Abstract
Type IV secretion systems (T4SS) mediate the transfer of DNA and protein substrates to target cells. TrwK, encoded by the conjugative plasmid R388, is a member of the VirB4 family, comprising the largest and most conserved proteins of T4SS. VirB4 was suggested to be an ATPase involved in energizing pilus assembly and substrate transport. However, conflicting experimental evidence concerning VirB4 ATP hydrolase activity was reported. Here, we demonstrate that TrwK is able to hydrolyze ATP in vitro in the absence of its potential macromolecular substrates and other T4SS components. The kinetic parameters of its ATPase activity have been characterized. The TrwK oligomerization state was investigated by analytical ultracentrifugation and electron microscopy, and its effects on ATPase activity were analyzed. The results suggest that the hexameric form of TrwK is the catalytically active state, much like the structurally related protein TrwB, the conjugative coupling protein.
Gram-negative bacteria use type IV secretion systems (T4SS) to transport protein and DNA substrates to phylogenetically diverse eukaryotic and prokaryotic target cells, resulting in pathogenesis and conjugative DNA transfer, respectively (2, 8, 9). Conjugation is a significant medical concern because it is responsible for the widespread transmission of antibiotic resistance genes and virulence factors among pathogenic bacteria (16, 42). A subfamily of T4SS transports only effector proteins to eukaryotic target cells, resulting in bacteria pathogenesis that causes many human diseases, such as gastric ulcers, Legionnaires' disease, brucellosis, and whooping cough (10).
T4SS in conjugative systems are macromolecular assemblies composed of 11 mating pair subunits (VirB1 to VirB11) and a coupling protein (VirD4) that span inner and outer bacterial membranes (18). Three of these subunits, VirB11, VirB4, and VirD4, are putative ATPases that energize DNA and protein substrate transfer as well as pilus assembly. In the conjugative (IncW) plasmid R388, the coupling protein is called TrwB and was shown to be a DNA-dependent ATPase (41). VirB11 (TrwD in R388) was also reported to display ATPase activity (32). However, there is no concluding evidence concerning the potential ATPase activity of VirB4-like proteins.
VirB4 proteins are the largest and most evolutionarily conserved proteins in T4SS (14). An important feature of VirB4 proteins is the presence of Walker A and Walker B motifs (23, 30), found to be essential for virulence and plasmid transfer (5). Interestingly, the VirB4 NTPase activity was reported to be dispensable for T4SS stabilization and pilus formation, suggesting that its role is to energize substrate translocation (45). However, there are conflicting experimental reports on the NTPase activity in VirB4 proteins (30, 38), and the unequivocal demonstration of such activity is needed.
Here, we demonstrate that TrwK, the VirB4 homologue in the conjugative plasmid R388, is able to hydrolyze ATP in vitro. The kinetic properties of this ATPase activity under the conditions tested (i.e., in the absence of macromolecular substrates and other T4SS subunits) were investigated. An analysis of the TrwK oligomeric state by analytical ultracentrifugation and electron microscopy has shown a dependency on salts, pH, and protein concentration. A fraction of the total protein present in the solution is in hexameric form, which is likely to be the catalytically active state, much like VirB11 (44) and VirD4 (17).
MATERIALS AND METHODS
Cloning of TrwK and mutants.
The R388 trwK gene was amplified by PCR and cloned into a pET3a expression vector (Novagen, Madison, WI). The Walker B mutant (D654A) was produced in two PCR steps, using the PCR product of the first reaction (174 bp) containing the mutation (TCCAGAACTCGGCAAACACGTACA) as a primer for a second PCR. The fragment was digested and ligated into the original vector containing the wild-type protein. The plasmids were transformed into Escherichia coli strain C41(DE3) (27).
In vivo complementation assays.
Derivatives of E. coli K12 strain DH5α carrying the plasmid pSU4133, a pR388 variant with a knockout mutation of the trwK gene (6), were transformed together with expression plasmid pET3a containing either the wild-type trwK gene or the trwK(D654A) mutant and mated with UB1637 as described by Moncalian et al. (28). Transconjugants were selected on trimethoprim and streptomycin plates.
Protein purification.
Protein expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 6 h of induction, cells were harvested and suspended in a buffer consisting of 20 mM spermidine, 200 mM NaCl, and 1 mM EDTA and stored at −20°C. Thawed cells were lysed as described by Tato et al. (41). Lysates were collected by centrifugation, diluted four times in buffer A (50 mM Tris-HCl [pH 7.6], 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride) and applied to a HiTrap SP-Sepharose (5-ml) column (Amersham, GE). TrwK-containing fractions were collected in the flowthrough and applied to a HiTrap Q-Sepharose (5-ml) column. TrwK was eluted from this column in a linear gradient of NaCl at a 250 mM salt concentration. The enriched fractions were pooled and diluted to a final concentration of 50 mM NaCl and applied to a second HiTrap Q-Sepharose (5-ml) column. TrwK-containing fractions were pooled, concentrated, and loaded onto a Superdex200 GL10_30 column. After isocratic elution in a buffer consisting of 50 mM HEPES-NaOH (pH 7.2), 150 mM NaCl, 10% (wt/vol) glycerol, 2 mM MgCl2, 0.1 mM EDTA, and 0.5 mM phenylmethylsulfonyl fluoride, the fractions with the richest content in TrwK were pooled and stored at −20°C. For the ATP hydrolysis assays, NaCl and MgCl2 were replaced with potassium acetate and magnesium acetate, respectively. The TrwK(D654A) mutant was purified exactly as the wild-type protein.
Mass spectrometry analysis.
