Abstract
Analysis of publicly available genomes of Streptococcus pneumoniae has led to the identification of a new genomic element containing genes typical of gram-positive pilus islets (PIs). Here, we demonstrate that this genomic region, herein referred to as PI-2 (consisting of pitA, sipA, pitB, srtG1, and srtG2) codes for a second functional pilus in pneumococcus. Polymerization of the PI-2 pilus requires the backbone protein PitB as well as the sortase SrtG1 and the signal peptidase-like protein SipA. Presence of PI-2 correlates with the genotype as defined by multilocus sequence typing and clonal complex (CC). The PI-2-positive CCs are associated with serotypes 1, 2, 7F, 19A, and 19F, considered to be emerging serotypes in both industrialized and developing countries. Interestingly, strains belonging to CC271 (where sequence type 271 is the predicted founder of the CC) contain both PI-1 and PI-2, as revealed by genome analyses. In these strains both pili are surface exposed and independently assembled. Furthermore, in vitro experiments provide evidence that the pilus encoded by PI-2 of S. pneumoniae is involved in adherence. Thus, pneumococci encode at least two types of pili that play a role in the initial host cell contact to the respiratory tract and are potential antigens for inclusion in a new generation of pneumococcal vaccines.
Streptococcus pneumoniae, also known as the pneumococcus, is a frequent colonizer of the human upper respiratory tract. However, this commensal pathogen can cause serious diseases such as pneumonia and meningitis and is known to be the most important vaccine-preventable cause of death in children under 5, affecting up to 1 million children each year (30, 39). Moreover, the World Health Organization estimates that over 90% of these deaths occur in developing countries (19). Despite this high burden of disease, the pathogenic mechanisms exploited by S. pneumoniae are not yet clear. A critical step is colonization of the nasopharynx and the initial interaction of pneumococci with host cells. Recently, pili were discovered in gram-positive bacteria (16, 20, 25, 46), mediating critical host-bacterial interactions, such as adherence to the epithelium and interaction with extracellular matrix proteins, and increasing virulence in mice (1, 4, 21-23, 28). Pili of gram-positive bacteria are composed of covalently linked subunits referred to as pilin subunits. Each pilin subunit has a C-terminal cell wall sorting signal (CWSS), consisting of an LPXTG-like motif required for the covalent attachment to either the cell wall or to other pilin subunits and a hydrophobic domain, followed by a stretch of basic amino acid residues. The pilus is assembled by specialized transpeptidases, also referred to as sortases (42, 43, 45), and is thought to ultimately be linked to the cell wall peptidoglycan by the housekeeping sortase (41).
In S. pneumoniae, the rlrA pilus is encoded by a 14-kb islet, herein referred to as pilus islet 1 (PI-1). PI-1 comprises seven genes: the rlrA transcriptional regulator, three pilus subunits with LPXTG-type CWSSs, and three sortase enzymes involved in synthesis of the pilus polymer and in the incorporation of ancillary pilus components. In PI-1, RrgB is the major subunit that forms the backbone of the structure, while the other two pilins, RrgA and RrgC, are ancillary proteins (4, 15, 18). Pilus backbone subunits of gram-positive bacteria may have two pilin-specific conserved sequences referred to as the pilin motif and the E box (46). The pilin motif is typically centrally located and contains a conserved lysine residue suggested to be critical for subunit cross-linking. Specifically, the lysine residue could provide the free amino group forming the iso-peptide bond that covalently links adjacent pilus subunits within the pilus filament. The E-box motif is another characteristic of certain pilus subunits (45). In Corynebacterium diphtheriae, the conserved glutamate residue in the SpaA backbone protein is required for the incorporation of SpaB (an ancillary protein) into the backbone (44). Recently, Nelson et al. showed that RrgA is the major rlrA pilus adhesin and that bacteria lacking RrgA are significantly less adherent to epithelial cells than wild-type organisms (29). In addition, purified RrgA protein binds to respiratory cells, while bacteria expressing RrgA with disrupted rrgB and rrgC genes exhibit wild-type adherence despite a failure to produce the polymerized pilus. Furthermore, RrgA mediates colonization of the pharyngeal epithelium of mice. Interestingly, similar observations have been made in Streptococcus agalactiae, indicating that rrgA homologues (gbs104, gbs1478, gbs1467, and sak1441, and san1519) are involved in pilus-mediated adherence to human cells (10, 21, 34), while in Streptococcus pyogenes (cpa) and C. diphtheriae both rrgA (spaC, spaF, and spaG) and rrgC (spaB, spaE, and spaI) homologues are defined as pilus-associated adhesins (1, 4, 36, 43, 45, 46).
Based on these data, PI-1 of the pneumococcus emerges as an important factor in colonization. However, PI-1 is not widely distributed in the 92 known pneumococcal serotypes (2, 5, 26, 32). Analysis of a global collection of S. pneumoniae isolates reported the frequency of PI-1 to be ∼30% overall and 50% among antibiotic-resistant strains, demonstrating a correlation between the presence of the islet and the genotype of the isolate (2, 26). Importantly, recent reports describe the spread of certain clones containing the PI-1, suggesting that the pilus may confer a selective fitness advantage (40).
Given that in S. agalactiae (group B streptococcus) and S. pyogenes (group A streptococcus) different pilus-encoding islet types have been identified (43), we investigated whether a similar situation was present in S. pneumoniae. In this report, we describe and characterize the genetic organization of a second PI, PI-2, that was identified in the partial genome sequence of a serotype 1 S. pneumoniae strain (INV104), and we provide evidence that PitB is the backbone protein, while both the sortase SrtG1 and the signal peptidase-related protein SipA, are necessary for assembly and polymerization of the pilus. Distribution of PI-2 in a global collection of clinical isolates was assessed to be 16%, with very high sequence conservation. Furthermore, our data suggest that this pneumococcal PI favors bacterial adhesion to host tissues. The presence of different pilus types reinforces the notion that these structures, being evolutionarily rewarded, may confer a critical selective advantage also for the pneumococcus.
MATERIALS AND METHODS
Strain collection.
A total of 305 isolates collected worldwide (Table 1) have been analyzed for the presence of PI-2. In detail, these isolates include the following: 26 clones from the Pneumococcal Molecular Epidemiological Network (PMEN) collection (24); 50 invasive pneumococcal isolates from the Centers for Disease Control and Prevention (CDC; Atlanta, GA) (6); 55 clinical isolates (28 carriage and 27 meningitis isolates) from Salvador, Brazil; 15 and 36 invasive clinical isolates from Ghana and Bangladesh, respectively (17); 40 clinical isolates from Italy; 5 invasive clinical isolates from Kenya; 8 clinical isolates from Sweden; 43 nasopharyngeal isolates from Norway (40a); and 11 laboratory strains from the Novartis collection and 13 from the University of Alabama. Additionally, data about PI-2 presence have been extrapolated for three isolates from sequence data available at the Sanger website (www.sanger.ac.uk).
TABLE 1.
Composition of the strain collection used to determine PI-2 distribution
| Strain source | Country | No. of isolates | Diagnosis (no. of isolates)a | Reference or source |
|---|---|---|---|---|
| PMEN | Various | 26 | ID | 24 |
| CDC | United States | 50 | ID | 6 |
| Oswaldo Cruz Foundation | Brazil | 55 | ID (27), C (28) | |
| ISS | Italy | 40 | ID (37), C (3) | |
| Swiss Tropical Institute | Ghana | 15 | ID | 17 |
| Dhaka Shishu Hospital | Bangladesh | 36 | ID | |
| Kenyan Medical Research Center | Kenya | 5 | ID | |
| Karolinska Institute | Sweden | 8 | ID | |
| Norwegian Institute of Public Health | Norway | 43 | C | 40a |
| University of Alabama | Various | 13 | ||
| Novartis | Various | 11 | ID | |
| Sanger Institute | Various | 3 | ID | www.sanger.ac.uk |
ID, invasive disease; C, carriage.
PI-2 detection and sequencing.
The INV104 (genomic sequence available at www.sanger.ac.uk) PI-2 nucleotide sequence (srtG2-pitA) was analyzed, and a set of 28 oligonucleotide primers was designed (Table 2): 24 were specific for regions inside the islet, while 2 annealed in two conserved genes (corresponding to SP1008-SP1009 of the S. pneumoniae TIGR4 genome sequence) that flank the islet. The set of primers was used to detect both the presence and location of PI-2 within all the isolates in the collection, as well as for sequence analysis of the entire locus. Briefly, the genomic location of the islet was determined by simultaneously assessing four PCR amplifications (Table 2) as follows: the first primer pairs (1008for and 1009rev) matched in regions flanking the operon (when PI-2 was absent, a lower fragment size was detectable); the second and third amplifications used the primer pairs 1008for-INTrev and INTfor-1009rev, where INTfor and INTrev annealed within the islet (fragments detectable only when the islet was present and inserted in the genomic region comprised between SP1008-SP1009); and the fourth amplified conserved regions within the islet (amplification detected whenever the islet was present in the genome).
