Abstract
Quinolone antibacterial drugs such as nalidixic acid target DNA gyrase in Escherichia coli. These inhibitors bind to and stabilize a normally transient covalent protein-DNA intermediate in the gyrase reaction cycle, referred to as the cleavage complex. Stabilization of the cleavage complex is necessary but not sufficient for cell killing—cytotoxicity apparently results from the conversion of cleavage complexes into overt DNA breaks by an as-yet-unknown mechanism(s). Quinolone treatment induces the bacterial SOS response in a RecBC-dependent manner, arguing that cleavage complexes are somehow converted into double-stranded breaks. However, the only proteins known to be required for SOS induction by nalidixic acid are RecA and RecBC. In hopes of identifying additional proteins involved in the cytotoxic response to nalidixic acid, we screened for E. coli mutants specifically deficient in SOS induction upon nalidixic acid treatment by using a dinD::lacZ reporter construct. From a collection of SOS partially constitutive mutants with disruptions of 47 different genes, we found that dnaQ insertion mutants are specifically deficient in the SOS response to nalidixic acid. dnaQ encodes DNA polymerase III ɛ subunit, the proofreading subunit of the replicative polymerase. The deficient response to nalidixic acid was rescued by the presence of the wild-type dnaQ gene, confirming involvement of the ɛ subunit. To further characterize the SOS deficiency of dnaQ mutants, we analyzed the expression of several additional SOS genes in response to nalidixic acid using real-time PCR. A subset of SOS genes lost their response to nalidixic acid in the dnaQ mutant strain, while two tested SOS genes (recA and recN) continued to exhibit induction. These results argue that the replication complex plays a role in modulating the SOS response to nalidixic acid and that the response is more complex than a simple on/off switch.
Type II DNA topoisomerases are essential enzymes involved in processes such as DNA replication, chromosome segregation, and transcription (7, 26, 55). Many clinically important antibacterial and antitumor agents target type II topoisomerases by altering the equilibrium of the topoisomerase reaction cycle to stabilize the normally transient intermediate called the cleavage complex (9, 11). Antibacterial quinolones such as nalidixic acid target DNA gyrase (and topoisomerase IV in certain bacteria) by stabilizing the cleavage complex, which consists of the topoisomerase linked via phosphotyrosine bonds to both 5′ ends of a staggered double-stranded DNA break (DSB). While the cleavage complex contains a latent DSB, this break is bridged by the covalently attached enzyme, and intact DNA is reconstituted when the enzyme completes the reaction cycle and dissociates. A variety of results argue that cleavage complex formation is necessary but not sufficient for cytotoxicity (reviewed in reference 11). Some cellular processing event apparently converts a subset of the cleavage complexes into cytotoxic lesions, because inhibition of RNA or protein synthesis can mitigate quinolone sensitivity (3, 10, 11, 28). However, the detailed mechanism(s) of cytotoxicity remains unclear.
A number of observations argue that quinolone treatment leads to frank DSBs that are likely involved in cytotoxicity. First, treatment of cells with bactericidal concentrations of a quinolone causes free rotation of chromosomal DNA in the presence of ethidium bromide, which is characteristic of broken DNA ends (3). Second, mutational inactivation of proteins involved in DSB repair (RecA and RecBC) leads to drug hypersensitivity (23, 30). Finally, nalidixic acid induces the bacterial SOS response in a manner that is dependent on the RecBC enzyme, which requires double-strand ends for activity (31, 36). SOS is the bacterial DNA damage response, in which more than 20 genes are upregulated after DNA damage (13, 54). Notably, mutations in recA, recB, or recC eliminate the nalidixic acid-induced SOS response and cause hypersensitivity to nalidixic acid, implicating drug-induced DSBs in both the SOS response and cytotoxicity.