Selected protein bands were excised manually from the gel and subjected to in-gel digestion with trypsin (Roche, Basel, Switzerland) according to the method of Shevchenko et al. (37) with minor modifications. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) spectra were acquired using a Q-Tof Micro mass spectrometer (Waters, Milford, MA) interfaced with a CapLC capillary chromatography system (Waters). Aliquots (8 μl) were loaded onto a Symmetry 300 C18 NanoEase Trap precolumn (Waters) connected to an XBridge BEH130 C18 column (Waters) equilibrated in 5% acetonitrile, 0.1% formic acid. Peptides were eluted with a linear gradient of 10 to 60% acetonitrile directly onto a NanoEase emitter (Waters). Obtained spectra were processed using the ProteinLynx global server (Waters) and searched against NCBI databases using the MASCOT search engine (Matrixscience).
ATP hydrolysis assays.
TrwK ATPase activity was measured by a coupled-enzyme assay (21). To analyze ATP concentration dependency, TrwK (1.85 μM) was incubated with 150 μl of ATP assay buffer, consisting of 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]-NaOH (pH 6.45), 75 mM potassium acetate, 5% (wt/vol) glycerol, 10 mM magnesium acetate, 1 mM potassium chloride, 1 mM dithiothreitol, 0.1 mM EDTA, 0.5 mM phosphoenolpyruvate, 0.25 mM NADH, 60 μg/ml pyruvate kinase, 60 μg/ml lactate dehydrogenase, and 0.1 to 10 mM ATP. The reactions were started by the addition of TrwK. Activity was measured by the decrease in NADH absorbance at 340 nm for 15 min at 37°C in a UV-1603 spectrophotometer (Shimadzu). The same procedure was repeated at pH 5.0 to pH 9.5 using as buffers sodium citrate (pH 5.0), MES (morpholineethanesulfonic acid)-NaOH (pH 5.5 to 5.9), PIPES-NaOH (pH 6.1 to 6.5), HEPES-NaOH (pH 6.8 to 7.25), Tris-HCl (pH 7.5 to 8.5), and sodium borate-NaOH (pH 9 to 9.5).
Analytical ultracentrifugation.
Analysis of the TrwK protein was performed at two protein concentrations (0.4 mg/ml and 0.2 mg/ml) in either HEPES (50 mM HEPES [pH 7.25], 300 mM KAc, 5 mM MgAc, 1 mM Tris[2-carboxyethyl]phosphine [TCEP], 0.1 mM EDTA) or PIPES buffer (50 mM PIPES [pH 6.45], 75 mM KAc, 5 mM MgAc, 1 mM TCEP, 0.1 mM EDTA). Sedimentation velocity runs were carried out at 40,000 rpm and 20°C in an XL-I analytical ultracentrifuge (Beckman-Coulter, Inc.) with UV-visible and interference optics detection systems, using an An50 rotor and 12-mm double-sector centerpieces. Sedimentation profiles were registered every 5 min at the appropriated wavelength (280 nm). The sedimentation coefficient distributions were calculated by least-squares boundary modeling of sedimentation velocity data using the sedimentation coefficient distribution method (36), as implemented in the SEDFIT program. Apparent molar masses associated with each peak were estimated based on the best-fit frictional ratio (f/f0 = 1.42).
Electron microscopy and image analysis.
Aliquots of TrwK (5 μl, 0.1 mg/ml) were applied onto freshly glow-discharged carbon-coated grids. Samples were negatively stained with 2% (wt/vol) uranyl acetate. Electron micrographs were recorded at ×50,000 nominal magnification on Kodak SO-163 film using a JEOL 1200EX-II electron microscope operated at 100 kV. The micrographs were digitized in a Zeiss SCAI scanner with a final sampling rate of 2.33 Å/pixel. A total of 4,907 TrwK particles were selected and normalized using XMIPP image processing software (39). The alignment and classification were performed by maximum likelihood multireference refinement methods (34). The final classes consisted of averages of 200 to 300 particles.
RESULTS
TrwK displays an ATPase activity in vitro.
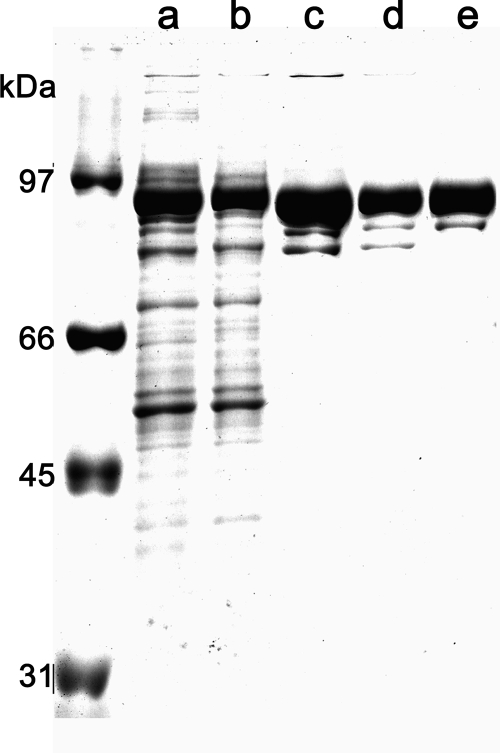
The TrwK protein was purified to homogeneity (Fig. 1). Sample purity was confirmed by MS/MS. Each of the two bands in Fig. 1, lane e, was extracted from the gel and subjected to MS analysis as described in Materials and Methods. The upper, more intense band with an apparent molecular weight (MWapp) of 94 was identified as TrwK (see Fig. S1 in the supplemental material). The band with an MWapp of 80 below the TrwK band was unambiguously identified by peptide fragment fingerprinting as a TrwK deletion product. Comparison of the sequence coverage of both bands revealed that the band with an MWapp of 80 was missing four peptides corresponding to the TrwK N terminus, suggesting an N-terminal deleted protein with a cleavage site in the region close to Arg173 (see Fig. S1 in the supplemental material). ATPase activity was analyzed by the coupled-enzyme method (Fig. 2A). We initially tested reaction conditions similar to those used to assay the ATPase activity of protein TrwB (40, 41), a structural homologue of VirB4 (26). However, because of the low velocity rates obtained in the presence of MgCl2 and NaCl (Fig. 2A, trace b), chloride salts were replaced by the corresponding acetate salts, magnesium acetate and potassium acetate (KAc), respectively. In the presence of these salts, the rate of ATP hydrolysis increased to values of 45 nmol ATP hydrolyzed min−1 mg−1 (Fig. 2A, trace c). ATP hydrolysis rates decreased with increasing concentrations of salt (Fig. 2B), being reduced to 50% at 300 mM KAc and to 5% at 600 mM KAc. In the presence of 75 mM NaCl, ATP hydrolysis rates were 85% lower than those at the same concentration of KAc and negligible at a high NaCl concentration. These findings suggest an inhibitory effect of chloride salts on TrwK ATP hydrolysis. TrwK ATPase activity was at its maximum at a pH of 6.5 and inhibited at a pH of <6 or >7.5 (Fig. 2C).
