCRII. A 3.3-kb fragment of PCRI -II::Hygr was inserted into the NotI site of pPR27 to make the conditional replication plasmid, pLL1, which contained the mycobacterial temperature-sensitive origin, counterselectable marker sacB, xylE reporter gene, and Genr cassette (23).
Purification of DNA restriction fragments and PCR fragments was performed using the QIAquick gel extraction kit (Qiagen, Chatsworth, CA). Plasmids were isolated from Escherichia coli TOP10 or XL1 Blue cells using the QIAprep miniprep kit (Qiagen). Molecular cloning and restriction endonuclease digestions were performed by standard techniques according to the manufacturer's recommendations.
Construction of the MSMEG_6386 conditional mutant.
To establish the essentiality of MSMEG_6386, a two-step recombination procedure was performed. In the first step, the plasmid pLL1 was electroporated into M. smegmatis using a Gene Pulser unit (Bio-Rad) with a single pulse (1.25 kV, 25 μF, 800 Ω). Transformants were grown in LB-Hyg-Gen plates at 30°C. A single colony was inoculated into LB-Hyg-Gen broth and incubated at 30°C, a permissive temperature for pLL1 replication, and then the cells were plated onto LB-Hyg-Gen plates and incubated at 42°C, a nonpermissive temperature for pLL1 replication. MSMEG_6386 conditional mutants with the single homologous recombination were selected using Southern blot analysis. In the second step, the single-homologous-recombination mutant was plated onto Hyg-Suc plates to select for mutants that underwent an intrachromosomal allelic exchange at the MSMEG_6386 locus.
pCG76, a Mycobacterium/E. coli shuttle plasmid harboring a mycobacterial temperature-sensitive origin of replication and a Strr cassette, was used as the rescue plasmid to carry a functional copy of MSMEG_6348 or Rv3792, respectively in the single homologous recombination mutant (14). MSMEG_6386 was amplified from M. smegmatis genomic DNA by PCR with rTth DNA polymerase. The upstream primer was CATATGCCGGTGGCGGCCAGGGTTCT (NdeI site underlined), and the downstream primer was GGATCCTCAGTGGCCATCGGTCTCCGGCTT (BamHI site underlined). The PCR product was cloned into pCR4Blunt-TOPO blunt vector, subcloned into the NdeI and BamHI sites of pET23b, and the Phsp60-MSMEG_6386 fragment was ligated into the XbaI and BamHI sites of pCG76 to generate the rescue plasmid, pCG76:MSMEG_6386. Rv3792 was amplified from M. tuberculosis H37Rv genomic DNA. The upstream primer was GATCGATCCATATGCCGAGCAGACGCAAAAGCCCCCAATTC (NdeI site underlined), and the downstream primer was GATCGATCAAGCTTCGCGCTCTCCTGCGGCTTGCGGATGGC (HindIII site underlined). The PCR product was cloned and subcloned in the similar way to that described above, and Phsp60-Rv3792 was ligated into the blunt-ended BamHI site of pCG76 to generate the rescue plasmid, pCG76:Rv3792.
Extraction of mycobacterial genomic DNA and Southern blot analysis.
Mycobacterial genomic DNA was isolated as follows. A single colony was inoculated into 10 ml of LB broth with appropriate antibiotics. Cells were harvested when the optical density at 600 nm (OD600) reached 1.0; the pellet was resuspended in 500 μl of TE buffer (10 mM Tris HCl, 1 mM EDTA [pH 8.0]) with 1 mg/ml lysozyme and 200 μg/ml RNase and incubated overnight at 37°C; the following day, 70 μl of 10% sodium dodecyl sulfate and 6 μl 10-mg/ml proteinase K was added and incubated for 20 min at 65°C, and then 100 μl 5 M NaCl and 80 μl cetyltrimethylammonium bromide-NaCl were added and incubated for 20 min at 65°C. DNA was extracted with chloroform and isoamyl alcohol (24:1 [vol/vol]), precipitated with 2-isopropanol, and washed with 75% ethanol. DNA probes were labeled with digoxigenin, and Southern blot analyses were performed as described for DIG High Prime DNA labeling and detection starter kit I (Roche).
Growth characteristics of the MSMEG_6386 conditional mutant with the intrachromosomal allelic exchange in the presence of rescue plasmids.
The M. smegmatis wild-type strain and MSMEG_6386 conditional mutants were inoculated in 20 ml LB broth (Lennox; Fisher Scientific), containing 0.05% Tween 80 and appropriate antibiotics and incubated at both 30°C and 42°C. OD600 was measured at intervals of 12 h for 4 days.
Overexpression of the Rv3792 in M. smegmatis.
Rv3792 was subcloned into the NdeI and HindIIII sites of pVV16 harboring the Kanr and Hygr cassettes (17) to generate plasmid pVV16:Rv3792, which allows Rv3792 to be constitutively expressed under the control of Phsp60. M. smegmatis was transformed with pVV16:Rv3792, and transformants were selected on LB-Kan-Hyg plates. The recombinant proteins carry a six-histidine tag at their C terminus.
Preparation of enzymatically active membrane and cell wall-enriched fractions.