DNA replication plays an important role in the cytotoxicity of antitumor agents that target type II topoisomerase in eukaryotes. Thus, eukaryotic cells are more sensitive to these inhibitors during S phase than during G1, and the DNA polymerase inhibitor aphidicolin abrogates sensitivity to topoisomerase inhibitors (8, 17, 57).
There are conflicting reports on whether DNA replication is important in the bacterial response to quinolones. In favor of a role for DNA replication, Gudas and Pardee (14) found that the SOS response to nalidixic acid was blocked after temperature shift-up of a temperature-sensitive dnaA mutant. Furthermore, we recently found that replication forks are blocked in vivo at the sites of quinolone-stabilized cleavage complexes on replicating plasmids (41). The blocked forks were sometimes broken in vivo, raising the possibility that replication-dependent breaks could be a cytotoxic lesion. On the other hand, two reports argue against a role for DNA replication in quinolone action. Sassanfar and Roberts (43) reported that SOS was not prevented after a temperature shift of a dnaC(Ts) uvrB mutant. In a direct measure of quinolone cytotoxicity, Zhao et al. (58) found that temperature shift-up of a dnaB(Ts) mutant does not protect against killing by nalidixic acid or ciprofloxacin (a fluoroquinolone). In summary, while it is clear that quinolones effectively block DNA replication, the physiological consequences of this blocked replication are still uncertain.
The only genes known to be required for the SOS response to nalidixic acid are recA and recBC (13, 54). In a prior study, we used an SOS reporter system to screen for Escherichia coli mutants specifically deficient in SOS induction upon nalidixic acid treatment, hoping to find additional genes involved in this process. However, all 18 mutants with this phenotype had insertions in recB or recC (37). Using the same SOS reporter construct, we also isolated a collection of mutants constitutive for the SOS response (39). Compared to a lexA mutant, all other insertion mutants were only partially constitutive for SOS. It is possible that a partially constitutive mutant would also be defective for SOS induction after nalidixic acid—such a mutant would probably have been missed in the Newmark et al. (37) screen for induction defects because it would be expressing the reporter construct regardless of the presence of nalidixic acid. We therefore examined the SOS response of each mutant in this collection, looking for mutants that are specifically defective in response to nalidixic acid. We found insertions in one gene, dnaQ, that cause this phenotype. The dnaQ gene encodes the ɛ subunit of DNA polymerase III, arguing that this subunit in particular, and the replication complex in general, is important in the nalidixic acid-induced SOS response. We also found that the dnaQ insertions are defective for induction of only a subset of SOS genes, implying greater complexity in the SOS response.
MATERIALS AND METHODS
Materials.
Kanamycin, mitomycin C, ο-nitrophenyl-β-d-galactopyranoside (ONPG), and nalidixic acid were purchased from Sigma; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was from Gold Biotechnology Inc.; and Taq DNA polymerase was from Invitrogen.
Bacterial strains and plasmids.
The wild-type dnaQ plasmid pMM5 was kindly provided by Roel Schaaper (NIEHS). pMM5 is a medium-copy-number plasmid with a pBR322-based origin that contains the 1.6-kb EcoRI genomic fragment surrounding dnaQ, including the rnhA and aspV genes (19, 27).
Genomic DNA isolation and PCR amplification.
Genomic DNA was purified using the MasterPure Complete DNA and RNA purification kit (Epicentre) essentially as described by the manufacturer, except that a phenol-chloroform extraction and ethanol precipitation were added after the RNase treatment. The final purified DNA pellet was resuspended in 30 μl of TE buffer (5 mM Tris-HCl, pH 7.8; 1 mM EDTA).