FIG. 1.
Purification of TrwK. Aliquots of pooled fractions corresponding to the different stages of the purification were resolved in sodium dodecyl sulfate polyacrylamide gels (10%) and stained with Coomassie blue. Lane a, cell lysates; lane b, SP-Sepharose flowthrough; lane c, Q-Sepharose elution; lane d, second Q-Sepharose elution; lane e, isocratic Superdex-200 elution. Both bands in lane e were excised manually from the gel and subjected to in-gel digestion with trypsin and analyzed by LC-MS/MS. Peptide fragment fingerprinting identified the larger band as TrwK and the smaller band as a TrwK N-terminal deletion (see the supplemental material).
FIG. 2.
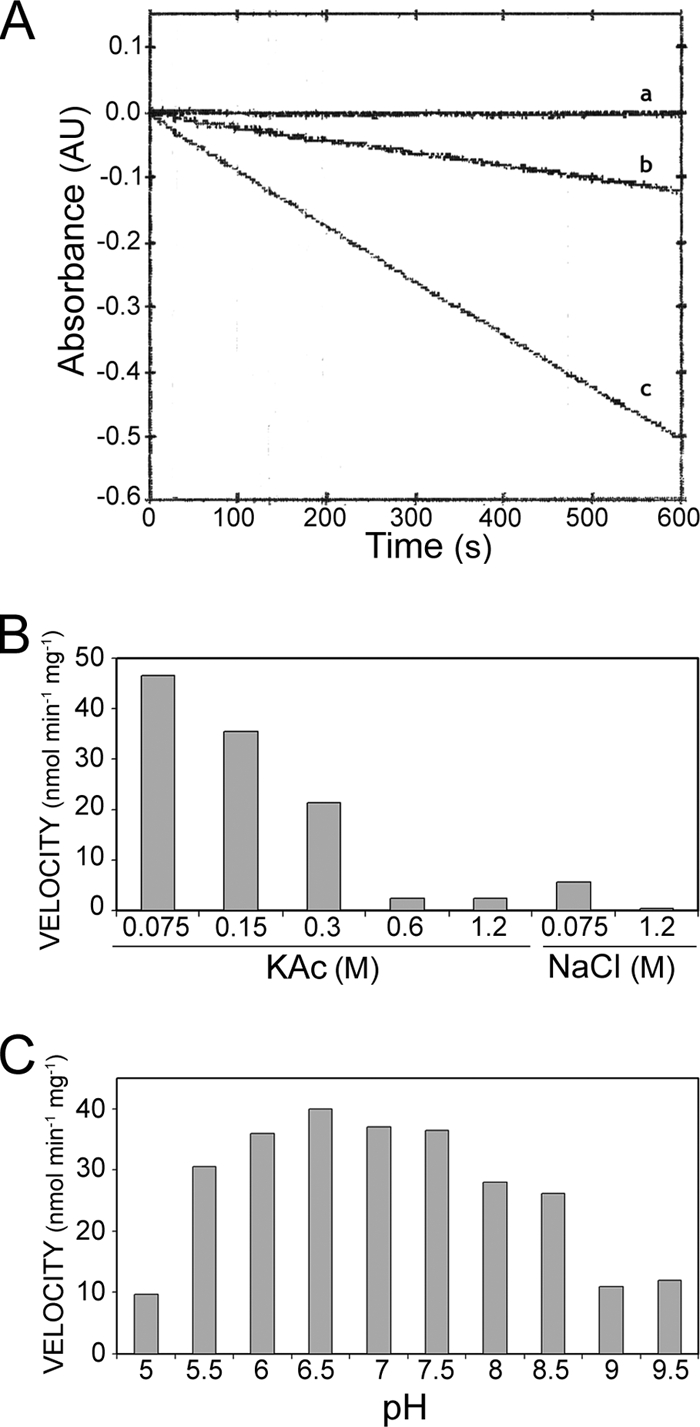
TrwK ATP hydrolase activity. TrwK ATPase activity was monitored by the decrease of NADH absorbance at 340 nm (A) in the presence of 75 mM NaCl (trace b) or 75 mM KAc (trace c). The control is shown in trace a. The effects of salt concentrations (B) and pH (C) are represented.
In vivo and in vitro analysis of a Walker B mutation in TrwK.