Cells (10 g) from a 2-liter culture of M. smegmatis with pVV16 or pVV16:Rv3792 were harvested and suspended in 40 ml of buffer A (50 mM morpholinepropanesulfonic acid [MOPS; pH 7.9], 5 mM 2-mercaptoethanol, 10 mM MgCl2), subjected to probe sonication (22), and centrifuged at 27,000 × g (Beckman Avanti HP-25I, JA25.50 rotor) for 20 min at 4°C. The pellet was resuspended in buffer A, and Percoll (Amersham Pharmacia Biotech) was added to achieve a 60% suspension, which was centrifuged at 27,000 × g for 60 min at 4°C. The white upper band, containing a particulate cell wall-enriched (P60) fraction, was isolated, and Percoll was removed by repeated rounds of suspension in buffer A and centrifugation. The P60 fraction was resuspended in buffer A to a protein concentration of 8 to 10 mg/ml. A membrane-enriched fraction was obtained by centrifuging the 27,000 × g supernatant at 100,000 × g (Beckman L7-80 ultracentrifuge, SW28 rotor) for 2 h at 4°C; the pellet was suspended in buffer A at a protein concentration of 15 to 20 mg/ml.
Synthesis of trisaccharide acceptors.
The oligosaccharides β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl and β-d-Galf-(1→6)-β-d-Galf-(1→5)-β-d-Galf-octyl were synthesized as described for the corresponding dec-9-enyl glycoside analogues (10). The synthesis of oligosaccharide α-d-Manp-(1→6)-α-d-Manp-(1→6)-α-d-Manp-(CH2)6SMe has previously been described (15).
Arabinosyltransferase assays using p[14C]Rpp.
Typical reaction mixtures contained 50 mM MOPS (pH 7.9), 5 mM 2-mercaptoethanol, 10 mM MgCl2, 1 mM ATP, 3.8 μM p[14C]Rpp (500,000 dpm), 60 μg (1 mM) acceptor, 500 μg membrane proteins, and 300 μg P60 proteins from either M. smegmatis with pVV16 or pVV16:Rv3792 in a total volume of 200 μl. The reaction mixtures were incubated at 37°C for 2 h, and then the reactions were terminated by addition of 200 μl of 100% ethanol. The resulting mixture was centrifuged at 14,000 × g for 5 min, and the supernatants were loaded onto prepacked strong-anion-exchange (SAX) columns (Burdick and Jackson). The columns were eluted sequentially with 2 ml water. The eluate was evaporated to dryness and partitioned between the two phases of water-saturated 1-butanol and water (1:1 [vol/vol]). The 1-butanol fraction was dried and resuspended in 200 μl of 1-butanol. The extracted radiolabeled material was quantitated by liquid scintillation counting in 5 ml of EcoScintA (National Diagnostics, Atlanta, GA). Aliquots of the radiolabeled material were also subjected to thin-layer chromatography (TLC) analysis using silica gel plates (silica gel 60F254; Merck) developed in CHCl3-CH3OH-1 M NH4OAc-NH4OH-H2O (180:140:9:9:23 [vol/vol/vol/vol/vol]). Autoradiograms of the TLC plates were obtained by exposure to Kodak X-Omat film at −70°C for 3 days.
Neutral sugar composition analysis.
Approximately 2,000 dpm of the 1-butanol-soluble enzymatic product was dried under a stream of air and hydrolyzed in 200 μl of 2 M trifluoroacetic acid (TFA) at 120°C for 2 h. The TFA was removed under a stream of air, and the hydrolysate was analyzed on a silica gel TLC plate developed in pyridine-ethyl acetate-acetic acid-water (5:5:1:3 [vol/vol/vol/vol]) followed by autoradiography as described above. Radioactive spots were identified by cochromatography with TFA-hydrolyzed 14C-labeled AG as standard.
MALDI-TOF MS and MALDI-TOF/TOF MS/MS analyses of enzymatic product.
In order to generate enzymatic product for characterization, synthetic nonradiolabeled DPA (provided by Avraham Liav, Colorado State University) was used in the cell-free assay. Typical reaction mixtures contained 200 μg of DPA, 60 μg of acceptor, and 1 mg of membrane proteins prepared from M. smegmatis with pVV16:Rv3792 in a total volume of 100 μl. Five reaction mixtures were set up for each acceptor, incubated at 37°C for 2 h, and then terminated by adding 100 μl 100% ethanol. The resulting mixture was centrifuged at 14,000 × g for 5 min, and the supernatants were loaded onto prepacked SAX columns. The columns were eluted sequentially with 2 ml water. The eluate was evaporated to dryness, resuspended in water, and allowed to run through a MixBed ion-exchange resin (Bio-Rad). Eluate (unbound) was subjected to TLC plate developed in CHCl3-CH3OH-1 M NH4OAc-NH4OH-H2O (180:140:9:9:23 [vol/vol/vol/vol/vol]). The radiolabled product using p[14C]Rpp was used as standard. The band corresponding to the radiolabled product was excised and eluted off silica gel with organic solvent. The dry residue was permethylated using the NaOH-dimethyl sulfoxide slurry method (9). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis was performed using an UltraFlex tandem-TOF (TOF/TOF) device (Bruker Daltonics, Billerica, MA), in which case the permethylation derivatives in acetonitrile were mixed 1:1 with 2,5-dihydroxybenzoic acid matrix (10 mg/ml in water) for spotting onto the target plate. MALDI-TOF/TOF collision-induced disocciation (CID) tandem-MS (MS/MS) sequencing of the permethyl derivatives using 2,5-dihydroxybenzoic acid as matrix was performed on a 4700 Proteomics analyzer (Applied Biosystems) as described previously (18).