PCR amplification of dnaE-containing DNA from dnaQ strains was performed with the following primers: dnaE-1 (5′-GACTTGCGTCCTGATTCTTG-3′), dnaE-4 (5′-CTGGAGATGATCAACAAGCG-3′), dnaE-5 (5′-TGATCAGGTCCTTCATGCC-3′), and dnaE-8 (5′-TAACCGCAGTCAGAGAATCG-3′). Sequencing was performed with dnaE-1, dnaE-2 (5′-AGGGTTGATCCTTCTTTCCG-3′), dnaE-3 (5′-ACATGAGCACCGAAGATTATCTG-3′), dnaE-4, dnaE-5, dnaE-6 (5′-GTTCTGTATTTGCTGAAGGTGC-3′), dnaE-7 (5′-TACCGACACCAAAAAGTTGAAC-3′), and dnaE-8.
PCR amplification of recQ-containing DNA from dnaQ strains was performed with the following primers: recQ-1 (5′-GTGGGGGTTATGCTAAACGA-3′) and recQ-2 (5′-GCCCACCATAATCAGCGTAT-3′). Sequencing was performed with recQ-1, recQ-2, recQ-3 (5′-GCGCGAAAGTAGAAGACACC-3′), recQ-4 (5′-CCATTGGTCGTGTGAATCAG-3′), and recQ-5 (5′-ATCAAAGTGGACCACGAAGC-3′).
Liquid β-galactosidase assay.
Overnight cultures were diluted 100-fold into LB and grown to mid-log phase (optical density at 600 nm [OD600] of approximately 0.5). The culture was then treated with either nalidixic acid (10 μg/ml), mitomycin C (1 μg/ml), or no drug for an additional 2 h, after which 1.0-ml samples were removed for analysis. β-Galactosidase assays were performed essentially by the method of Miller (34). Briefly, the cells were spun down and resuspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 10 mM dithiothreitol). Two drops of chloroform and 1 drop of 0.1% sodium dodecyl sulfate were added to the cell suspension followed by vigorous vortexing for 10 seconds. Appropriate amounts of the lysate were then mixed with Z buffer to a final volume of 1.0 ml. To start the reactions, 200 μl of 4 mM ONPG was added and the reaction mixtures were incubated at 30°C until a moderate yellow color was observed. The reactions were stopped with 0.5 ml of 1 M sodium bicarbonate, and the cellular debris was pelleted. The optical density was recorded using an Anthos 2001 plate reader with a 405-nm filter. Miller units were calculated as follows: units = 1,000 × OD405/(t × v × OD600), where OD405 is the optical density of the reaction mixture, OD600 reflects the cell density, t is the reaction time in minutes, and v is the volume of culture (in ml) used in the assay.
RNA isolation.
Overnight cultures were diluted 100-fold into LB and grown until the OD600 was 0.5. The log-phase culture was then treated with nalidixic acid (10 μg/ml) or no drug (control) for 120 min. Each sample was then diluted with two volumes of RNAprotect bacterial reagent (Qiagen), vortexed for 5 s, and incubated at room temperature for 5 min. The cells were then collected by centrifugation for 10 min at 5,000 × g, and the pellet was frozen at −20°C. At the end of 120 min, 1 ml of each culture was also used to perform β-galactosidase assays, which confirmed the expected phenotypes for SOS induction (data not shown). The following day, the frozen pellets were processed using the RNeasy minikit (Qiagen), and the RNA was eluted with 40 μl of RNase-free water. Total RNA concentration was determined by measuring the A260 of RNA samples.
Quantitative real-time PCR.