In order to discriminate the observed ATPase activity from possible contaminants that could be present in the purified protein sample, an essential aspartic residue in the TrwK Walker B motif was mutated. This mutant [TrwK(D654A)] was subjected to the same purification protocol, and the mutant enzyme behaved exactly as the wild-type enzyme. The ATPase activity of the TrwK(D654A) mutant protein, measured under the same conditions as that of the wild-type TrwK, was undetectable (<1 nmol ATP hydrolyzed min−1 mg−1). These results were corroborated by in vivo complementation assays of cells carrying the plasmid pSU4133 containing the knockout mutation of the trwK gene (6). As shown in Table 1, while wild-type trwK complemented the trwK knockout mutation at a frequency similar to the wild-type situation, the plasmid containing the trwK(D654) mutation was unable to complement the mutation at all (at least a 4-log decrease). This result indicates that the TrwK mutant protein is inactive in conjugation because it lost its ATPase activity.
TABLE 1.
Conjugation frequencies of R388 and trwK mutants in the presence of complementing proteins
| Plasmids in donora | Variant of TrwK in donor | Conjugation frequenciesb |
|---|---|---|
| R388 | TrwK | 4 × 10−1 |
| pSU4133 | R388::ΔtrwK | 1 × 10−6 |
| pSU4133 + pET3a | None | 4 × 10−7 |
| pSU4133 + pET3a_trwK | TrwK | 2 × 10−2 |
| pSU4133 + pET3a_trwK_D654A | TrwK(D654A) | 7 × 10−6 |
Donor cells (E. coli K12 strain DH5α) carrying the plasmids shown in the first column were mated with strain UB1637.
Transfer frequencies of transconjugants selected on streptomycin and trimethoprim plates.
Kinetic analysis of TrwK ATPase activity.
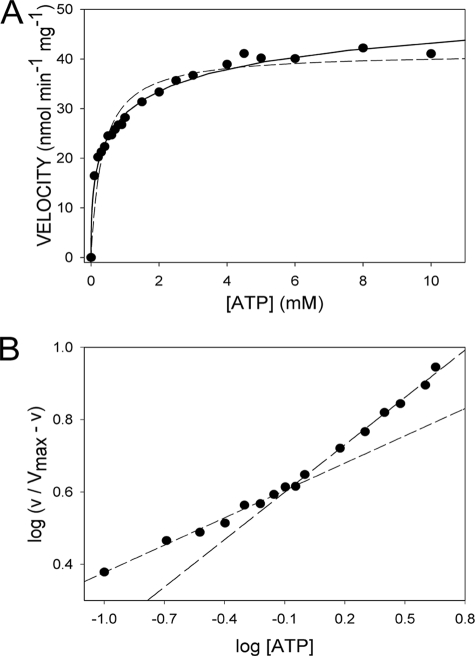
TrwK kinetic parameters for ATP hydrolysis were determined by analyzing the effect of ATP concentration on ATPase activity rates (Fig. 3). Fitting the data to either a Michaelis-Menten (Fig. 3A, dashed line) or a Hill (Fig. 3A, solid line) equation resulted in a better fit for the latter. An empirical Hill equation with an apparent negative cooperativity (Hill, n = 0.5) accounts for the dependence of the ATP hydrolysis rate on ATP concentration. A rough estimate of Vmax, calculated by plotting the double reciprocal of 1/v versus 1/[ATP]0.5, was 48.4 nmol min−1 mg−1, and the K0.5 was 0.82 mM (Km app = 0.7 mM). Then, assuming that the reaction rate is proportional to the fractional saturation of the enzyme, the Hill plot (log[v/(Vmax − v) versus log[ATP]) was drawn and Hill coefficients (nH) of 0.5 and 0.75 were calculated for low (<1 mM) and high (>1 mM) ATP concentrations, respectively (Fig. 3B). This could be an indication of negative cooperativity for ATP hydrolysis if all the binding sites were identical and the quaternary structure was unique.
FIG. 3.
Kinetic analysis of TrwK ATP hydrolysis. (A) ATP hydrolysis rates represented as a function of ATP concentration fit better with a Hill equation (solid line) than with a Michaelis-Menten equation (dashed line). (B) Hill plot for ATP hydrolysis by TrwK. The slope of the line for the low ATP concentrations (<1 mM) and medium ATP concentrations (>1 mM) were estimated as apparent Hill coefficients of 0.5 and 0.75, respectively.
Effect of salts and pH on the ATPase activity and oligomeric state of TrwK.
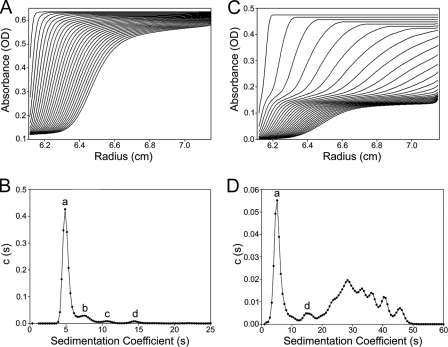
The oligomeric state of TrwK and its relationship with its ATPase activity was investigated. The state of association and degree of homogeneity of TrwK was determined by sedimentation velocity. In HEPES buffer (pH 7.25), the most predominant TrwK species (84.1% of the loading concentration) sedimented with an s value of 5.1 S and an estimated molar mass of ∼89,000, which is compatible with the monomer mass (Fig. 4A and B). The remaining species observed had s values of 8 S (9.6% of the total protein), 11 S (∼3%), and ∼15 S (∼3%), with estimated masses of ∼176,000, ∼282,000, and ∼452,000, that could be identified as dimers, trimers, and hexamers of TrwK, respectively. Interestingly, a sedimentation velocity analysis of TrwK in PIPES buffer (pH 6.45) under conditions similar to those used in the ATPase assays (Fig. 4C and D) revealed the formation of large-size oligomers. Under these conditions only, ∼32% of the protein remained as monomers and ∼5% as hexamers, which could explain the relatively low ATP hydrolysis rates. On the other hand, this polydispersity could also explain the apparent negative cooperativity observed in the kinetic analysis of TrwK ATPase activity.