RESULTS
Construction of MSMEG_6386 conditional mutant.
A two-step homologous recombination procedure was used to achieve intrachromosomal allelic exchange at the MSMEG_6386 locus in M. smegmatis (14, 16, 17). In the first step, the conditional replication plasmid pLL1 was constructed, which harbors the xylE reporter gene encoding catechol 2,3-dioxygenase, and a yellow color develops in colonies expressing xylE when sprayed with catechol. pLL1 was introduced into M. smegmatis by electroporation, and the Hygr Genr transformants at 30°C were then selected on LB-Hyg-Gen plates at 42°C. Hygr Genr and XylE-positive colonies were the candidates for selecting the mutants with the single homologous recombination. Southern blot analysis on the chromosomal DNA from nine candidates indicated that three resulted from a single homologous recombination (data not shown), while the other six arose from illegitimate recombination.
In the second step, a single homologous recombination mutant M. smegmatis LL1, was grown in LB-Hyg-Gen broth and then plated onto LB-Hyg-Suc plates to select for mutants that had undergone intrachromosomal allelic exchange. The candidates for the allelic exchange are expected to be Sucr Hygr and remain white (xylE negative) when sprayed with catechol, while the yellow colonies (xylE positive) on the Suc-Hyg plates are likely to be sacB mutants. Spraying thousands of Sucr Hygr colonies with catechol revealed that none of them exhibited the expected phenotype, suggesting that the MSMEG_6386 gene is essential for M. smegmatis.
M. smegmatis LL1 was transformed with the temperature-sensitive rescue plasmid pCG76:MSMEG_6386 or pCG76:Rv3792, and transformants were plated on LB-Hyg-Str-Suc plates at 30°C to select for mutants with allelic exchange. Southern blot analysis showed that all Hygr Sucr Strr colonies with the XylE-negative phenotype had undergone intrachromosomal allelic exchange (Fig. 2). Thus, allelic exchange at the chromosomal MSMEG_6386 locus was achievable only in the presence of the rescue plasmid carrying a functional copy of MSMEG_6386 or Rv3792, suggesting that MSMEG_6386 is essential and that these two proteins have similar functions.
FIG. 2.
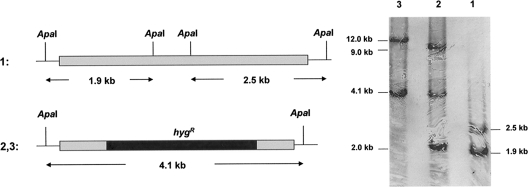
Allelic exchange at the MSMEG_6386 locus. Southern blot analysis and expected hybridization profiles of M. smegmatis chromosomal DNA (lane 1) and the conditional mutant M. smegmatis LL2 and LL3 chromosomal DNA (lanes 2 and 3). ApaI was used to digest the chromosomal DNA. The probe used corresponds to the 2.1-kb fragment of PCRI -II generated with the primers used in the initial step. The signal detected corresponds to the 2.0-kb and 9.0-kb fragments (lane 2) and the 12.0-kb fragment (lane 3), which were carried by the rescue plasmid.
Growth characteristics of the MSMEG_6386 conditional mutant.
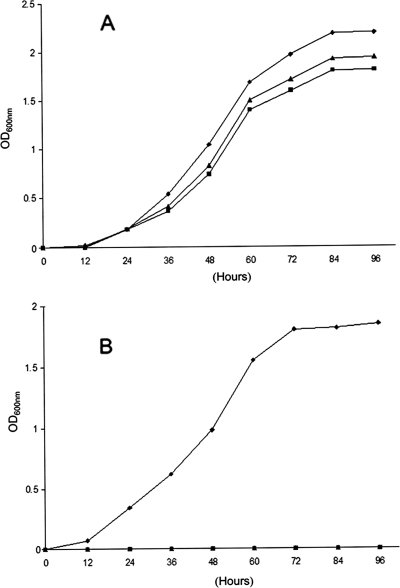
To conclusively confirm that MSMEG_6386 is essential for growth of M. smegmatis, we investigated the ability of the mutants with allelic exchange in the presence of pCG76:MSMEG_6386 (M. smegmatis LL2) or pCG76:Rv3792 (M. smegmatis LL3) to survive at 42°C, a temperature at which the rescue plasmid is unable to replicate. The growth characteristics of M. smegmatis mutants LL2 and LL3 and the wild-type M. smegmatis strain at 30°C and 42°C are presented in Fig. 3. As expected, M. smegmatis LL2 and LL3 exhibited the same growth characteristics as wild-type M. smegmatis at 30°C, as shown in Fig. 3A. After a temperature shift to 42°C, M. smegmatis LL2 and LL3 were unable to grow, although the wild-type M. smegmatis strain continued to grow exponentially, as shown in Fig. 3B. These results indicate that MSMEG_6386 is essential for M. smegmatis.
FIG. 3.