Total RNA (see above) was treated with DNase I (Promega) at 1 U per microgram of RNA. cDNA synthesis was performed using random hexamer primers (Invitrogen) and Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer's protocol, including the use of RNase Out (Invitrogen) to inhibit RNase activity. As a control, DNase-treated samples and completed cDNA reaction mixtures were amplified by PCR with each primer pair to check for the absence and presence of appropriate DNA products, respectively. Quantitative real-time PCRs were then performed with Sybr green (ABgene) according to the manufacturer's protocol on an ABI7300 thermal cycler. Primers specific to each target gene were identical to those used by Robbins-Manke et al. (42) except for the dinD primer pair: dinD-left (5′-ATTGGCTCAAATGCCAAAAG-3′) and dinD-right (5′-AAATGCTTCATCGTCAGCAA-3′). Negative control assays without template cDNA were performed to ensure that primers alone did not yield amplification product. Denaturation curves were performed at the end of real-time PCRs to confirm the identity of amplification products. Relative gene expression values were measured by including a standard curve for each gene assay. JH39 wild-type genomic DNA was used to make a serial dilution for quantification of the relative amounts of product in each experimental sample. To account for variation in real-time PCRs, expression of the constitutively expressed gapA (d-glyceraldehyde 3-phosphate dehydrogenase) gene was determined for each sample and used to normalize gene expression values. Due to the mutator phenotype of the dnaQ strain, gene expression values were determined from three separate experiments started from different colonies.
RESULTS
Identification of mutants with reduced SOS response to nalidixic acid.
DSBs are likely generated from nalidixic acid-stabilized cleavage complexes as a necessary step in SOS induction (see the introduction). In a prior search for genes involved in DSB formation, the EZ::TN <KAN-2> Tnp Transposome kit (Epicentre) was used to create a library of transposon mutants in the JH39 strain of E. coli (37). This strain has a dinD::lacZ fusion that provides a convenient reporter for the SOS response, allowing mutants to be identified by simple color screens on plates containing X-Gal (15). The strain also has an sfiA (sulA) mutation, which reduces filamentation and inhibition of cell division upon SOS induction (13). Eighteen mutants were found with a strongly reduced SOS response to nalidixic acid but a normal response to UV. However, each had an insertion in recB or recC, and therefore we failed to uncover any new genes required for DSB formation (37).
One limitation of the above screen is that mutants that are partially constitutive for the SOS response would produce β-galactosidase regardless of the presence of the drug. If any of these partially constitutive mutants had a defect in SOS induction following nalidixic acid treatment, they could easily be missed in the screen due to the constitutive level of β-galactosidase. For this and other reasons, we performed a screen for constitutive SOS mutants using the same starting strain. We found insertions into 47 different genes that caused some level of constitutive SOS response that was reproducibly detected in plate assays, and a large majority of these also showed significant levels of constitutive SOS expression in quantitative measures from mid-log-phase culture (39).
We investigated the response of all 47 mutants to nalidixic acid, reasoning that even the subset that showed apparent constitutive expression only on plates might still have a defect in SOS induction after nalidixic acid. Quantitative β-galactosidase assays were performed on samples of cultures that had been incubated with or without nalidixic acid (10 μg/ml) for 2 hours. Induction levels are presented in the rightmost column of Table 1 as the difference in β-galactosidase levels with and without nalidixic acid. The parental JH39 strain is included as a control, and the remaining strains are presented in order of decreasing levels of induction. Transposon insertions in recA and recB were also included as controls and, as expected, showed no induction in this assay (Table 1).
TABLE 1.