FIG. 4.
Sedimentation velocity analysis of TrwK. The sedimentation velocity profiles and distribution of sedimentation coefficients were obtained from experiments conducted either in HEPES buffer at pH 7.25 (A, B) or PIPES buffer at pH 6.45 (C, D). The labeled peaks in panels B and D correspond to monomers (a), dimers (b), trimers (c), and hexamers (d). OD, optical density [c(s)].
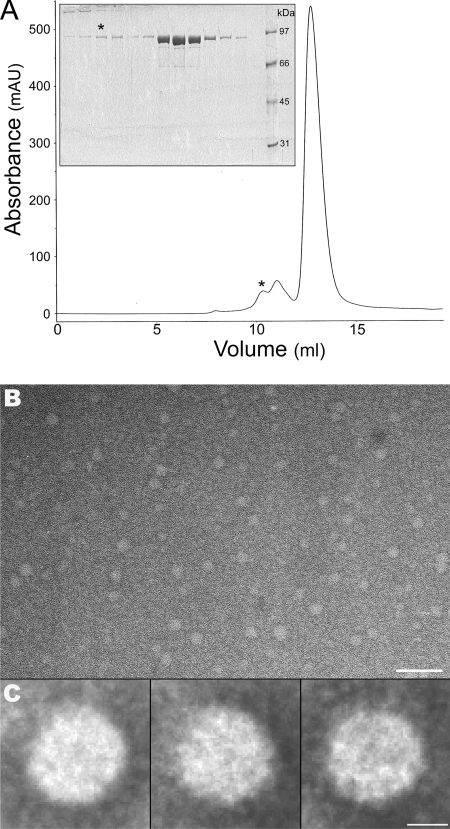
A size exclusion analysis of TrwK by chromatography in a Superdex200 column (Fig. 5A) showed the existence of a peak of MWapp of ∼500, which fits the expected mass of a TrwK hexamer. Aliquots of TrwK corresponding to this fraction were analyzed by electron microscopy. As shown in Fig. 5B, TrwK does in fact form oligomers of round shape and similar size. Image analysis and classification of 4,907 images in class averages of 200 to 300 particles (Fig. 5C) allowed the visualization of ring-shaped structures with dimensions of ∼113 Å in diameter consistent with a hexameric ring similar to that formed by TrwB (17, 19).
FIG. 5.
Electron microscopy of TrwK oligomers. Protein eluted from a size exclusion chromatography column (GL 10/30 Superdex200) at an MWapp of ∼500 (indicated by the asterisk in panel A; insert, sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of fractions from the chromatogram) were stained with uranyl acetate and analyzed by electron microscopy (B; scale bar, 50 nm). Particles (4,907) were selected, aligned, and classified by maximum likelihood multireference alignment methods. Three of the class averages (200 to 300 particles) are represented in panel C (scale bar, 5 nm).
TrwK does not behave as an integral membrane protein.
Since VirB4 proteins have been proposed to be integral inner membrane proteins (11), we decided to study the effect of phospholipids on TrwK ATP hydrolase activity under conditions similar to those described for TrwD (VirB11), another hexameric ATPase in T4SS (32). TrwK (1.8 μM) was incubated with phosphatidylcholine-cardiolipin-Triton X-100 (1:1:3) ternary mixtures (50 μM), and its ATPase activity was measured by the coupled-enzyme assay. No effect was observed on the rate of ATPase hydrolysis after the addition of lipids or detergents (0.01 to 2 mM Triton X-100) (data not shown). These results and, moreover, the fact that no detergent was needed to maintain TrwK in solution suggest that TrwK might not be an integral membrane protein.
To further investigate the membrane association of VirB4 proteins, the amino acid sequences of various members of the VirB4 family were analyzed by several programs that predict transmembrane segments (Sosui, HMMTOP, DAS, TMHMM, and TopPred). Proteins corresponding to widely spread branches of the phylogenetic tree of the VirB4 family (14) were chosen for this analysis. As shown in Table 2, with the exception of IncX members and TrbE of the IncP plasmid RP4, all the analyzed sequences were negative for predicted transmembrane regions. These findings underscore the fact that TrwK behaves as a soluble protein in solution, suggesting that most TrwK-like proteins might not be integral membrane proteins, as previously reported.
TABLE 2.