TABLE 2.
Nucleotide sequences of the primers used in this study for either detection or sequencing of PI-2a
| Forward primer
|
Reverse primer
|
Purpose | ||
|---|---|---|---|---|
| Name | Sequence (5′-3′) | Name | Sequence (5′-3′) | |
| 1008for | GCTGGATCGAGTTTGAAACCAGAA | 1009 rev | TAAGGATCACCAAAGTCCAAGGCA | PI-2 detection |
| INTfor | ATGGCTTCAGGGGCTATGTTCGGTG | INTrev | TTTCAGTGTATGTTTTAGTGCTTCA | PI-2 detection |
| P01 for | GCTGGATCGAGTTTGAAACCAGAA | P01 rev | TGAACGTTCATGCTATCTCCCTTG | PI-2 sequencing |
| P02 for | TCAAACACATTAATTGACACGAAG | P02 rev | GCAATTGTTAACGAACTAACAATA | PI-2 sequencing |
| P03 for | TTGGCCAAATTGAGCTGGTTGGTA | P03 rev | CTGTTCGTTCTTCTTAGTTTCAATC | PI-2 sequencing |
| P04 for | TGTAACTATATCAGATGTCTTGTCC | P04 rev | AGATTTATTAGGTGATTGTTCACC | PI-2 sequencing |
| P05 for | ACCAGACGCTCCGGGATACACAGT | P05 rev | CCTAGAGGAAGAACGACTCCAAGC | PI-2 sequencing |
| P06 for | CGTGGGTATCAGGTGTCCTATGATAA | P06 rev | GCCTCGTCTTCTAATGACTGTTAC | PI-2 sequencing and detection |
| P07 for | GCAACCTTCCCGATGAAAGTCCCA | P07 rev | TGGTTCTACTTTAGGGGCGTTAG | PI-2 sequencing |
| P08 for | CATATGAAATGCACGTTATTGTTA | P08 rev | GCAAGAACGAGACGCCTAGACAGAC | PI-2 sequencing |
| P09 for | ACGTTACTCCTACTGGTCTCTTGA | P09 rev | TAGATGTTAAAGTCATAGGCATCAA | PI-2 sequencing |
| P10 for | GGGTCATGTTTGGTGATGTGGCTAA | P10 rev | CTGTGCTCGAAGCATCCTTTTCAAT | PI-2 sequencing |
| P11 for | ATGACGGTTCAAAAAAGAGCGCGAT | P11 rev | TAAGGAGTAGAAAGGCTAGAATGT | PI-2 sequencing |
| P12 for | AATAACGGATGCACCGGTGAAAG | P112rev | TAAGGATCACCAAAGTCCAAGGCA | PI-2 sequencing |
Primers are paired by rows.
MLST.
Multilocus sequence typing (MLST) was performed as previously described (12). Briefly, internal fragments of the aroE, gdh, gki, recP, spi, xpt, and ddl genes were amplified by PCR directly from the bacteria using the primer pairs indicated at http://spneumoniae.mlst.net/misc/info.asp#experimental. Sequences were obtained on both DNA strands by use of an ABI 3730xl DNA Analyzer. Alleles from the MLST website (http://spneumoniae.mlst.net) were downloaded for alignment analyses and sequence type (ST) determination. In MLST, an ST is uniquely determined by the allelic profile. New allelic profiles have been submitted to the MLST database for ST assignment.
eBURST analysis.
Clonal complexes (CCs) are groups of STs sharing a recent common ancestor. The eBURST algorithm defines CCs by partitioning the MLST data set into groups of single-locus variants (SLV), i.e., profiles that differ at one of the seven MLST loci (13). This partitioning associates each ST with a CC and also identifies the most likely founder ST, defined as being the ST with the greatest number of SLV within the CC. To explore the relationship between PI-2 presence in our data set and CC, we ran eBURST with default settings on the entire MLST database and subsequently assigned each ST within our data set to a CC. In this work we have named CCs according to the ST number of the eBURST predicted founder.
Gene prediction and multiple alignment.
The Smith-Waterman algorithm with INV104 PI-2 gene sequences was used to search for genes in the previously obtained PI-2 sequences (see “PI-2 detection and sequencing” above). Multiple alignment of the entire islet was obtained with Clustal W, version 1.83 (8). Alignment of the PI-2 locus was performed in nine isolates.
Production of purified proteins and antisera.
pitB and pitA were amplified from chromosomal DNA of serotype 1 strain SPPD by PCR with the following primers: pitBfor, GTGCGTGCTAGCGATGATAATTCAGCAATAACCAAA; pitBrev, CAGCGTCTCGAGGTCGTCGATTTTGTTAGTAACTTT; pitAfor, GTGCGTGCTAGCATGGACGCGGGCATTGGCACTGGG; and pitArev, CAGCGTCTCGAGGCTGTTTTTATTATTCGTGACTGT. PCR products were inserted into the Escherichia coli expression plasmid, pET-21b+ (Invitrogen). The expression vector was transformed into E. coli BL21 Star(DE3), induced with isopropyl-β-d-thiogalactopyranoside, and the protein was purified on a HisTrap column according to the manufacturer's instructions (GE Biotech). Purified recombinant His6-tagged proteins were subsequently used to immunize BALB/c mice (20 μg) (Charles River Laboratory). Polyclonal antisera against His-tagged pneumococcal PI-1 proteins RrgA and RrgB were produced as previously described (4).
Generation of S. pneumoniae mutants.
Serotype 1 PN110 (Istituto Superiore di Sanita [ISS] collection) and serotype 19F Taiwan-14 (PMEN) isogenic mutants described in Table 3 were made by PCR-based overlap extension. Briefly, fragments of approximately 500 bp upstream and downstream of the target gene were amplified by PCR and spliced to an antibiotic cassette (kanamycin or erythromycin); the PCR fragments were then cloned into pGEMt (Promega) and transformed in the appropriate S. pneumoniae strain by conventional methods (3). To select the bacteria in which the target gene was replaced with the antibiotic cassette, bacteria were plated on blood-agar plates with erythromycin (1 μg/ml) or kanamycin (500 μg/ml). Mutants were confirmed by PCR, sequencing and Western blot analysis. Primers used are described in Table 4.
TABLE 3.
Definition of isogenic knock-out mutants generated on PN110 and 19F Taiwan14 in this study
| Straina | Relevant characteristic(s) |
|---|---|
| PN110 wt | Serotype 1 PN110 strain |
| PN110ΔPI2 | PI-2::Kan |
| PN110ΔpitA | pitA::Kan |
| PN110ΔsipA | sipA::Kan |
| PN110ΔpitB | pitB::Kan |
| PN110ΔsrtG1 | srtG1::Kan |
| PN110ΔsrtG2 | srtG2::Kan |
| 19F Taiwan-14 wt | Serotype 19F Taiwan-14 strain (PMEN clone) |
| 19F Tw14ΔPI1 | rlrA::Erm |
| 19F Tw14ΔPI2 | PI-2::Kan |
| 19F Tw14ΔPI1ΔPI2 | rlrA::Erm PI-2::Kan |
wt, wild type.
TABLE 4.