Growth characteristics of the M. smegmatis wild type and the MSMEG_6386 conditional mutants at 30°C (A) and 42°C (B). Shown are growth curves for wild-type M. smegmatis (⧫), M. smegmatis LL2 (▪), and M. smegmatis LL3 (▴) cultivated in LB-Tween 80 and LB-Tween 80-Hyg broth, respectively.
In vitro arabinosyltransferase assay.
We expressed MSMEG_6386 and Rv3792 using the same expression vector, pVV16, in M. smegmatis; however, only the recombinant Rv3792 could be detected using the anti-His monoclonal antibody (data not shown); therefore, M. smegmatis with overexpressed Rv3792 was used in vitro arabinosyltransferase assay.
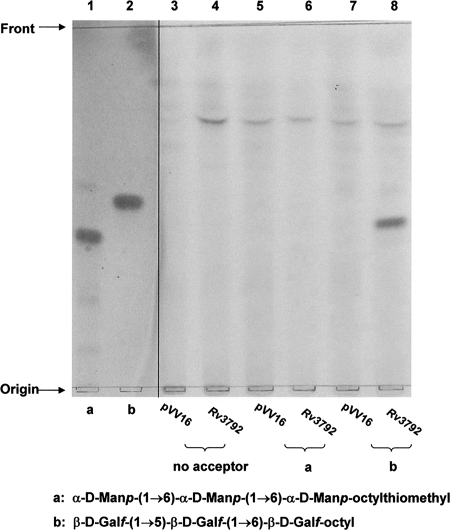
We uitilized exogenous, chemically synthesized acceptors α-d-Manp-(1→6)-α-d-Manp-(1→6)-α-d-Manp-octylthiomethyl (Fig. 4, lane 1), β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl (Fig. 4, lane 2), and p[14C]Rpp as the Araf donor. Analysis of the products from M. smegmatis with overexpressed Rv3792, resulted in the formation of a single product for the trigalactan acceptor (Fig. 4, lane 8). No product was observed with either the control reaction that did not include the acceptors (Fig. 4, lane 4), with trimannan acceptor (Fig. 4, lane 6), or with M. smegmatis with vector control (Fig. 4, lanes 3, 5, and 7). With β-d-Galf-(1→6)-β-d-Galf-(1→5)-β-d-Galf-octyl as the acceptor, a single product was also observed with M. smegmatis with overexpressed Rv3792 (data not presented).
FIG. 4.
Effect of overexpressed Rv3792 in M. smegmatis on the in vitro incorporation of [14C]Araf into the synthetic trigalactan or trimannan acceptors. In lanes 1 and 2, the acceptors α-d-Manp-(1→6)-α-d-Manp-(1→6)-α-d-Manp-(CH2)6SMe (lane 1) and β-d-Gal-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl (lane 2) were visualized by α-naphthol-sulfuric acid. Lanes 3 and 4 show the control reaction (no acceptor) from M. smegmatis with pVV16 (lane 3) and overexpressed Rv3792 (lane 4). Lanes 5 and 6 show use of α-d-Manp-(1→6)-α-d-Manp-(1→6)-α-d-Manp-(CH2)6SMe and M. smegmatis with pVV16 (lane 5) and overexpressed Rv3792 (lane 6). Lanes 7 and 8 show the incorporation of [14C]Araf into β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl from M. smegmatis with pVV16 (lane 7) and overexpressed Rv3792 (lane 8). The partially purified labeled products were applied to TLC plates and developed in CHCl3-CH3OH-1 M NH4OAc-NH4OH-H2O (180:140:9:9:23 [vol/vol/vol/vol/vol]) and subjected to autoradiography.
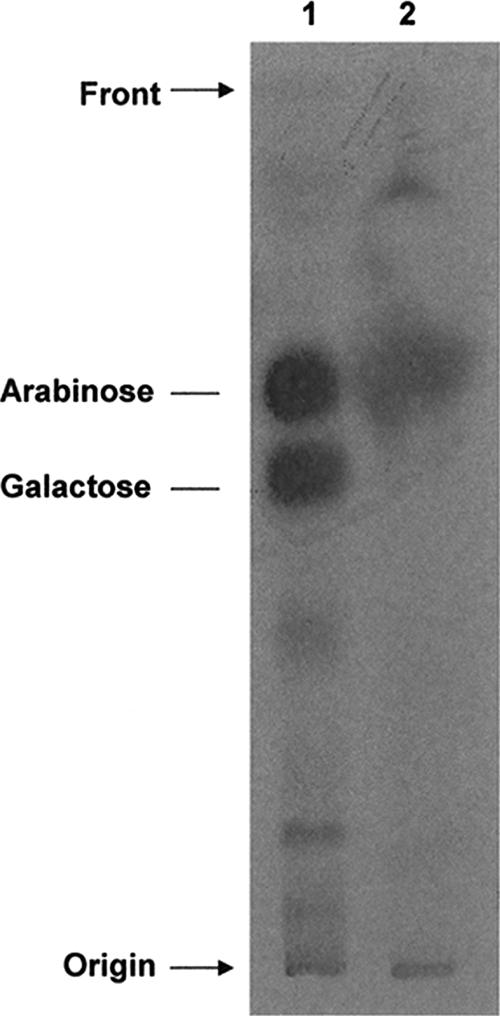
When assays were performed in the presence of various concentrations of EMB, product formation remained unaffected (data not shown). A cell-free assay was also performed with DP[14C]A, and a band was obtained akin to that of product from the reaction utilizing p[14C]Rpp (data not shown). An aliquot of the 1-butanol-soluble material (∼2,000 dpm) was hydrolyzed with 2 M TFA, and the hydrolysate was subjected to TLC. Autoradiography revealed that the radioactivity was associated with arabinose (Fig. 5), which confirmed that [14C]Araf has been added to the trigalactan acceptor.