Membrane topology predictions for VirB4 proteinsa
| Family or source | Protein | Organism | Predicted transmembrane region | Size (aa) |
|---|---|---|---|---|
| PilF/PilH | TrhC R27 | Salmonella enterica serovar Typhi | No | 887 |
| TraC F | Escherichia coli | No | 875 | |
| TraC R100 | E. coli | No | 876 | |
| TrbE family | TrbE RP4 (IncP) | E. coli | Yes (2) | 852 |
| TrbE pTi | Agrobacterium tumefaciens | No | 822 | |
| LvhB4 | Legionella pneumophila | No | 826 | |
| VirB4 | Caulobacter crescentus | No | 794 | |
| Tol plasmids, Xanthomonas | MpfC | Pseudomonas putida | No | 894 |
| VirB4 pXcB | Xanthomonas citri | No | 877 | |
| VirB4 | Wolbachia endosymbiont | No | 801 | |
| VirB4 | Rickettsia felis | No | 810 | |
| Agrobacterium VirB4 | VirB4 | A. tumefaciens | No | 789 |
| VirB4 p42D | Rhizobium etli | No | 795 | |
| VirB4 pSYMA | Sinorhizobium meliloti | No | 792 | |
| VirB4 | Bartonella henselae | No | 784 | |
| PilW/PilN | TraB pKM101 | E. coli | No | 866 |
| TrwK R388 | E. coli | No | 823 | |
| TrwK | Bartonella quintana | No | 823 | |
| PilX, Helicobacter | CagE | Helicobacter pylori | Yes (2) | 981 |
| VirB4 pCL46 | Campylobacter lari | Yes (1) | 924 | |
| CmgB3/4 | Campylobacter jejuni | Yes (1) | 922 | |
| TriC | Yersinia enterocolitica | Yes (2) | 915 | |
| VirB4 pCRY | Yersinia pestis | Yes (1) | 894 | |
| PilX3/4 R6K | E. coli | Yes (1) | 919 | |
| Brucella and cryptic plasmids from soil | PtlC | Bordetella pertussis | No | 824 |
| TraC pIPO2T | No | 826 | ||
| VirB4 | Brucella suis | No | 831 | |
| VirB4 | Xanthomonas campestris | No | 817 | |
| VirB4 | Dichelobacter nodosus | No | 796 | |
| VirB4 pXF51 | Xylella fastidiosa | No | 815 | |
| TraE pF3028 | Haemophilus influenzae | No | 807 |
Amino acid sequences of representative members of widely separated branches of the VirB4 phylogenetic tree analyzed by transmembrane-predicting programs (Sosui, HMMTOP, DAS, TMHMM, and TopPred). DNA sequencing of the intergenic region between PilX3 and PilX4 has revealed that both proteins are fused together in plasmid R6K (unpublished data). Proteins listed below those of a given family but separated from them by a space are evolutionarily close to that family but not members of it.
DISCUSSION
This study demonstrates that TrwK, the VirB4 homologue of the conjugative plasmid R388, displays an ATP hydrolytic activity in the absence of other T4SS subunits or additional cofactors. The similarity among members of the VirB4 protein family is striking (14), with four conserved motifs (including the Walker A and B motifs) and one conserved domain (23, 30). However, previous reports on the biochemical analysis of RP4 TrbEHNΔ2 and R388 TrwK concluded that these proteins were not able to hydrolyze ATP or GTP (30), which is in contrast to the very weak ATPase activity reported for Agrobacterium tumefaciens pTiC58 VirB4 (38). Here, we present strong evidence that demonstrates that TrwK is able to hydrolyze ATP. TrwK ATPase activity cannot be due to a contaminant present in the purified protein preparation, since a protein containing a D654A mutation in the Walker B motif, purified in exactly the same way, was void of in vitro ATPase activity (as well as in vivo complementing activity). A previous report showed that the equivalent residue to D654 in TrbE of RP4 (D694) is essential for in vivo function (30). Interestingly, the choice of magnesium and potassium acetate proved to be essential for the determination of ATPase activity. It is worth noting that in the E. coli cytoplasm, potassium is at high concentrations, whereas sodium amounts are negligible (31). On the other hand, chloride anions can significantly perturb biopolymer processes in E. coli (35), hence, the convenience of replacing them with other univalent anions, such as acetate.
Characterization of the ATPase activity of TrwK revealed that this activity did not fit with classical Michaelis-Menten kinetics but with a Hill equation. Analysis of the kinetic parameters of the reaction showed an nH of less than one, which could imply negative cooperativity for ATP hydrolysis (15, 24, 25). However, a mixture of nonidentical binding sites will also yield nH values of <1. Therefore, negative cooperativity systems should not be identified using only the fact that nH is <1, unless evidence is obtained to prove that all the binding sites are identical (1, 43). To explore the relationship between the oligomeric state of TrwK and its ATPase activity, we analyzed the aggregation properties of the purified protein by analytical ultracentrifugation and electron microscopy. These analyses revealed the presence of a mixture of oligomeric species in the preparation, which could explain the kinetic properties determined for TrwK ATP hydrolysis, rather than negative cooperativity.
VirB4 proteins have been previously reported to form dimers or higher aggregates (12). A model, based on the structural similarities among the Agrobacterium VirB4 C terminus and TrwB, suggests that VirB4 assembles as a hexamer (26). Our experimental analysis indicates that purified TrwK is present mainly as a monomer in solution, which is in agreement with previously reported results (30). However, our sedimentation velocity analysis also revealed the presence of dimers, trimers, and hexamers. Interestingly, an analysis of the oligomeric state of TrwK under conditions similar to those of the ATPase enzymatic assays favored the formation of higher-order oligomers. These multimeric forms have also been observed in analyses by gel filtration and blue native polyacrylamide gel electrophoresis of A. tumefaciens VirB4 (45). It is thus likely that VirB4 proteins function as homohexameric complexes much like VirB11 (33, 44) and VirD4 (17).
VirB4 proteins have been reported to contain four transmembrane domains (11). However, one of the most surprising findings in our studies was the observation that no detergent was needed in the purification of TrwK, neither in the analytical centrifugation analysis nor in the ATP hydrolysis assays. Since the presence of detergents is essential to prevent the aggregation of membrane proteins, we decided to further investigate the putative membrane nature of TrwK. Previous sequence analysis predictions for VirB4 have found from zero (26) to four transmembrane domains (7). Our analyses indicate that with the exception of VirB4 homologues belonging to the IncX branch and TrbE of RP4 plasmid, all the VirB4 proteins were negative for transmembrane segment predictions. Interestingly, a recent sequence analysis in related T4SS identified genes in members of this branch of the VirB4 family codifying a unique polypeptide composed of the VirB3 and VirB4 domains fused together (3), and so far, eight of such chimeric proteins have been found in the databases (9). On the other hand, it is worth noting the absence of VirB3-like protein homologues among members of this subgroup of proteins. Furthermore, we resequenced the pilX3 to pilX4 region of the most representative member of the IncX branch, conjugative plasmid R6K, and found that the sequences of PilX3 and PilX4 correspond to a single polypeptide (see Fig. S2 in the supplemental material), in contrast to the reported R6K annotation (29).