Nucleotide sequences of the primers used to create the knockout mutants
| Genetic region | Primera | Sequence (5′-3′)b |
|---|---|---|
| Kan cassette | Kan 3F | gtcatgatggcctaagtggccaacCTGCAGGAACAGTGAATTGGAGTT |
| Kan 4R | cgatgcaaatttaaatgccggctagCTGCAGCGTTGCGGATGTACTTCA | |
| Erm cassette | Erm 3F | gtcatgatggcctaagtggccaacTCGATACAAATTCCCCGTAGGCGC |
| Erm 4R | cgatgcaaatttaaatgccggctagGAAACAGCAAAGAATGGCGGAAAC | |
| rlrA PI-1 | rlrA 1F | aattgtcgacTATAATCTCCACAGTGGGATTTAC |
| rlrA 2R | gttggccacttaggccatcatgacCAGATGTAAACTTAATAAAGTCCA | |
| rlrA 5F | ctagccggcatttaaatttgcatcgCAGGGATTCGCTCAGTGATTGCTG | |
| rlrA 6R | tttagcggccgcACAAAGAGCCGGAAAAAGGAACAG | |
| PI-2 | pilus-2 1F | aattgtcgacCATAAAATAGAATACAAAACTTC |
| pilus-2 2R | gttggccacttaggccatcatgacTCCAGTGACAACTCATACTGGTCT | |
| pilus-2 5F | ctagccggcatttaaatttgcatcgTTCTATCTACTACAATGGTCGAGA | |
| pilus-2 6R | tttagcggccgcTATTATTTTCTCTATCCATTCTAT | |
| pitA | pitA 1F | aattgtcgacCATAAAATAGAATACAAAACTTC |
| pitA 2R | gttggccacttaggccatcatgacTCCAGTGACAACTCATACTGGTCT | |
| pitA 5F | ctagccggcatttaaatttgcatcgATCGTATAGTGCAATAGTAAATAAT | |
| pitA 6R | tttagcggccgcGGCTAGTACAAATCCAACCATCAG | |
| sipA | sipA 1F | aattgtcgacATCGTATAGTGCAATAGTAAATAAT |
| sipA 2R | gttggccacttaggccatcatgacGGCTAGTACAAATCCAACCATCAG | |
| sipA 5F | ctagccggcatttaaatttgcatcgGACAATCGAACAACGGCTGTAGAC | |
| sipA 6R | tttagcggccgcAGCATGGCCATAAGCAACTCCCGA | |
| pitB | pitB 1F | aattgtcgacAATGACGATGGCATGAAGCCCGCC |
| pitB 2R | gttggccacttaggccatcatgacTGAATACGGACAAAGCGGTTATAG | |
| pitB 5F | ctagccggcatttaaatttgcatcgCTCTTGATTGATAACCTTCCATTC | |
| pitB 6R | tttagcggccgcTGGTGCCCGTAGATAATGGTATTA | |
| srtG1 | srtG1 1F | aattgtcgacTCCAATTCCATATCAAGATTCAAC |
| srtG1 2R | gttggccacttaggccatcatgacATGAGCAGCAAGTGATTGATCACC | |
| srtG1 5F | ctagccggcatttaaatttgcatcgCCTCTACTTGATCCTACGTCGTAG | |
| srtG1 6R | tttagcggccgcAGAGTGTCCGTAGATGATCGTATT | |
| srtG2 | srtG2 1F | aattgtcgacCTATCTAGACCACCTGCTCTCAGT |
| srtG2 2R | gttggccacttaggccatcatgacCACAGAAGAATACCAGAAATACGT | |
| srtG2 5F | ctagccggcatttaaatttgcatcgTGTTTCCTTAACATAACTAATGGA | |
| srtG2 6R | tttagcggccgcTGGTCTTGGGGTCTAACTCCACGA |
Primers F3 and R4 were used to amplify the antibiotic cassettes while F1-R2 and F5-R6 were used to amplify the regions upstream and downstream of the gene of interest.
Uppercase letters represent bases complementary to S. pneumoniae sequence. Lowercase letters represent bases added to facilitate mutagenesis.
Biochemical analysis of S. pneumoniae pili and immunoblotting.
Mutanolysin extracts for pilus polymerization analysis were obtained as previously described (4). Samples were loaded onto 4 to 12% or 3 to 8% NuPage Tris gels (Invitrogen) and transferred to nitrocellulose membranes. The membranes were then immunoblotted with serum against the pneumococcal antigens and with secondary antibodies conjugated to alkaline phosphatase (Promega). Membranes were developed according to manufacturer's instructions.
Flow cytometry.
Bacteria were grown in Todd-Hewitt broth containing yeast extract to exponential phase (optical density at 600 nm [OD600] of 0.2), fixed with 2% paraformaldehyde, and then treated with mouse antisera raised against PitB recombinant protein (anti-PitB antibody, dilution 1:100). After samples were labeled with a fluorescein isothiocyanate-conjugated secondary antibody (Jackson Laboratories), bacterial staining was analyzed using a FACScan flow cytometer (Becton Dickinson). Sera from mice immunized with phosphate-buffered saline (PBS) plus adjuvant were used as negative controls.
Cell culture techniques.
A549 (respiratory epithelium), 16 HBE 14o− (bronchial epithelium), Detroit (nasopharyngeal epithelium), and Hep-2C (laryngeal epithelium) cells were grown on coverslips to confluence in polystyrene 12-well plates (Corning). Cell culture medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with 10% heat-inactivated fetal calf serum and 2 mM glutamine) was produced by the Novartis Vaccines and Diagnostics laboratories.
Adherence assays.
Before use, the monolayers were washed two times with PBS. S. pneumoniae cells grown to early log phase (OD600 of 0.2) in Todd-Hewitt broth containing yeast extract were resuspended in cell culture medium without serum. Bacterial suspensions were applied to cell monolayers at a multiplicity of infection of approximately 100, centrifuged at 1,000 rpm for 2 min at 4°C, and subsequently incubated for 1 h at 37°C in 5% CO2-95% air atmosphere. After incubation, the infected monolayers were washed three times with PBS to remove nonadherent bacteria, fixed in 2.5% paraformaldehyde for 15 min, washed three times with PBS, and stained. For fluorescence microscopy, the coverslips were incubated in PBS with 1% saponin and 3% bovine serum albumin (BSA) and then labeled for 1 h with primary antibodies in the same solution. After the cells were washed twice, secondary antibodies and phalloidin were added for 30 min. Bacteria per 100 epithelial cells were counted by fluorescence microscopy, and three independent determinations were made. Significant differences were detected by a Student's t test. The number of cells that was counted in total for each individual experiment was ≥400. For inhibition experiments using antiserum, bacteria were incubated for 15 min at 37°C with anti-PitB, anti-green fluorescent protein (GFP), or anti-RrgA antiserum or medium alone. Then, bacteria were added to A549 cells and incubated for 1 h at 37°C in a 5% CO2-95% air atmosphere.
Protein binding assays.
A549 cells were nonenzymatically detached from the support by using cell dissociation solution (Sigma), harvested, and resuspended in DMEM supplemented with 1% BSA in the absence of serum and antibiotics. The cells were mixed with either medium alone or four concentrations (5, 50,100, and 200 μg/ml) of the purified proteins, resuspended in medium, and incubated for 2 h on ice. A549 cells were then washed twice with 1% BSA in PBS and incubated with antibodies against each protein for 1 h on ice. After two additional washes, the preparations were incubated with Alexa Fluor 488 secondary antibodies (Molecular Probes), and 10,000 cells were analyzed with a FACSCalibur flow cytometer. Additionally, binding assays were performed on adherent A549 respiratory epithelial cell monolayers grown on 13-mm glass coverslips. A DMEM-based solution containing 100 μg/ml of purified protein in the absence of serum and antibiotics was added to cell monolayers and incubated for 2 h on ice. Negative control wells were treated and analyzed in parallel with medium in the absence of exogenous proteins. Subsequently, cells were washed three times with PBS and fixed with 2.5% paraformaldehyde. Cells were then incubated with antibodies against each protein for 1 h at room temperature. After two additional washes, the preparations were incubated with Alexa Fluor 488 secondary antibodies and phalloidin (Molecular Probes) and visualized by confocal microscopy (Bio-Rad Radiance 2100). Antibody cross-reactivity was negative by immunofluorescence and fluorescence-activated cell sorting (FACS) analysis (data not shown).
Immunogold labeling and electron microscopy.
Immunogold electron microscopy was performed as previously described (4). Briefly, bacteria were grown overnight on blood-agar plates, resuspended in PBS, charged onto Formvar-coated nickel grids, allowed to stand for 5 min, and subsequently fixed with 2% paraformaldehyde and 1× PBS before being labeled with 1:10 dilutions of polyclonal anti-PitB and/or anti-RrgB antiserum in blocking buffer (1% BSA in PBS). Samples were then washed with blocking buffer and subsequently incubated with a 1:20 dilution of a secondary goat anti-mouse immunoglobulin G (5 nm or 20 nm) or goat anti-guinea pig immunoglobulin G (10 nm) conjugated to gold particles (BB International). Finally, samples were washed with 5 drops of distilled water and stained with 1% phosphotungstic acid before analysis in a CM10 transmission electron microscope (Philips Electronic Instruments, Inc.) operating at 80 kV.
Statistical analyses.