FIG. 5.
Monosaccharide analysis of the radiolabeled product after TFA hydrolysis. Radioactive AG (2,000 dpm; lane 1) and the radiolabeled product from the reaction of the incorporation of [14C]Araf into β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl (lane 2) from M. smegmatis with overexpressed Rv3792 were hydrolyzed with 2 M TFA, applied to the TLC plate, developed in pyridine-ethyl acetate-acetic acid-water (5:5:1:3 [vol/vol/vol/vol]), and subjected to autoradiography.
MALDI-TOF MS and MALDI-TOF/TOF MS/MS analysis of the enzymatic products.
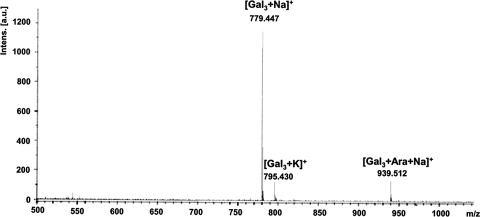
Products formed with both galactan acceptors were extracted from a preparative TLC plate. The products were methylated, and MALDI-TOF MS analysis of revealed a strong molecular ion at m/z 939.5 (M + Na+) for Araf-(1-?)- added to the acceptors. Ions for unreacted acceptor m/z 779.4 (M + Na+) and 795.4 (M + K+) were also observed, indicating that some starting material cochromatographed even after TLC separation. Both trigalactan acceptors provided same molecular ion profiles: only one is shown (Fig. 6).
FIG. 6.
MALDI-TOF analysis of enzymatic product. Product formed with β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl extracted from a preparative TLC was methylated, and MALDI-TOF analysis of this revealed a strong molecular ion at m/z 939.5 (M + Na+) for Araf-(1-?)- added to the acceptors. Intens., intensity; a.u., arbitrary units.
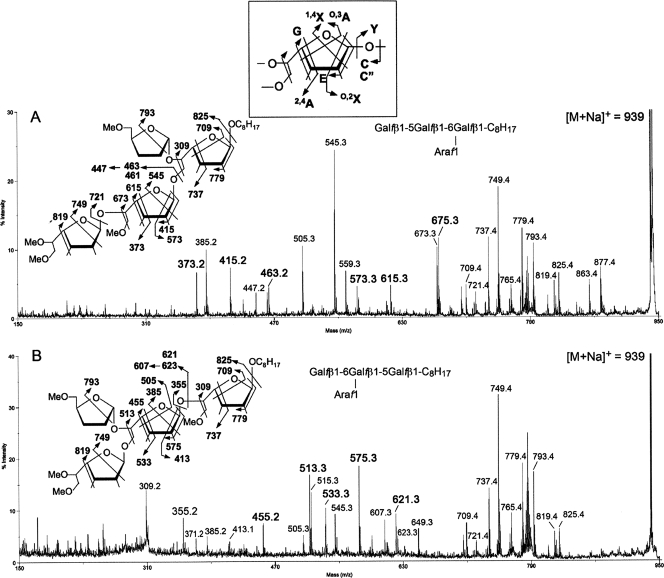
From the MALDI-TOF MS analysis of the permethyl derivatives, it is clear that both Gal3-C8H17 synthetic acceptors can have only a single Ara residue added on to give the common product Gal3Ara1-C8H17. To further define the location of the newly added Ara, the respective sodiated molecular ions were selected for MALDI MS/MS analysis. It was found that under high-energy CID MS/MS, as performed on a MALDI-TOF/TOF MS, the observed fragmentation pattern for the Galf is similar to that established for the Araf (18, 32). The original Galf3-C8H17 synthetic acceptor (data not shown) gave the expected series of 1,4X, O,2X, O,3A, 2,4A, C, and Y, as well as the E and G ions representing concerted elimination of substituents around the ring. (The nomenclature for the ions formed is widely used for all glycan MS/MS and refers to formation of satellite ions due to cleavages across the ring; this is shown in the inset in Fig. 7). Importantly, the Galf can further give a reducing terminal ion derived from loss of both C-5 and C-6 substituents, with and without creating a double bond (see illustration on Fig. 7). Based on these series of characteristic ions, the MS/MS pattern of the Gal3Ara1-C8H17 products (Fig. 7) can be interpreted to be consistent with the single Ara being mostly added onto the C-5 of a 6-linked Galf.
FIG. 7.