Induction of β-galactosidase by nalidixic acid in selected SOS mutants
| Genea | nb | Uninducedc | Nalidixic acidd | Differencee |
|---|---|---|---|---|
| JH39 | 51 | 8.5 ± 2.1 | 98.7 ± 41.4 | 90.2 ± 41.1 |
| recN | 4 | 15.5 ± 1.1 | 243 ± 75.2 | 228 ± 74.5 |
| cvpA | 3 | 17.6 ± 1.4 | 184 ± 9.1 | 166 ± 10.5 |
| folA | 3 | 16.7 ± 0.8 | 142 ± 11.8 | 126 ± 11.1 |
| tdcE | 3 | 28.7 ± 2.1 | 143 ± 19.4 | 115 ± 17.5 |
| uvrD | 3 | 22.5 ± 1.3 | 129 ± 30.7 | 107 ± 29.4 |
| purL | 3 | 10.6 ± 0.9 | 115 ± 3.3 | 104 ± 2.5 |
| htrA | 3 | 7.1 ± 0.8 | 105 ± 37.9 | 97.9 ± 37.3 |
| ompA | 3 | 7.2 ± 1.1 | 99.4 ± 36.4 | 92.2 ± 35.3 |
| dam | 3 | 55.1 ± 15.9 | 140 ± 18.2 | 85.3 ± 29.5 |
| lexA | 3 | 202 ± 84.2 | 287 ± 144 | 84.7 ± 99.1 |
| yfgM | 3 | 7.5 ± 0.2 | 92.0 ± 15.6 | 84.4 ± 15.5 |
| yfgL | 3 | 7.7 ± 0.3 | 88.1 ± 19.1 | 80.4 ± 19.1 |
| thyAf | 4 | 15.5 ± 3.5 | 92.7 ± 56.1 | 77.2 ± 52.7 |
| polA | 3 | 33.0 ± 4.2 | 107 ± 8.3 | 74.4 ± 4.3 |
| acrA | 3 | 7.4 ± 1.1 | 73.3 ± 19.0 | 65.9 ± 19.6 |
| acrB | 3 | 7.4 ± 0.6 | 72.7 ± 9.9 | 65.3 ± 9.4 |
| yncD | 3 | 17.4 ± 4.7 | 78.8 ± 7.4 | 61.5 ± 9.4 |
| rpoZ | 3 | 26.5 ± 0.4 | 87.7 ± 5.9 | 61.3 ± 5.9 |
| tolC | 3 | 8.0 ± 1.3 | 69.2 ± 13.2 | 61.2 ± 12.0 |
| purF | 3 | 43.1 ± 1.1 | 103 ± 3.8 | 59.4 ± 4.3 |
| rluD | 3 | 7.7 ± 0.5 | 65.2 ± 8.4 | 57.5 ± 8.0 |
| ruvB | 3 | 18.3 ± 1.7 | 74.4 ± 15.1 | 56.1 ± 13.4 |
| recGg | 4 | 22.8 ± 1.9 | 79.6 ± 50.1 | 56.8 ± 49.3 |
| ruvA | 3 | 17.0 ± 1.8 | 72.9 ± 9.0 | 55.9 ± 7.6 |
| yebC | 3 | 15.0 ± 0.2 | 70.5 ± 8.3 | 55.5 ± 8.1 |
| ruvC | 4 | 21.2 ± 6.2 | 73.9 ± 16.6 | 52.7 ± 19.1 |
| xerDh | 6 | 43.1 ± 3.0 | 95.2 ± 30.9 | 52.2 ± 32.0 |
| dcd | 3 | 26.8 ± 0.8 | 77.8 ± 2.8 | 51.0 ± 2.1 |
| damX | 3 | 28.3 ± 0.7 | 78.3 ± 0.8 | 50.0 ± 0.8 |
| rep | 4 | 19.8 ± 3.3 | 67.8 ± 3.2 | 48.0 ± 3.2 |
| dsbB | 3 | 6.1 ± 0.2 | 53.9 ± 6.1 | 47.8 ± 5.9 |
| xerCi | 6 | 46.9 ± 3.2 | 91.8 ± 32.4 | 44.9 ± 29.3 |
| arpB | 3 | 9.6 ± 0.6 | 53.7 ± 19.9 | 44.1 ± 19.4 |
| surA | 3 | 6.3 ± 0.7 | 47.9 ± 7.5 | 41.6 ± 6.8 |
| ftsEj | 4 | 14.5 ± 4.9 | 53.8 ± 22.0 | 39.3 ± 26.2 |
| envC | 3 | 8.3 ± 0.7 | 45.4 ± 5.6 | 37.1 ± 5.6 |
| ftsXk | 5 | 14.1 ± 7.2 | 50.8 ± 25.9 | 36.7 ± 32.5 |
| yehBl | 3 | 10.6 ± 0.2 | 41.6 ± 5.3 | 30.9 ± 5.1 |
| spoTm | 6 | 29.8 ± 4.8 | 59.3 ± 22.5 | 29.5 ± 18.4 |
| purE | 3 | 10.4 ± 3.0 | 35.8 ± 3.3 | 25.4 ± 6.3 |
| tynA | 3 | 30.1 ± 1.4 | 55.3 ± 4.0 | 25.2 ± 2.9 |
| yejM | 3 | 7.6 ± 1.4 | 32.1 ± 1.1 | 24.4 ± 1.7 |
| ftsKn | 10 | 37.8 ± 4.9 | 61.6 ± 24.9 | 23.8 ± 23.6 |
| yhcBo | 7 | 10.1 ± 3.2 | 27.5 ± 17.4 | 17.4 ± 14.6 |