For adherence assays, data were analyzed by a Student's t test.
Nucleotide sequence accession numbers.
The following GenBank accession numbers were assigned to S. pneumoniae PI-2: serotype 7F strain 32_14, EU311532; serotype 19F strain 5167-99, EU311533; serotype 2 strain 31620, EU311534; serotype 1 strain PN110, EU311535; serotype 19F strain pgx1416, EU311536; serotype 19F strain SP231, EU311537; serotype 1 strain SPPD, EU311538; and serotype 19F strain Taiwan-14, EU311539.
RESULTS
Genomic organization of PI-2 in S. pneumoniae.
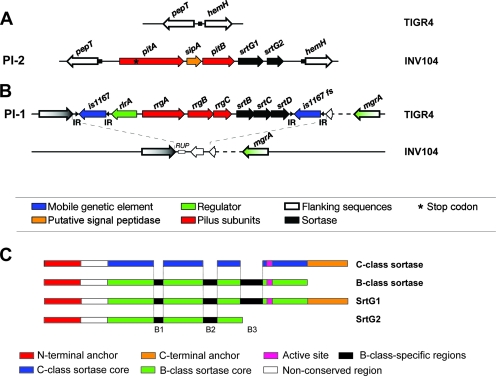
Previously, our laboratory demonstrated that the rlrA pilus proteins RrgA, RrgB, and RrgC are covalently linked to form a high-molecular-weight (HMW) pilus structure, encoded by PI-1 (4, 18). In the search for additional pilus structures, we analyzed all the publicly available S. pneumoniae genome sequences and found a region whose organization resembled the pilus-encoding islets in S. pyogenes (group A streptococcus) (25) in the serotype 1 INV104 strain (www.sanger.ac.uk). This newly identified genetic region, herein named PI-2, is composed of five genes: two genes encoding products displaying similarity to sortases (srtG1 and srtG2), a gene encoding a signal peptidase-related product (sipA), and two genes coding for putative LPXTG-type surface-anchored proteins (pitB and pitA, the latter containing a stop codon) (Fig. 1A). PI-2 is 6,575 bp in length and has a G+C content of 38.5%, similar to the overall G+C content of S. pneumoniae. In the strains where the islet is absent, the flanking genes pepT and hemH are separated by a noncoding region of approximately 140 bp that contains a putative 7-bp insertion site (TCCTTTT), which is found duplicated at the boundaries of PI-2 (Fig. 1A). INV104 does not contain any remnants of PI-1 at the locus that corresponds to the site where PI-1 is inserted in TIGR4 (Fig. 1B). Homology searches revealed that PitB and PitA share 27% and 25% sequence conservation, respectively, with the backbone and ancillary proteins of a pilus produced by S. pyogenes. These data suggest that PitB is the most probable backbone of the pilus and PitA, a putative ancillary protein. Detailed investigation of PitB and PitA amino acid sequences highlighted the presence of motifs characteristic of gram-positive pilus subunits: a signal peptide and a noncanonical CWSS containing the sequences VTPTG (PitB) or VPETG (PitA). In addition, PitB is predicted to have both a pilin motif, FKENNKSNAPKV, found at residue 200 and an E-box motif, YTVTETGVAGY, at the C-terminal region (highly conserved residues are indicated in boldface).
FIG. 1.
The genomic organization of pilus-encoding islets in S. pneumoniae. Schematic representation of PI-2 (A) and PI-1 (B) genomic regions in TIGR4 and INV104 strains. TIGR4 is positive for PI-1 and negative for PI-2, whereas INV104 is positive for PI-2 and negative for PI-1 presence. Genes coding for proteins with different roles are represented with different colors, as shown. (C) Schematic representations of the architectures of class B and C sortases and comparison with those of SrtG1 and SrtG2. Functional elements are shown in different colors, as indicated.
Distribution of PI-2 in a global collection of clinical isolates.
In order to determine the PI-2 prevalence, a global collection of 305 clinical S. pneumoniae isolates was analyzed (Table 1). The collection represents 43 serotypes and 148 diverse STs, which can be grouped into 70 CCs (by eBURST analyses) and which include isolates from invasive disease as well as nasopharyngeal carriage. The prevalence of PI-1 in this collection is 31.5% (96 positive isolates), confirming previously published data (2, 5, 26). PI-2 presence was determined by PCR analyses as described in the Materials and Methods section. Overall, 50 isolates contained PI-2 (16.4%), which was found consistently inserted in the same genomic region. As previously demonstrated for PI-1(2, 26), presence of PI-2 correlates with the genotype defined by CC. Furthermore, prevalence of the pilus is the same in both invasive disease and carriage isolates. When the collection was stratified by serotype, the serotypes tested were not homogeneous for PI-2 presence (data not shown), confirming that there is no correlation between the PI-2 and serotype. Of all the strains tested, PI-2 is present in six different CCs (74, 191, 251, 271, 304, and 306) out of 70 tested (8.6%), corresponding to serotypes 2, 7F, 19A, 19F, and 1 (Table 5). Interestingly, only CC271 from this strain collection contains both PI-2 and PI-1.
TABLE 5.
Distribution of PI-2 in the S. pneumoniae strain collectiona
| CC | Serotype | Disease outcomec | PI-2 | PI-1 | Strain name(s) (serotype, ST) |
|---|---|---|---|---|---|
| 74 | 2 | ID | Yes | No | 31620 (2, 74) |
| 191 | 7F | C, ID | Yes | No | 32_14 (7F, 191) |
| 251 | 19F | Yes | No | 5167-99 (19F, 251) | |
| 271b | 19A, 19F | C, ID | Yes | Yes | 19F Taiwan-14 (19F, 236), pgx1416 (19F, 271), sp231 (19F, 271) |
| 304 | 1 | ID | Yes | No | PN110 (1, 304) |
| 306 | 1 | ID | Yes | No | INV104 (1, 227), SPPD (1, 306) |
CCs positive for PI-2, the serotype to which they are associated, and their provenance are shown (n = 305 isolates).
CC271 was associated with both pilus-encoding islets.
ID, invasive disease; C, carriage.
Conservation of PI-2 sequences.
Nine isolates representing the six positive CCs were analyzed for sequence variability of PI-2: INV104, SPPD, PN110, 31620, 32_14, 5167-99, SP231, PGX1416, and 19F Taiwan-14 (Table 5). Multiple sequence alignment of the nine PI-2s by Clustal W revealed an overall conservation of ∼99% along the entire islet. Further analysis of the pitA sequences revealed the presence of the previously mentioned stop codon (UGA) in all the isolates, as well as an additional frameshift following the stop codon in strain 5167-99. Moreover, the putative sortase srtG2 (99.2% conserved) is complete only in isolate 5167-99, whereas in the other isolates tested either the C-terminal part of the gene product containing the active cysteine residue is missing or a dinucleotide insertion in the 5′ region of srtG2 results in a nonfunctional pseudogene. Recently, a classification system for the vast variety of sortases that emerge from genomic sequencing of various microorganisms has been proposed, resulting in the definition of four major classes, designated A to D (11). Interestingly, the srtG1 gene product cannot be easily classified: while the major part of the sequence is highly similar to B-type sortases, SrtG1 possesses the N- and C-terminal transmembrane regions typical for C-type sortases (Fig. 1C). Moreover, the LPXTG-like motif of its putative substrate PitB, VTPTG (see above), does not correspond to any known consensus sequence as described by Comfort and Clubb (9). For this reason we propose the name srtG for the sortase genes encoded by PI-2.
The backbone protein PitB is surface exposed and assembled into a pilus.