MALDI-TOF/TOF MS/MS analysis. Shown are the enzymatic product Gal3Ara1-C8H17 obtained from using the synthetic acceptor β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl (A) and β-d-Galf-(1→6)-β-d-Galf-(1→5)-β-d-Galf-octyl (B). The [M + Na]+ molecular ions afforded by the permethylated sample at m/z 939 (Fig. 6) were selected for high-energy CID MS/MS. Assignment of the key fragment ions were as schematically illustrated. In addition to the well-established A, X, C, and Y ions, the G and E ions were named by Spina et al. and have been described in full in the context of furanoses (18, 32). (Note that the 1,4X ions give linkage information only pertaining to the C-1 position. The 0,2X ions provide information about whether C-2 contains a glycosidic bond. 0,3A and 2,4A ions provide the C-5 and C-3 linkage information. C and Y ions, as well as the E and G ions, represent concerted elimination of substituents around the ring.) The two specific ion series corresponding to E − 30 mass units and C − 16 mass units (m/z 447 and 607, respectively) were likewise identified previously for the arabinan and not further drawn out here. The concerted cleavages corresponding to loss of both C-5 and C-6 substituents on the Galf have not been reported before and were noted to produce a pair of ions differing in having either unsaturated or saturated bonds (e.g., m/z 675/673 in panel A and 513/515 in panel B). The 1,4X ions at m/z 385 in panel A and 545 in panel B coincide, respectively, with the E ions at m/z 415 and 575 − 30 mass units and thus may not be taken as sequence-specific ions to indicate presence of isomeric products. In contrast, other specific ions as described in the text are present only in either and not both spectra. The ion at m/z 505 in panel A may be assigned as an O,3A ion, which is indicative of possible presence of Araf on the nonreducing terminal Gal2, as in panel B, but other supporting ions that are specific to this isomeric structure are lacking.
In the case of Ara1(Gal-5Gal-6Gal-C8H17) (Fig. 7A), the C or C‴, E, and 2,4A ions at m/z 463/461, 415, and 373, respectively, indicate a nonreducing end Gal2 that was not arabinosylated and thus localize the Ara to the innermost 6-linked Gal, and the O,3A ion at m/z 709 places the Ara on no other position but C-5. In comparison, the corresponding C or C‴, E, and 2,4A ions for Ara1(Gal-6Gal-5Gal-C8H17) were shifted to m/z 623/621, 575, and 533, respectively (Fig. 7B), localizing the Ara at the nonreducing end Gal2 instead. In this case, the 1,4X ion at m/z 749, coupled with the cleavage ions at m/z 513/515 and G ion at m/z 455, would define an Ara substituent at the C-5 of the middle 6-linked Galf. The corresponding ions at m/z 673/675 and 615, which indicate substitution elsewhere, were not detected. In addition, lack of arabinosylation at the nonreducing end Galf in both cases is supported by the absence of a prominent 1,4X ion at m/z 589, which would arise from loss of a nonreducing terminal Ara-Gal-. It should, however, be noted that the MS/MS data cannot rule out the coexistence of a minor amount of isomeric products corresponding to Ara substituent at other positions. More importantly, the data positively established that the preferred site of single arabinosylation is on the C-5 of a 6-linked Gal and not a terminal Gal. This finding is consistent with previously defined arabinosylation position on the galactans of AG (1).
DISCUSSION
The distinct structural aspects of the mycobacterial cell wall and their importance in the viability of the organism suggest that the search for novel drug targets directed toward its inhibition may prove fruitful. As proof of principle, many of the current frontline drugs such as isoniazid (inhibits mycolic acid synthesis) (25) and EMB (targets arabinosyltransferases) act directly on cell wall synthesis (13, 30). Despite the direct association of EMB resistance and arabinosyltransferases (2, 29), it is interesting to note that there are distinct arabinosyltransferases which are both essential and not sensitive to EMB (1, 32).
It was predicted years ago, that a cluster comprising 31 genes are perhaps involved in AG biosynthesis (3). The embCAB, Rv3792, Rv3805c, and glf genes (galactan polymerization) and fbpA (mycolyltransferase) are all present in this large gene cluster. Interspersed throughout this gene cluster are genes encoding proteins with similarity to other polysaccharide biosynthetic proteins. Several genes with unknown function are arranged in potential operons and could very well be involved in arabinan synthesis. Although, broadly speaking, only three linkages are involved—β1→2, α1→3, and α1→5—logically, the arabinan domain of AG and LAM must utilize additional arabinofuranosyltransferases for assembly and polymerization.
Unlike the emb genes that are nonessential for M. smegmatis (12, 33), in our present study, we show that the homolog of Rv3792 in M. smegmatis (MSMEG_6386) is an essential gene. The inability to form an intrachromosomal allelic exchange event at the MSMEG_6386 locus in the absence of the rescue plasmid, coupled with the inability of M. smegmatis LL2 and LL3 to grow at 42°C, a temperature at which the rescue plasmid has been lost, conclusively demonstrated that transfer of the first arabinose to galactan is necessary for the viability of M. smegmatis, although the Corynebacterium glutamicum mutant with its orthologue deletion has been reported to be viable (1). In addition, Rv3792 has been predicted to be essential by Himar1-based transposon mutagenesis of M. tuberculosis (26). Comparison of protein sequences (BLAST in NCBI; www.ncbi.nlm.nih.gov/BLAST/bl2seq/wblast2.cgi) suggested that Rv3792 shares 78% and 68% identity to the homologous proteins in M. leprae (ML0107) and M. smegmatis (MSMEG_6386), while having only 37% identity to that of C. glutamicum (Ncg0185). One possibility is that a proper, fully complemented cell wall synthesis is not required in Corynebacterium. This is reflected in the fact that the overall cell wall architecture of corynebacteria is distinctly different from that of mycobacteria. The mycolic acids in corynebacteria are much simpler in structure, with only C-32 to C-36 carbon atoms, and the arabinogalactan-bound mycolates are not present in sufficient quantity to form a complete monolayer around the cell as is seen in mycobacteria (6). Studies have shown that the glycosyl linkage profile of corynebacterium AG was broadly similar to that of M. tuberculosis/M. smegmatis (24). However, the AG lacks terminal Ara6 motifs and has a less elaborate linear terminal Ara4 for corynomycolate deposition. Lack of a terminal Ara6 appears to be consistent with the lack of EmbAB functions i.e., deposition of the β-d-Araf-(1→2)-α-d-Araf disaccharide to the C-3 position of the 3,5-linked Araf residue as in mycobacteria (12). It is noteworthy, that disruption of the single emb gene in C. glutamicum (embCg) led to depletion of all but three Araf residues in AG which are deposited by an orthologue of Rv3792 in Corynebacterium (1).