| folK | 25 | 20.5 ± 3.9 | 22.5 ± 8.5 | 2.9 ± 5.9 |
| purA | 6 | 6.3 ± 0.3 | 5.8 ± 0.5 | 0.5 ± 0.4 |
| dnaQ | 18 | 18.9 ± 11.0 | 19.3 ± 9.7 | 0.4 ± 9.4 |
| recBp | 4 | 4.8 ± 0.2 | 4.4 ± 1.5 | −0.4 ± 1.5 |
| recAp | 4 | 4.1 ± 0.1 | 3.2 ± 0.5 | −1.0 ± 0.5 |
Alternate gene names: arpB, b1720/b1721; ecfD, b2595; envC, yibP; rluD, sfhB; yfgL, b2512; yfgM, b2513; yncD, b1451; yddW, b1491.
Number of colonies tested from the same strain. The particular transposon insertion strain tested for each mutant is the same as that in the work of O'Reilly and Kreuzer (39).
β-Galactosidase units expressed after 2 hours of growth in the absence of nalidixic acid. The data in this column are from the same measurements as those in Table 1 of reference 39.
β-Galactosidase units expressed after 2 hours of growth in the presence of nalidixic acid.
Average of the differences between β-galactosidase units in the presence of nalidixic acid and those in the absence of nalidixic acid.
Three thyA trials had an average difference of 103.6 ± 0.5, while one trial had a difference of −1.8.
The four recG trials had differences of 98.9, 95.7, 35.5, and −2.82.
Three xerD trials had an average difference of 23.0 ± 2.2, while three trials had a difference of 81.4 ± 1.6.
Three xerC trials had an average difference of 18.8 ± 5.2, while three trials had a difference of 71.0 ± 8.7.
Three ftsE trials had an average difference of 50.9 ± 14.8, while one trial had a difference of 4.4.
The five trials of ftsX appeared to fall into two groups: the inducible group had an average difference of 59.4 ± 12.7 (n = 3), while the uninducible group had an average difference of 2.7 ± 6.6 (n = 2).
Two of the six spoT trials had an average difference of 50.9 ± 16.4, while the other four had an average difference of 18.8 ± 4.4.
The original isolate of yehB was deficient for nalidixic acid induction, but the three transductants (shown here) were not.
The 10 trials of ftsK appeared to fall into three groups: an inducible group with an average difference of 48.6 ± 12.1 (n = 4), a slightly inducible group with an average difference of 15.3 ± 4.6 (n = 3), and an uninducible group with an average difference of −0.6 ± 5.3 (n = 3).
The seven trials of yhcB appeared to fall into two groups: an inducible group with an average difference of 30.9 ± 5.1 (n = 3) and an uninducible group with an average difference of −0.4 ± 3.5 (n = 4).
Data for the recA (I78) and recB (I40) strains are shown as controls.