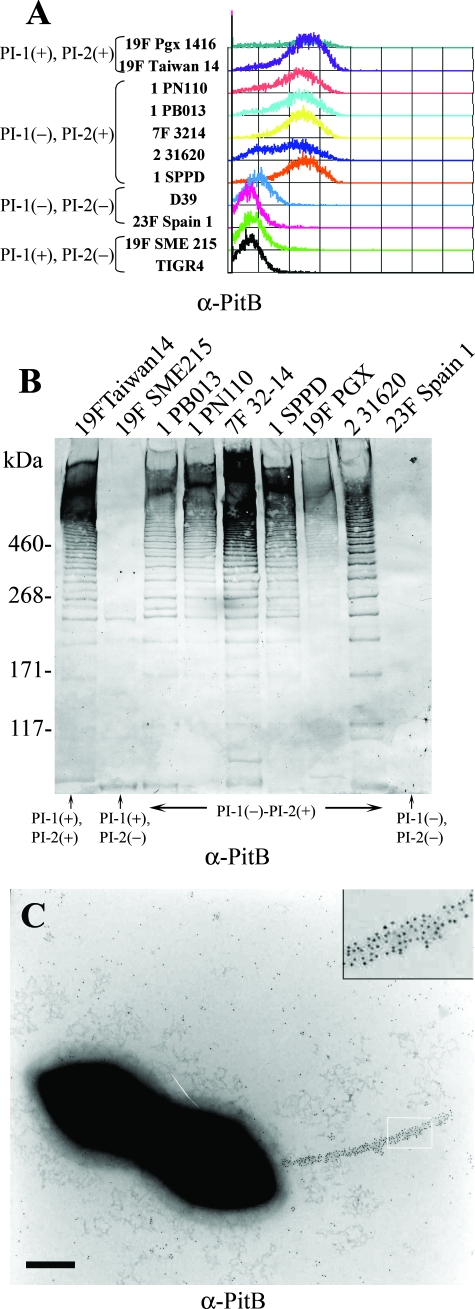
To determine if the putative cell wall-anchored PitB was expressed and assembled into a pilus structure, mouse serum was raised against a His-tagged PitB recombinant protein and used to label bacteria grown in liquid culture at mid-log phase. The PI-2-positive isolates were specifically recognized by anti-PitB antibody upon FACS analyses, whereas the isolates lacking PI-2 showed a background level of staining (Fig. 2A). As expected on the basis of the high level of sequence conservation, the antibody was able to detect efficiently the protein expressed by all the isolates. In addition, immunoblotting with anti-PitB antibody on mutanolysin cell wall extracts was performed on several strains (both PI-2 positive and negative). As shown in Fig. 2B, a typical HMW ladder was detected only in PI-2-positive isolates by immunoblot analysis with anti-PitB antibody. Detection of an HMW ladder is indicative for the presence of extended pili, resulting from the fact that in pili of gram-positive bacteria there is always a statistical distribution of covalently assembled pilus filaments of various lengths, with the shorter ones entering the gel while the longer ones remain close to the starting point. Furthermore, immunogold electron microscopy, with anti-PitB antibody on the wild-type serotype 1 strain PN110 showed localization of the protein along a pilus structure (typically one to two per cell). As shown in Fig. 2C, pili were present on the surface of the bacteria and were decorated by 5-nm gold particles.
FIG. 2.
Detection of a functional surface-exposed pilus structure encoded by PI-2. (A) FACS analysis performed on S. pneumoniae clinical isolates (OD600 of 0.2) containing PI-1, PI-2, or both (presence or absence indicated by a plus or minus sign, respectively) labeled with mouse polyclonal PitB antiserum (secondary antibody was fluorescein isothiocyanate labeled). (B) Immunoblot analysis of different S. pneumoniae mutanolysin extracts reacted with mouse polyclonal anti-PitB antiserum. (C) Immunogold localization of PitB in pili of S. pneumoniae PN110 whole cells. Inset shows an enlarged portion of the pilus. α, anti.
PitB is polymerized into the pilus structure in an SrtG1- and SipA-dependent manner.
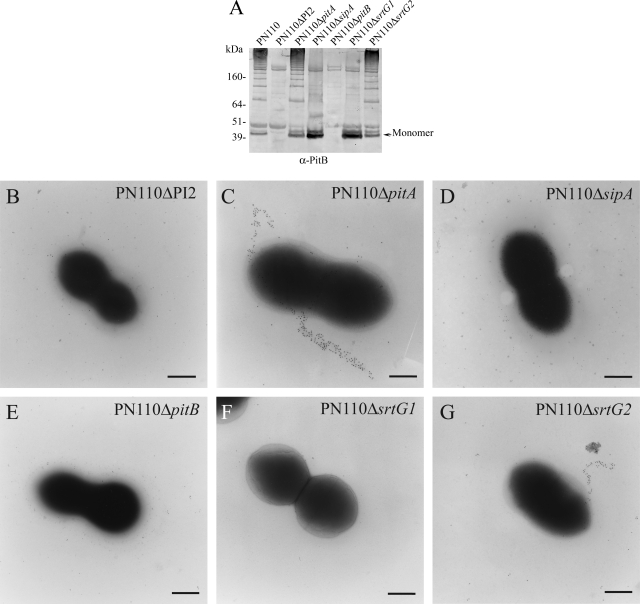
In order to demonstrate the requirement of PI-2 and the islet-specific genes in the assembly of the pilus, a panel of mutants was constructed by insertional mutagenesis in the strain PN110 (Table 3). Specifically, isogenic mutants containing deletions of the entire PI-2 or the individual open reading frames contained in the islet were created. Figure 3 shows the typical HMW ladder detected by Western blotting of the mutanolysin extract in the mutants (panel A), and immunogold electron microscopy (panel B) performed with anti-PitB antibody on the strain lacking PI-2 (PN110ΔPI2), confirming that the presence of the islet is necessary for pilus assembly. Accordingly, the pilus was not formed by a mutant strain containing a deletion of pitB (Fig. 3A and E). Furthermore, lack of the putative sortase SrtG1 impairs polymerization of PitB without affecting its expression (Fig. 3A and F). Indeed, in the PN110ΔsrtG1 mutant, the pilus backbone protein is found in mutanolysin cell extracts as a monomer, suggesting that SrtG1 acts as the sortase involved in the pilus polymerization.
FIG. 3.
Effect of PI-2 gene deletions on PI-2 polymerization in S. pneumoniae Serotype 1. (A) Western blot performed with anti-PitB antibodies on mutanolysin extracts of PN110 knockout isogenic mutants (Table 3). The PitB monomer is indicated. (B to G) Immunogold labeling with anti-PitB antibodies of whole-cell PN110 deletion mutants. Mutants PN110ΔPI2 (B), PN110ΔsipA (D), PN110ΔpitB (E), and PN110ΔsrtG1 (F) show lack of pilus formation. Panels C and G represent PN110ΔpitA and PN110ΔsrtG2, respectively. Bacteria were charged on Formvar carbon grids and immunogold decorated with mouse anti-PitB (gold particle size, 5 nm). Scale bar, 0.2 μm. α, anti.
We observed the same results with the isogenic mutant of sipA, confirming recent data obtained in S. pyogenes, where the essentiality of the signal peptidase-like protein for pilus assembly has been demonstrated (Fig. 3A and D) and an alternative function as a chaperone has been proposed (47). In contrast, the expression or polymerization of PitB was not affected in isogenic mutants of the putative ancillary pilus subunit PitA and the sortase-like SrtG2 (Fig. 3A, C, and G). These observations demonstrate that PitA and SrtG2 are dispensable for pilus polymerization.
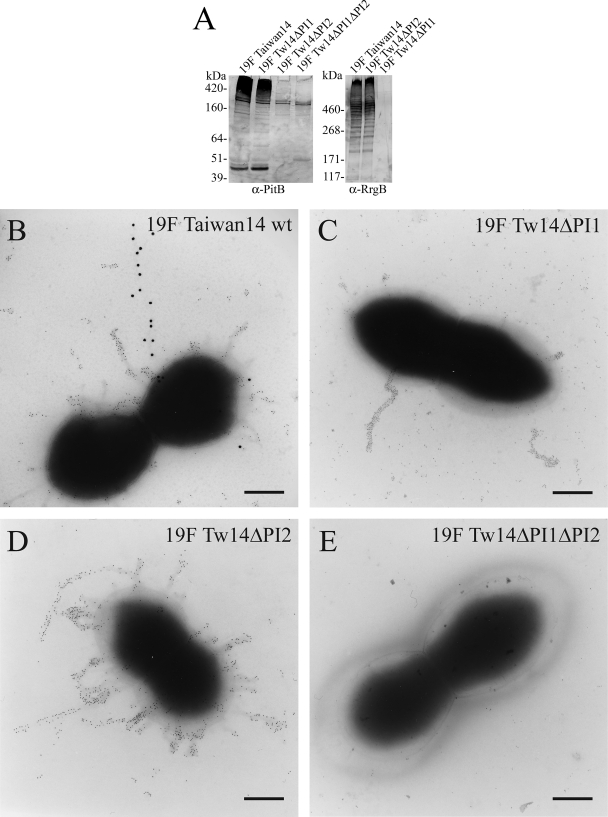
Additionally, insertional mutants were generated on the 19F Taiwan-14 strain containing both PIs by alternatively deleting PI-1, PI-2, or both. Western blot analysis and immunogold electron microscopy (Fig. 4) performed with anti-PitB and anti-RrgB antibodies showed that both pili are assembled on the surface of 19F Taiwan-14 and that the assembly of the two pili is independent.