Rv3792 is organized immediately upstream of embC. With this in mind and in order to determine if Rv3792 could also transfer Araf to the mannose acceptors, we attempted to conduct a cell-free assay using a mannose acceptor. Failure of the overexpressed strain to generate product with the mannose acceptor suggests that it is perhaps not involved in initiating arabinosylation of the mannan in LAM synthesis. Herein, we have also established an in vitro arabinosyltransfearase assay using β-d-Galf-(1→5)-β-d-Galf-(1→6)-β-d-Galf-octyl and β-d-Galf-(1→6)-β-d-Galf-(1→5)-β-d-Galf-octyl as the acceptor and pRpp and DPA as the Araf donor. MALDI-TOF MS/MS analysis of the enzymatic product formed helped to localize the Araf at the nonreducing end Gal2. More importantly, the data positively established that the preferred site of single arabinosylation is on C-5 of a 6-linked Galf and not a terminal Galf. This finding is consistent with previously defined arabinosylation position on the galactans of AG (11). Although Rv3792 shows no significant sequence similarity to the Emb proteins, the predicted topology shows similar N-terminal transmembrane domains and C-terminal region outside of periplasm. Furthermore, there are conserved negatively charged residues, such as D and R, located in the second loop outside of periplasm in the N terminus, which are predicted to be involved in the transfer of Araf using DPA as the donor.
We conclude that transfer of the first Araf residue to the galactan backbone is essential in M. smegmatis, and specificity of Rv3792 function makes it a potential target for developing novel class of inhibitors along the line of EMB to disrupt the cell wall assembly. Also, the arabinosyltransferase required to initiate the arabinan chain from the mannan still remains to be identified and should also be essential, considering the role of LAM in M. tuberculosis. Current efforts are now concentrated on this aspect.
Acknowledgments
We thank Anita G. Amin for generously providing p[14C]Rpp and Jian Zhang and Mary Jackson for helpful discussions. We thank Avraham Liav for providing DPA.
This work was supported by grant AI 37139 from the National Institutes of Health to D.C. and Taiwan NSC grant 95-2311-B-001-031 to K.K. We thank the ETH Zürich and the New Zealand Foundation for Research, Science & Technology. High-energy CID MALDI MS/MS analyses were performed at the National Core Facilities for Proteomics located at the Institute of Biological Chemistry, Academia Sinica, supported by Taiwan NSC grant 95-3112-B-001-014).
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Alderwick, L. J., M. Seidel, H. Sahm, G. S. Besra, and L. Eggeling. 2006. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 28115653-15661. [DOI] [PubMed] [Google Scholar]
- 2.Belanger, A. E., G. S. Besra, M. E. Ford, K. Mikusova, J. T. Belisle, P. J. Brennan, and J. M. Inamine. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. USA 9311919-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belanger, A. E., and J. I. Inamine. 2000. Genetics of cell wall biosynthesis, p. 191-202. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, DC.
- 4.Berg, S., D. Kaur, M. Jackson, and P. J. Brennan. 2007. The glycosyltransferases of Mycobacterium tuberculosis; roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 1735R-56R. [DOI] [PubMed] [Google Scholar]
- 5.Berg, S., J. Starbuck, J. B. Torrelles, V. D. Vissa, D. C. Crick, D. Chatterjee, and P. J. Brennan. 2005. Roles of conserved proline and glycosyltransferase motifs of EmbC in biosynthesis of lipoarabinomannan. J. Biol. Chem. 2805651-5663. [DOI] [PubMed] [Google Scholar]
- 6.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 7.Briken, V., S. A. Porcelli, G. S. Besra, and L. Kremer. 2004. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol. Microbiol. 53391-403. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, D., and K. H. Khoo. 1998. Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology 8113-120. [DOI] [PubMed] [Google Scholar]
- 9.Ciucanu, I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131209-217. [Google Scholar]
- 10.Completo, G. C., and T. L. Lowary. 20 May 2008, posting date. Synthesis of galactofuranose-containing acceptor substrates for mycobacterial galactofuranosyltransferases. J. Org. Chem. [Epub ahead of print.] doi: 10.1021/jo800457. [DOI] [PubMed]
- 11.Daffe, M., M. McNeil, and P. J. Brennan. 1993. Major structural features of the cell wall arabinogalactans of Mycobacterium, Rhodococcus, and Nocardia spp. Carbohydr. Res. 249383-398. [DOI] [PubMed] [Google Scholar]
- 12.Escuyer, V. E., M. A. Lety, J. B. Torrelles, K. H. Khoo, J. B. Tang, C. D. Rithner, C. Frehel, M. R. McNeil, P. J. Brennan, and D. Chatterjee. 2001. The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J. Biol. Chem. 27648854-48862. [DOI] [PubMed] [Google Scholar]
- 13.Forbes, M., N. A. Kuck, and E. A. Peets. 1962. Mode of action of ethambutol. J. Bacteriol. 841099-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guilhot, C., I. Otal, I. Van Rompaey, C. Martin, and B. Gicquel. 1994. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J. Bacteriol. 176535-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holemann, A., B. L. Stocker, and P. H. Seeberger. 2006. Synthesis of a core arabinomannan oligosaccharide of Mycobacterium tuberculosis. J. Org. Chem. 718071-8088. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, M., L. R. Camacho, B. Gicquel, and C. Guilhot. 2001. Mycobacterium tuberculosis protocols, vol. 54. Humana Press, Totowa, NJ.