VirB3 is an integral membrane protein originally suggested to be located at the periplasmic face of the outer membrane (20). However, recent analyses demonstrate that VirB3 is located at the inner bacterial membrane (4, 18, 22). Therefore, it is possible that VirB4-like proteins lacking membrane-spanning segments will localize at the inner membrane by interactions with VirB3. In the present work, as TrwK was overproduced in large quantities, most of the recombinant protein was recovered in the soluble fraction. However, in physiological conditions, it is likely that TrwK will be anchored to the inner membrane by interactions with TrwM (VirB3).
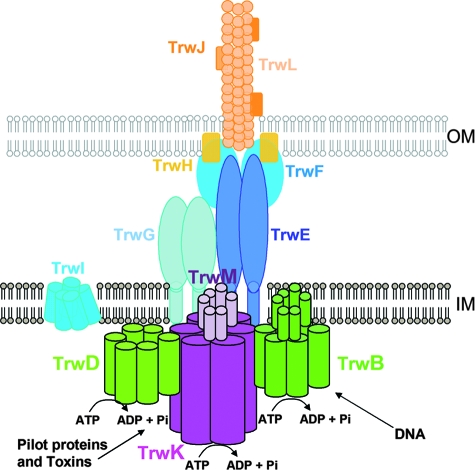
According to our model (Fig. 6), TrwK (VirB4) will be localized at the cytosolic side of the inner membrane assembled as a functional complex with TrwM (VirB3). TrwK oligomerization will promote pilus assembly and stabilization of the core components, TrwG and TrwE (VirB8 and VirB10, respectively), in an ATPase-independent process (45). Once anchored to the membrane, VirB4 (TrwK) will interact with VirB11 (TrwD) (13, 45). TrwD (VirB11) is a hexameric ATPase that could be involved in the unfolding of the proteins or toxins to be transported (33, 44), whereas TrwK will energize substrate transport (45). In conjugative T4SS, the coupling protein TrwB (VirD4) will pump DNA in an ATP-dependent fashion (41).
FIG. 6.
A model showing the three hexameric ATPases in conjugative T4SS. T4SS in conjugative bacteria contain three hexameric ATPases on the cytoplasmic side of the inner membrane. According to our model, TrwK (VirB4) will be anchored to the membrane by interactions with TrwM (VirB3) and will be involved in energizing substrate transport. TrwK oligomerization will also promote pilus assembly and stabilization of the core components, TrwG and TrwE (VirB8 and VirB10, respectively). TrwD (VirB11) might interact directly with the membrane and/or with TrwK (VirB4) and will provide energy to unfold protein substrates so they can be transported through the membrane. TrwB (VirD4) will be directly attached to the membrane by its transmembrane region at its N-terminal domain and will provide the energy to pump DNA. OM, outer membrane; IM, inner membrane.
In summary, this report is the first rigorous demonstration of an ATP hydrolase activity for a VirB4-like protein. This ATPase activity might be essential to energize substrate translocation. On the other hand, our work also provides important insights into the dynamics of TrwK oligomerization, which could be essential in pilus assembly and T4SS stabilization. Finally, the fact that no detergent is needed to keep TrwK in solution suggests that it does not contain membrane-spanning segments and, therefore, under physiological conditions will be anchored to the membrane by interactions with other components of the T4SS.
Supplementary Material
Acknowledgments
We are very grateful to J. M. Arizmendi and K. Aloria for MS analysis performed in the proteomics facility at the University of the Basque Country and to R. Fernández-López and M. P. Garcillán-Barcia for their help with the amino acid sequence analysis.
This work was supported by grants BFU2005-03477/BMC (Spanish Ministry of Education) and LSHM-CT-2005_019023 (European VI Framework Program) to F.C.
Footnotes
Published ahead of print on 6 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abeliovich, H. 2005. An empirical extremum principle for the hill coefficient in ligand-protein interactions showing negative cooperativity. Biophys. J. 8976-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 431359-1365. [DOI] [PubMed] [Google Scholar]
- 3.Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 1503507-3517. [DOI] [PubMed] [Google Scholar]
- 4.Beijersbergen, A., S. J. Smith, and P. J. Hooykaas. 1994. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid 32212-218. [DOI] [PubMed] [Google Scholar]
- 5.Berger, B. R., and P. J. Christie. 1993. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J. Bacteriol. 1751723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolland, S., M. Llosa, P. Avila, and F. de la Cruz. 1990. General organization of the conjugal transfer genes of the IncW plasmid R388 and interactions between R388 and IncN and IncP plasmids. J. Bacteriol. 1725795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, T. B., and M. H. Saier, Jr. 2001. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 1473201-3214. [DOI] [PubMed] [Google Scholar]
- 8.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, T. A., and P. J. Christie. 1997. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang, T. A., X. R. Zhou, B. Graf, and P. J. Christie. 1999. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol. Microbiol. 321239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draper, O., R. Middleton, M. Doucleff, and P. C. Zambryski. 2006. Topology of the VirB4 C terminus in the Agrobacterium tumefaciens VirB/D4 type IV secretion system. J. Biol. Chem. 28137628-37635. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Lopez, R., M. P. Garcillan-Barcia, C. Revilla, M. Lazaro, L. Vielva, and F. de la Cruz. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30942-966. [DOI] [PubMed] [Google Scholar]
- 15.Fersht, A. 1999. Structure and mechanism in protein science. W.H. Freeman & Company, New York, NY.