FIG. 4.
S. pneumoniae 19F Taiwan-14 strain expresses two independent pili. (A) Western blotting performed with polyclonal anti-PitB and anti-RrgB antibodies on mutanolysin extracts of 19F Taiwan-14 wild-type and knockout isogenic mutants lacking PI-1 and/or PI-2 (19F Tw14ΔPI1, Tw14ΔPI2, and Tw14ΔPI1ΔPI2). (B and E) Double immunogold labeling performed with mouse anti-PitB (gold particle size, 20 nm) and guinea pig anti-RrgB (gold particle size, 5 nm) on 19F Taiwan-14 wild type (wt) and 19F Tw14ΔPI1ΔPI2. (C) Deletion mutant 19F Tw14ΔPI1 labeled with mouse anti-PitB (gold particle size, 5 nm). (D) 19F Tw14ΔPI2 labeled with mouse anti-RrgB (gold particle size, 5 nm). Scale bar, 0.2 μm. α, anti.
PI-2 pilus mediates adhesion to respiratory cells.
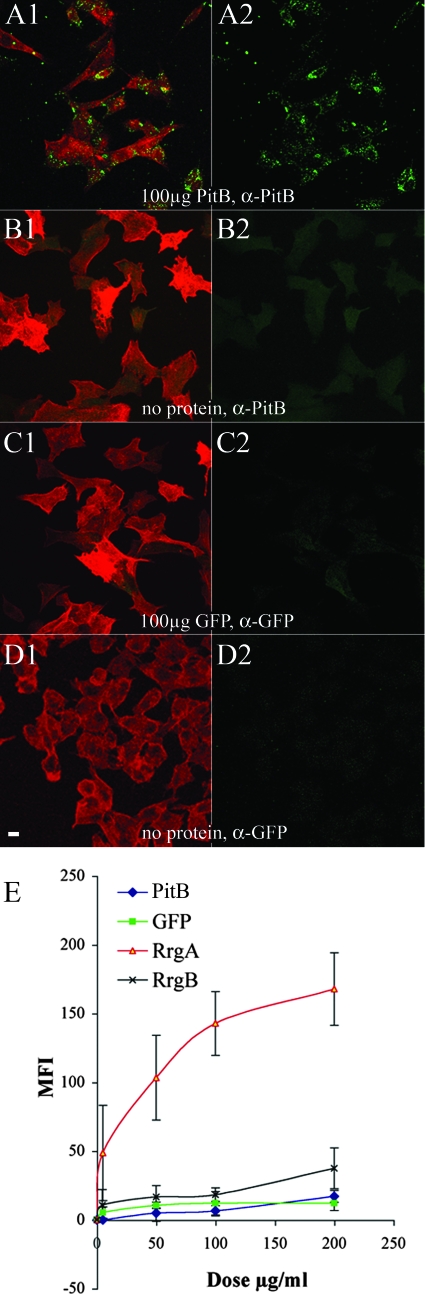
Several lines of evidence indicate that pili play a major role in adhesion to host epithelial cells. To address whether the pilus backbone protein PitB was involved in adherence to host cells, we incubated recombinant PitB protein with A549 human respiratory epithelial cells. Adherent A549 cell monolayers were incubated with medium containing 100 μg/ml of PitB in the absence of serum. The cells were then washed in PBS, and the protein was subsequently detected immunologically by confocal microscopy. The pilus backbone protein PitB showed low affinity for the cells with a diffuse binding pattern (Fig. 5A1 and A2). However, GFP did not bind to A549 monolayers at a dose equivalent to that used for PitB. Binding of the pilus backbone was also evaluated by incubating the purified protein with A549 cells in suspension. The level of adherence was determined by FACS analysis at a range of protein doses (5, 50, 100, and 200 μg/ml). For comparison, we used the purified ancillary protein RrgA as well as the backbone subunit RrgB of the PI-1 pilus. In this assay, cell suspensions were incubated with the purified proteins. Levels of adherence determined by FACS showed that PitB, RrgB, and GFP, at previously described doses, had comparable mean fluorescence intensity (MFI) values, which were significantly lower than the MFI of RrgA (Fig. 5E).
FIG. 5.
(A1 to D2) Adherence assays of purified backbone protein to A549 cells. Cells grown on coverslips were incubated with 100 μg/ml of purified PitB (A1 and A2) or GFP (C1 and C2) in DMEM (B1 and B2) or with medium alone (D1 and D2) for 2 h at 4°C. Cells were labeled with anti-PitB (A1, A2, B1, and B2) and anti-GFP antibodies (C1, C2, D1, and D2) and phalloidin (A1, B1, C1, and D1). Imaging was performed with a confocal microscope. Scale bar, 10 μm. (E) Binding of purified proteins to A549 cells in suspension. The level of adherence was determined by FACS analysis at a range of protein doses (0, 5, 50, 100, and 200 μg/ml) of RrgA (triangles), RrgB (crosses), PitB (diamonds), and GFP (squares). Proteins were incubated for 2 h at 4°C, labeled with antiserum specific to each protein, and detected with Alexa Fluor 488-conjugated secondary antibodies. RrgA and GFP were used, respectively, as positive and negative controls. Cells were analyzed with a FACSCalibur flow cytometer, and the net MFI for each population was calculated from three independent experiments. Nonsignificant differences were detected between PitB, GFP, and RrgB (P > 0.05), and only RrgA was significantly different from GFP binding (P < 0.05). α, anti.
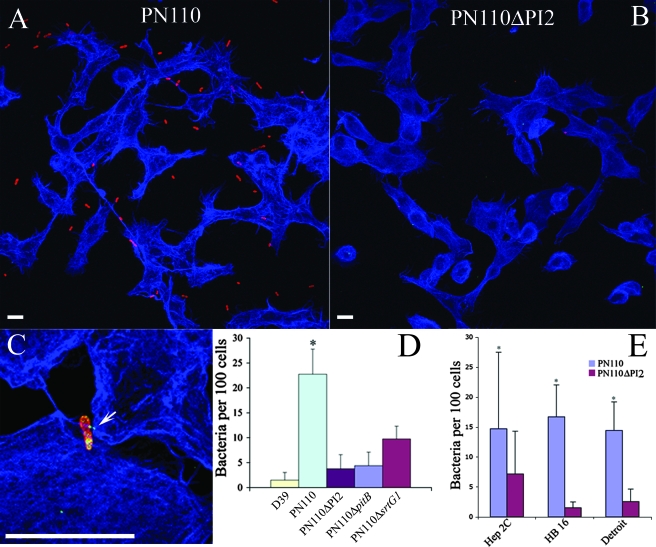
In order to confirm the adhesive properties of the backbone protein, we infected A549 cell monolayers with serotype 1 S. pneumoniae PN110. After 1 h of infection (multiplicity of infection of 100), strain PN110 showed a significant increase in the level of adherence, greater than the insertion-deletion mutant lacking the entire PI (PN110ΔPI2). Interestingly, PN110ΔPI2 bound at levels comparable to the nonpiliated serotype 2, strain D39. Furthermore, PN110 lacking pitB or the sortase srtG1 showed levels of adherence significantly lower than the wild-type parental strain (Fig. 6A to D).
FIG. 6.
Cocultivation experiments. (A and B) A549 cells cocultivated with pneumococcal strains labeled with Omniserum primary antibodies and analyzed by confocal microscopy. (A) PN110 wild type. (B) PN110ΔPI2. (C) Confocal three-dimensional reconstruction of A549 cells cocultivated with PN110 wild type labeled with Omniserum and mouse anti-PitB antibodies. The arrow indicates the location of an extended pilus contacting host cells. In panels A to C, bacteria were visualized with Alexa Fluor 568-conjugated secondary antibodies (red), A549 cells were visualized with phalloidin conjugated to Alexa Fluor 647 secondary antibodies (blue), and PitB was visualized with Alexa Fluor 488-conjugated secondary antibodies (green). Scale bar, 10 μm. (D) Adherence quantification of different strains on A549 cells. (E) Adherence quantification of PN110 wild type and PN110ΔPI2 on various cell lines. For both panels D and E, bacteria were counted in five different microscope fields, and average results of three independent experiments are shown. Significant differences were detected by a Student's t test (*P < 0.05).