- 17.Jackson, M., D. C. Crick, and P. J. Brennan. 2000. Phosphatidylinositol is an essential phospholipid of mycobacteria. J. Biol. Chem. 27530092-30099. [DOI] [PubMed] [Google Scholar]
- 18.Lee, A., S. W. Wu, M. S. Scherman, J. B. Torrelles, D. Chatterjee, M. R. McNeil, and K. H. Khoo. 2006. Sequencing of oligoarabinosyl units released from mycobacterial arabinogalactan by endogenous arabinanase: identification of distinctive and novel structural motifs. Biochemistry 4515817-15828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNeil, M., M. Daffe, and P. J. Brennan. 1991. Location of the mycolyl ester substituents in the cell walls of mycobacteria. J. Biol. Chem. 26613217-13223. [PubMed] [Google Scholar]
- 20.McNeil, M. R. 1999. Arabinogalactan in mycobacteria: structure, biosynthesis, and genetics, p. 207-223. In J. B. Goldberg (ed.), Genetics of bacterial polysaccharides. CRC Press, Boca Raton, FL.
- 21.Migliori, G. B., R. Loddenkemper, F. Blasi, and M. C. Raviglione. 2007. 125 years after Robert Koch's discovery of the tubercle bacillus: the new XDR-TB threat. Is “science” enough to tackle the epidemic? Eur. Respir. J. 29423-427. [DOI] [PubMed] [Google Scholar]
- 22.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 2717820-7828. [DOI] [PubMed] [Google Scholar]
- 23.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 9410955-10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puech, V., M. Chami, A. Lemassu, M. A. Laneelle, B. Schiffler, P. Gounon, N. Bayan, R. Benz, and M. Daffe. 2001. Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology 1471365-1382. [DOI] [PubMed] [Google Scholar]
- 25.Quémard, A., S. Mazères, A. Sut, G. Lanéelle, and C. Lacave. 1995. Certain properties of isoniazid inhibition of mycolic acid synthesis in cell-free systems of M. aurum and M. avium. Biochim. Biophys. Acta Lipids Lipid Metab. 125498-104. [DOI] [PubMed] [Google Scholar]
- 26.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 4877-84. [DOI] [PubMed] [Google Scholar]
- 27.Shi, L., S. Berg, A. Lee, J. S. Spencer, J. Zhang, V. Vissa, M. R. McNeil, K. H. Khoo, and D. Chatterjee. 2006. The carboxy terminus of EmbC from Mycobacterium smegmatis mediates chain length extension of the arabinan in lipoarabinomannan. J. Biol. Chem. 28119512-19526. [DOI] [PubMed] [Google Scholar]
- 28.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 41911-1919. [DOI] [PubMed] [Google Scholar]
- 29.Sreevatsan, S., K. E. Stockbauer, X. Pan, B. N. Kreiswirth, S. L. Moghazeh, W. R. Jacobs, Jr., A. Telenti, and J. M. Musser. 1997. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob. Agents Chemother. 411677-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takayama, K., E. L. Armstrong, K. A. Kunugi, and J. O. Kilburn. 1979. Inhibition by ethambutol of mycolic acid transfer into the cell wall of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 16240-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Telenti, A., W. J. Philipp, S. Sreevatsan, C. Bernasconi, K. E. Stockbauer, B. Wieles, J. M. Musser, and W. R. Jacobs, Jr. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 3567-570. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, J., K. H. Khoo, S. W. Wu, and D. Chatterjee. 2007. Characterization of a distinct arabinofuranosyltransferase in Mycobacterium smegmatis. J. Am. Chem. Soc. 1299650-9662. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, N., J. B. Torrelles, M. R. McNeil, V. E. Escuyer, K. H. Khoo, P. J. Brennan, and D. Chatterjee. 2003. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol. Microbiol. 5069-76. [DOI] [PubMed] [Google Scholar]
- 34.Zignol, M., M. S. Hosseini, A. Wright, C. L. Weezenbeek, P. Nunn, C. J. Watt, B. G. Williams, and C. Dye. 2006. Global incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 194479-485. [DOI] [PubMed] [Google Scholar]