- 16.Fischer, W., R. Haas, and S. Odenbreit. 2002. Type IV secretion systems in pathogenic bacteria. Int. J. Med. Microbiol. 292159-168. [DOI] [PubMed] [Google Scholar]
- 17.Gomis-Ruth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409637-641. [DOI] [PubMed] [Google Scholar]
- 18.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. 2000. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 1821564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hormaeche, I., I. Alkorta, F. Moro, J. M. Valpuesta, F. M. Goni, and F. De La Cruz. 2002. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J. Biol. Chem. 27746456-46462. [DOI] [PubMed] [Google Scholar]
- 20.Jones, A. L., K. Shirasu, and C. I. Kado. 1994. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J. Bacteriol. 1765255-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreuzer, K. N., and C. V. Jongeneel. 1983. Escherichia coli phage T4 topoisomerase. Methods Enzymol. 100144-160. [DOI] [PubMed] [Google Scholar]
- 22.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2241-15. [DOI] [PubMed] [Google Scholar]
- 23.Lessl, M., D. Balzer, W. Pansegrau, and E. Lanka. 1992. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J. Biol. Chem. 26720471-20480. [PubMed] [Google Scholar]
- 24.Levitzki, A., and D. E. Koshland, Jr. 1969. Negative cooperativity in regulatory enzymes. Proc. Natl. Acad. Sci. USA 621121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levitzki, A., and D. E. Koshland, Jr. 1976. The role of negative cooperativity and half-of-the-sites reactivity in enzyme regulation. Curr. Top. Cell Regul. 101-40. [DOI] [PubMed] [Google Scholar]
- 26.Middleton, R., K. Sjolander, N. Krishnamurthy, J. Foley, and P. Zambryski. 2005. Predicted hexameric structure of the Agrobacterium VirB4 C terminus suggests VirB4 acts as a docking site during type IV secretion. Proc. Natl. Acad. Sci. USA 1021685-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260289-298. [DOI] [PubMed] [Google Scholar]
- 28.Moncalian, G., E. Cabezon, I. Alkorta, M. Valle, F. Moro, J. M. Valpuesta, F. M. Goni, and F. de La Cruz. 1999. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 27436117-36124. [DOI] [PubMed] [Google Scholar]
- 29.Nunez, B., P. Avila, and F. de la Cruz. 1997. Genes involved in conjugative DNA processing of plasmid R6K. Mol. Microbiol. 241157-1168. [DOI] [PubMed] [Google Scholar]
- 30.Rabel, C., A. M. Grahn, R. Lurz, and E. Lanka. 2003. The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J. Bacteriol. 1851045-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Record, M. T., Jr., E. S. Courtenay, S. Cayley, and H. J. Guttman. 1998. Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments. Trends Biochem. Sci. 23190-194. [DOI] [PubMed] [Google Scholar]
- 32.Rivas, S., S. Bolland, E. Cabezon, F. M. Goni, and F. de la Cruz. 1997. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 27225583-25590. [DOI] [PubMed] [Google Scholar]
- 33.Savvides, S. N., H. J. Yeo, M. R. Beck, F. Blaesing, R. Lurz, E. Lanka, R. Buhrdorf, W. Fischer, R. Haas, and G. Waksman. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 221969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheres, S. H., M. Valle, R. Nunez, C. O. Sorzano, R. Marabini, G. T. Herman, and J. M. Carazo. 2005. Maximum-likelihood multi-reference refinement for electron microscopy images. J. Mol. Biol. 348139-149. [DOI] [PubMed] [Google Scholar]
- 35.Schleich, T., and P. H. von Hippel. 1970. Ion-induced water-proton chemical shifts and the conformational stability of macromolecules. Biochemistry 91059-1066. [DOI] [PubMed] [Google Scholar]
- 36.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 781606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 38.Shirasu, K., Z. Koukolikova-Nicola, B. Hohn, and C. I. Kado. 1994. An inner-membrane-associated virulence protein essential for T-DNA transfer from Agrobacterium tumefaciens to plants exhibits ATPase activity and similarities to conjugative transfer genes. Mol. Microbiol. 11581-588. [DOI] [PubMed] [Google Scholar]
- 39.Sorzano, C. O., R. Marabini, J. Velazquez-Muriel, J. R. Bilbao-Castro, S. H. Scheres, J. M. Carazo, and A. Pascual-Montano. 2004. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148194-204. [DOI] [PubMed] [Google Scholar]
- 40.Tato, I., I. Matilla, I. Arechaga, S. Zunzunegui, F. de la Cruz, and E. Cabezon. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 28225569-25576. [DOI] [PubMed] [Google Scholar]
- 41.Tato, I., S. Zunzunegui, F. de la Cruz, and E. Cabezon. 2005. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl. Acad. Sci. USA 1028156-8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waters, V. L. 1999. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 4D433-D456. [DOI] [PubMed] [Google Scholar]
- 43.Weiss, J. N. 1997. The Hill equation revisited: uses and misuses. FASEB J. 11835-841. [PubMed] [Google Scholar]
- 44.Yeo, H. J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 61461-1472. [DOI] [PubMed] [Google Scholar]
- 45.Yuan, Q., A. Carle, C. Gao, D. Sivanesan, K. A. Aly, C. Hoppner, L. Krall, N. Domke, and C. Baron. 2005. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 28026349-26359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.