Furthermore, we tested the ability of piliated pneumococcus to adhere to three additional cell lines: 16 HBE 14o− (bronchial epithelium), Detroit (nasopharyngeal line), and Hep-2C (laryngeal line). Strain PN110 adhered significantly better than the pilus knockout mutant PN110ΔPI2 to all the cell lines tested (Fig. 6E). To further confirm the role of the pilus in adhesion to epithelial cells, we incubated PN110 bacteria with anti-PitB antibody and then infected A549 cell monolayers. Presence of the antiserum significantly inhibited adhesion to the cells, decreasing the number of bacteria from about 20 per 100 cells without antiserum or with anti-GFP or anti-RrgA antiserum to 4 per 100 cells in the presence of anti-PitB antibody (P < 0.05) (data not shown).
DISCUSSION
In the present work, we report the identification of a second PI, PI-2, in S. pneumoniae, a major cause of disease in children (19, 30). Pili or fimbriae are extracellular organelles located on the surface of bacteria. In gram-negative pathogens, pili are important virulence factors involved in conjugation between bacteria, adhesion to the host, motility, and transfer of effector molecules (27, 35, 37). In gram-positive organisms, pili are not as well studied. However, reports indicate that C. diphtheriae, as well as Streptococcus spp. and Actinomyces spp. have elaborate pili composed of LPXTG-type protein subunits. These subunits are covalently linked to one another by sortase-mediated transpeptidation reactions. Lack of pilus-specific sortases abolishes the polymerization of the pilus (16, 25, 45). In S. pneumoniae PI-2 is similar to previously described gram-positive pili in its global genetic organization and in its sequence homology with the FCT-3 pilus from S. pyogenes (25, 43). Indeed, the islet contains five genes coding for two putative sortases (srtG1 and srtG2), a signal peptidase-related protein (sipA), and two LPXTG-type surface-anchored proteins (pitB and pitA). Interestingly, through BLAST iterative searches with the PI-2 proteins, we identified a group of conserved hypothetical proteins, organized as a locus in the genome of the human intestinal gram-positive bacterium Ruminococcus gnavus; the homology between S. pneumoniae backbone protein PitB and R. gnavus was 27%, while for SipA and SrtG1 similarity increased to 43% and 41%, respectively. This finding suggests that horizontal transfer may occur between bacteria of different genera and that the human commensal Ruminococcus spp. could also encode pilus structures.
The results presented in this work demonstrate that PitB is the backbone subunit, as already suggested by the sequence analysis. In fact, deletion of the other CWSS-containing protein PitA does not affect pilus polymerization. Immunogold detection with anti-PitB antiserum shows elongated structures that resemble the previously described RrgB pilus filaments (4, 18) although many fewer filaments were present per bacterium (Fig. 2C and 4B). In the case of isolate 19F Taiwan-14, it is evident that the PI-1 pili cover most of the surface of the pneumococcus, while PI-2 pili are found in single copies and extend further from the cell. Furthermore, polymerization of PitB requires sortase SrtG1 and the LepA-homologue SipA. This latter result confirms the importance of the signal peptidase-like protein and corroborates its hypothesized role as a chaperone in pilus polymerization, recently shown in S. pyogenes (47). In addition, protein sequence alignment performed between SipA and other known and hypothetical signal peptidases shows the lack in SipA of the two conserved residues, serine and lysine, required for peptidase activity. An analogous situation was found in SipA1 and SipA2 of S. pyogenes (47).
We next investigated the role of the hypothetical ancillary protein encoding gene pitA and the putative sortase gene srtG2; we found that pitA contains a stop codon in all nine sequences analyzed, that srtG2 contains a frameshift in all but one isolate, and that the lack of these genes does not affect the pilus assembly. These observations suggest that the PI-2 pilus may be composed of only the pilus backbone PitB. To verify this possibility, we raised antibodies against a portion of the PitA starting after the internal stop codon. Western blot analysis of cell wall mutanolysin extracts with anti-PitA antiserum did not reveal the typical HMW ladder or the monomeric protein. Furthermore, mass spectrometric analysis of the mutanolysin extracts was not able to detect the PitA protein (data not shown). At this moment we cannot exclude the possibility that pitA is expressed through an alternative translation of the UGA stop codon located inside the open reading frames. Indeed, UGA can be translated into tryptophan at a very low frequency (31), and a low level of the protein could be present but undetectable by our assays. Therefore, srtG2 and pitA could be considered pseudogenes that may have lost their ability to encode functional proteins.
To date, all pili of gram-positive bacteria have at least one functional ancillary protein, and several groups of investigators have demonstrated that ancillary proteins play a major role in adhesion (1, 29). Here, we provide evidence that this newly identified pilus favors adherence to the host cells. Interestingly, PI-2-mediated adherence appears to rely solely on the pilus backbone protein. Indeed, study of isogenic mutants of the islet genes has shown that expression of pitB is strictly required for adhesion to respiratory cells, since the PN110ΔpitB strain is completely impaired, as is the PN110ΔPI2 strain, in this property. On the other hand, deletion of srtG1 did not result in the complete loss of binding capacity. PN110ΔsrtG1 may still express and translocate a monomeric form of PitB to the bacterial surface. Therefore, adhesion mediated by a monomeric form of PitB may explain the adhesive properties of the sortase mutant. In this regard, we evaluated adherence properties of the PitB purified protein by immunofluorescence microscopy on adherent A549 cells and by FACS analysis. Both assays showed low binding capacity of the protein, confirming our expectations for a pilus backbone subunit. However, experiments on adherent cells show a diffuse binding pattern, which suggests that PitB may recognize a cellular protein expressed at low levels. We believe that the difference in adhesion observed with the two approaches may be due to the redistribution of proteins recognized by the pilus backbone protein (PitB) over the cell surface upon detachment from the support. Given that the polymerized pilus is able to mediate stronger binding, it is likely that both the sum of the binding of the PitB subunits and the structural conformation of the pilus are important for its adhesive mechanism. However, this novel pilus is not as effective in mediating attachment to host cells as the rlrA pilus of S. pneumoniae (data not shown), and this phenomenon may be explained by the lack of a detectable expression of ancillary subunits.
Moreover, the low distribution of PI-2 among clinical isolate strains (16%), the low level of PI-2 expression in all the isolates tested, and the presence of frameshift mutations and stop codons, taken together, lead to the hypothesis that this second PI may not be required but could be an additional factor important for survival in the host.
Lastly, the PI-2-positive CCs have been found associated with nonvaccine serotypes lacking PI-1, including 1, 2, 7F, and 19A, considered to be emerging serotypes in both industrialized and developing countries. Serotype replacement and vaccine evasion are phenomena predicted to lead to a decrease in strain coverage of polysaccharide-based vaccines and an increase in nonvaccine serotypes (NVT) (14, 33, 38). In fact, certain NVT drug-resistant clones, such as ST156, are increasing in prevalence (40) while the well-known vaccine type serotype 4 was recently described to undergo capsular switching by recombination to an NVT 19A capsule (7). Therefore, a serotype-independent vaccine that includes pilus antigens may avoid these limitations. The role of pili in pneumococcal pathogenesis, in addition to strain coverage, suggests that these antigens are eligible for inclusion in the next generation of protein-based vaccines.