Two insertion mutants, with a transposon in recN or cvpA, showed significantly higher levels of induction than did the JH39 control (Table 1). The molecular function of CvpA is not well understood, but RecN is an SMC (structural maintenance of chromosomes) family protein known to be involved in DSB repair (12, 32, 40). The finding that the recN mutant is hyperinducible lends support to the hypothesis that nalidixic acid induces DSBs. Previously, recN mutants were shown to be hyperinducible after treatment with gamma rays, which induce DSBs (2). Furthermore, recN mutants are hypersensitive to nalidixic acid, at least in Neisseria gonorrheae (47).
Most of the constitutive strains tested showed a response to nalidixic acid that was not statistically distinct from that of the JH39 wild-type strain (Table 1). However, a number of mutants appeared to have a reduced response, and the corresponding proteins could be involved in multiple partially redundant pathways for nalidixic acid induction of the SOS response. However, it also seems possible that the response in some of these mutants is reduced for trivial reasons (e.g., reduced growth rate/metabolism). It should be noted that a few of the mutants showed high variability in SOS response from different cultures for unknown reasons (Table 1, footnotes f to l, n, and o).
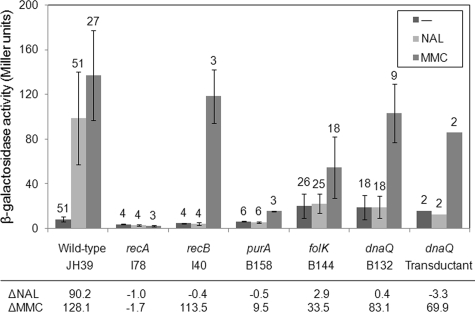
The most striking finding was that mutants with insertions in dnaQ, purA, or folK showed little or no induction in response to nalidixic acid in all trials (Table 1). These three mutants, along with wild-type and recA and recB mutant controls, were therefore analyzed for SOS response with mitomycin C treatment to see whether the induction defect is specific for nalidixic acid. Like UV, mitomycin C induces SOS by a mechanism that is RecFOR dependent but RecBC independent (56) and is a convenient inducer for liquid assays. As expected, the recA mutant was uninducible by nalidixic acid or mitomycin C, while the recB mutant was inducible by only mitomycin C (Fig. 1). Remarkably, the purA insertion mutant was not inducible by either nalidixic acid or mitomycin C, suggesting a general defect in the SOS response. The SOS response to mitomycin C was quite variable in parallel cultures of the folK insertion mutant (large standard deviation in Fig. 1), but most cultures showed a significant induction (data not shown). Further analysis will be necessary to determine the nature of the variability in the folK strain and also to investigate the apparent SOS defect in the purA insertion strain. One mutant, the dnaQ insertion strain (B132), behaved much like the recB mutant, reproducibly showing induction by mitomycin C but not nalidixic acid (Fig. 1). We therefore focused our attention on dnaQ in our search for a gene product involved in DSB production after nalidixic acid treatment.
FIG. 1.
dnaQ strains are defective in the SOS response to nalidixic acid. β-Galactosidase activity levels (Miller units) are shown for uninduced control samples (−), nalidixic acid-induced samples (NAL), and mitomycin C-induced samples (MMC) for each strain. Error bars indicate ±1 standard deviation, with the number of trials indicated above the error bar. With only two trials, no standard deviation is listed for the dnaQ transductants; individual values were 19.0 and 13.5 for uninduced samples, 11.4 and 14.3 for nalidixic acid samples, and 94.8 and 77.4 for mitomycin C samples. The differential responses to nalidixic acid (ΔNAL) and mitomycin C (ΔMMC) are listed at the bottom.
Further analysis of the dnaQ mutant.
The dnaQ gene encodes DNA polymerase III ɛ subunit, which is the 3′-to-5′ proofreading exonuclease of the holoenzyme (45). Previous studies of dnaQ concluded that the gene is nearly essential, with knockout mutants forming very small colonies and rapidly accumulating suppressor mutations that restore robust growth (49). The accumulation of suppressor mutations is particularly rapid because of the mutator phenotype of dnaQ mutants (20, 38, 49).