Acknowledgments
We thank Julian Parkhill from the Sanger Institute for INV104 sequence information and the Novartis Sequencing Facility, Beatrice Rogolino, Silvia Guidotti, and Monica Giraldi for technical assistance. We acknowledge the following for providing S. pneumoniae clinical isolates for this study: the Active Bacterial Core surveillance (ABCs; collaboration between the CDC, state health departments, and universities http://www.cdc.gov/ncidod/dbmd/abcs/index.htm) and Bernard Beall from the CDC for supplying genotyped and serotyped ABCs strains, Annalisa Pantosti (ISS, Italy), Albert Ko and Mitermayer G. Reis (Oswaldo Cruz Foundation, Brazil), Gerd Plushke (Swiss Tropical Institute), Samir Saha (Dhaka Shishu Hospital, Bangladesh), Anthony Scott (Kenyan Medical Research Center, Kenya), and Birgitta Henriques-Normark and Staffan Normark (Karolinska Institute, Sweden). Finally, we are indebted to David Markovitz (University of Michigan) for all his suggestions and critical comments.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Abbot, E. L., W. D. Smith, G. P. Siou, C. Chiriboga, R. J. Smith, J. A. Wilson, B. H. Hirst, and M. A. Kehoe. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell Microbiol. 91822-1833. [DOI] [PubMed] [Google Scholar]
- 2.Aguiar, S. I., I. Serrano, F. R. Pinto, J. Melo-Cristino, and M. Ramirez. 2008. The presence of the pilus locus is a clonal property among pneumococcal invasive isolates. BMC Microbiol. 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 2975-83. [DOI] [PubMed] [Google Scholar]
- 4.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 1032857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basset, A., K. Trzcinski, C. Hermos, L. O'Brien, K., R. Reid, M. Santosham, A. J. McAdam, M. Lipsitch, and R. Malley. 2007. Association of the pneumococcal pilus with certain capsular serotypes but not with increased virulence. J. Clin. Microbiol. 451684-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beall, B., M. C. McEllistrem, R. E. Gertz, Jr., S. Wedel, D. J. Boxrud, A. L. Gonzalez, M. J. Medina, R. Pai, T. A. Thompson, L. H. Harrison, L. McGee, and C. G. Whitney. 2006. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J. Clin. Microbiol. 44999-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brueggemann, A. B., R. Pai, D. W. Crook, and B. Beall. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 313497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comfort, D., and R. T. Clubb. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 722710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 601401-1413. [DOI] [PubMed] [Google Scholar]
- 11.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of gram-positive bacteria. Res. Microbiol. 156289-297. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 1443049-3060. [DOI] [PubMed] [Google Scholar]
- 13.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks, L. A., L. H. Harrison, B. Flannery, J. L. Hadler, W. Schaffner, A. S. Craig, D. Jackson, A. Thomas, B. Beall, R. Lynfield, A. Reingold, M. M. Farley, and C. G. Whitney. 2007. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J. Infect. Dis. 1961346-1354. [DOI] [PubMed] [Google Scholar]
- 15.Hilleringmann, M., F. Giusti, B. C. Baudner, V. Masignani, A. Covacci, R. Rappuoli, M. A. Barocchi, and I. Ferlenghi. 2008. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PLoS Pathog. 4e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B streptococcus. Science 309105. [DOI] [PubMed] [Google Scholar]
- 17.Leimkugel, J., A. Adams Forgor, S. Gagneux, V. Pfluger, C. Flierl, E. Awine, M. Naegeli, J. P. Dangy, T. Smith, A. Hodgson, and G. Pluschke. 2005. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J. Infect. Dis. 192192-199. [DOI] [PubMed] [Google Scholar]
- 18.LeMieux, J., D. L. Hava, A. Basset, and A. Camilli. 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 742453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine, O. S., K. L. O'Brien, M. Knoll, R. A. Adegbola, S. Black, T. Cherian, R. Dagan, D. Goldblatt, A. Grange, B. Greenwood, T. Hennessy, K. P. Klugman, S. A. Madhi, K. Mulholland, H. Nohynek, M. Santosham, S. K. Saha, J. A. Scott, S. Sow, C. G. Whitney, and F. Cutts. 2006. Pneumococcal vaccination in developing countries. Lancet 3671880-1882. [DOI] [PubMed] [Google Scholar]
- 20.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. Nardi Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B streptococcus vaccine by multiple genome screen. Science 309148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maisey, H. C., M. Hensler, V. Nizet, and K. S. Doran. 2007. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 1891464-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2007. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 64111-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manetti, A. G., C. Zingaretti, F. Falugi, S. Capo, M. Bombaci, F. Bagnoli, G. Gambellini, G. Bensi, M. Mora, A. M. Edwards, J. M. Musser, E. A. Graviss, J. L. Telford, G. Grandi, and I. Margarit. 2007. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64968-983. [DOI] [PubMed] [Google Scholar]
- 24.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 392565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA 10215641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moschioni, M., C. Donati, A. Muzzi, V. Masignani, S. Censini, W. P. Hanage, C. J. Bishop, J. N. Reis, S. Normark, B. Henriques-Normark, A. Covacci, R. Rappuoli, and M. A. Barocchi. 2008. Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J. Infect. Dis. 197888-896. [DOI] [PubMed] [Google Scholar]
- 27.Mota, L. J., and G. R. Cornelis. 2005. The bacterial injection kit: type III secretion systems. Ann. Med. 37234-249. [DOI] [PubMed] [Google Scholar]
- 28.Nallapareddy, S. R., K. V. Singh, J. Sillanpaa, D. A. Garsin, M. Hook, S. L. Erlandsen, and B. E. Murray. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 1162799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, A. L., J. Ries, F. Bagnoli, S. Dahlberg, S. Falker, S. Rounioja, J. Tschop, E. Morfeldt, I. Ferlenghi, M. Hilleringmann, D. W. Holden, R. Rappuoli, S. Normark, M. A. Barocchi, and B. Henriques-Normark. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortqvist, A., J. Hedlund, and M. Kalin. 2005. Streptococcus pneumoniae: epidemiology, risk factors, and clinical features. Semin. Respir. Crit. Care Med. 26563-574. [DOI] [PubMed] [Google Scholar]
- 31.Parker, J. 1989. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 53273-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paterson, G. K., and T. J. Mitchell. 2006. The role of Streptococcus pneumoniae sortase A in colonisation and pathogenesis. Microbes Infect. 8145-153. [DOI] [PubMed] [Google Scholar]
- 33.Pichichero, M. E., and J. R. Casey. 2007. Emergence of a multiresistant serotype 19A pneumococcal strain not included in the 7-valent conjugate vaccine as an otopathogen in children. JAMA 2981772-1778. [DOI] [PubMed] [Google Scholar]
- 34.Rosini, R., C. D. Rinaudo, M. Soriani, P. Lauer, M. Mora, D. Maione, A. Taddei, I. Santi, C. Ghezzo, C. Brettoni, S. Buccato, I. Margarit, G. Grandi, and J. L. Telford. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61126-141. [DOI] [PubMed] [Google Scholar]
- 35.Sauer, F. G., M. A. Mulvey, J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 365-72. [DOI] [PubMed] [Google Scholar]
- 36.Scott, J. R., and D. Zähner. 2006. Pili with strong attachments: gram-positive bacteria do it differently. Mol. Microbiol. 62320-330. [DOI] [PubMed] [Google Scholar]
- 37.Silverman, P. M. 1997. Towards a structural biology of bacterial conjugation. Mol. Microbiol. 23423-429. [DOI] [PubMed] [Google Scholar]
- 38.Singleton, R. J., T. W. Hennessy, L. R. Bulkow, L. L. Hammitt, T. Zulz, D. A. Hurlburt, J. C. Butler, K. Rudolph, and A. Parkinson. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 2971784-1792. [DOI] [PubMed] [Google Scholar]
- 39.Sinha, A., O. Levine, M. D. Knoll, F. Muhib, and T. A. Lieu. 2007. Cost-effectiveness of pneumococcal conjugate vaccination in the prevention of child mortality: an international economic analysis. Lancet 369389-396. [DOI] [PubMed] [Google Scholar]
- 40.Sjostrom, K., C. Blomberg, J. Fernebro, J. Dagerhamn, E. Morfeldt, M. A. Barocchi, S. Browall, M. Moschioni, M. Andersson, F. Henriques, B. Albiger, R. Rappuoli, S. Normark, and B. Henriques-Normark. 2007. Clonal success of piliated penicillin nonsusceptible pneumococci. Proc. Natl. Acad. Sci. USA 10412907-12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Sogstad, M. K., I. S. Aaberge, J. O. Sørdal, E. A. Høiby, L. O. Frøholm, A. R. Alme, and D. A. Caugant. 2006. Carriage of Streptococcus pneumoniae in healthy Norwegian children attending day-care centres. Eur. J. Clin. Microbiol. Infect. Dis. 25510-514. [DOI] [PubMed] [Google Scholar]
- 41.Swaminathan, A., A. Mandlik, A. Swierczynski, A. Gaspar, A. Das, and H. Ton-That. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 66961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swierczynski, A., and H. Ton-That. 2006. Type III pilus of corynebacteria: pilus length is determined by the level of its major pilin subunit. J. Bacteriol. 1886318-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4509-519. [DOI] [PubMed] [Google Scholar]
- 44.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53251-261. [DOI] [PubMed] [Google Scholar]
- 45.Ton-That, H., and O. Schneewind. 2004. Assembly of pili in gram-positive bacteria. Trends Microbiol. 12228-234. [DOI] [PubMed] [Google Scholar]
- 46.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 501429-1438. [DOI] [PubMed] [Google Scholar]
- 47.Zähner, D., and J. R. Scott. 2008. SipA is required for pilus formation in Streptococcus pyogenes serotype M3. J. Bacteriol. 190527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]