Because dnaQ mutants have a strong mutator phenotype, we were concerned that the SOS induction phenotype might be a result of a second-site mutation caused by the increased mutation frequency, rather than by the transposon insertion mutation in dnaQ. We therefore performed phage P1 transduction using the dnaQ insertion mutant as a donor and JH39 as recipient, selecting for the kanamycin resistance gene of the transposon. The transductants reproduced the specific defect in SOS induction after nalidixic acid treatment, arguing that the phenotype is caused by the dnaQ insertion or a nearby (cotransducible) secondary mutation (Fig. 1). The original screen for SOS constitutive mutants turned up two independent dnaQ insertion mutants, the B132 mutant analyzed here and a mutant designated B119 (39). Strain B119 showed very little SOS induction after nalidixic acid treatment but also appeared to have a reduced response to mitomycin C (see Fig. S1 in the supplemental material). We therefore repeated the P1 transduction test using B119 as the donor, and the transductants displayed a phenotype much closer to that of the B132 strains—inducible by mitomycin C but not by nalidixic acid (see Fig. S1 in the supplemental material). We infer that the two dnaQ insertions cause the same phenotype but that the original B119 strain had a secondary mutation that reduces the SOS response to mitomycin C. Overall, these results argue that the specific defect in SOS induction with nalidixic acid treatment is caused by the dnaQ insertion mutation.
The fact that the two dnaQ insertion mutants grew quite well suggested that they had already acquired a suppressor mutation. Previous work has identified a dnaQ suppressor mutation that substitutes glycine for valine at residue 832 of the DNA polymerase III catalytic subunit (α) encoded by the dnaE gene (21, 22, 49). The P1 transduction results do not rule out the possibility that our dnaQ strains carry a dnaE suppressor mutation, because dnaE is quite close to dnaQ and therefore cotransducible. Sequencing of the entire dnaE gene in five different dnaQ isolates (the two original insertions and three transductants) did not reveal a common suppressor mutation in any of the strains. In fact, the dnaE sequence was wild type in four strains, while the fifth strain contained a single point mutation that would cause a change in the penultimate amino acid of the protein (data not shown).
Another gene that could conceivably be involved in suppressing dnaQ deficiency is recQ. Knockout mutations of recQ were previously found to suppress the SOS induction caused by incubating temperature-sensitive dnaE mutants at a semipermissive temperature (16). Indeed, these authors proposed that the wild-type RecQ protein generates single-stranded DNA as an SOS-inducing signal at the debilitated forks under the semipermissive condition. Because the products of dnaQ and dnaE function together at the replisome, it would not be farfetched to imagine that inactivation of RecQ would likewise suppress growth defects of a dnaQ knockout and potentially also contribute to an inability to induce SOS after nalidixic acid treatment. We therefore sequenced the entire recQ gene in the two original dnaQ insertion mutants and in four of the P1 transductants carrying dnaQ (see above). All six recQ genes had the wild-type sequence, demonstrating that our dnaQ strains do not carry suppressor mutations in this gene.
We next analyzed the phenotype of a pure dnaQ knockout in the JH39 background by transducing JH39 with a duplication construct that has an intact dnaQ gene flanked by a dnaQ::tet disruption, with an intervening counterselectable sacB marker (48, 49). Upon selection for loss of sacB (on sucrose-containing plates), survivors must have undergone homologous recombination to lose the duplication and restore a single copy of dnaQ. In previous studies of this duplication in different genetic backgrounds, selection for the haploid dnaQ::tet segregant (on sucrose tetracycline plates) resulted in very small and irregular colonies, which developed faster-growing sectors upon prolonged incubation (49; R. Schaaper, personal communication). When JH39 with this duplication was plated onto sucrose plates lacking tetracycline, predominantly large colonies were obtained at a frequency of roughly 6 × 10−4, while the same culture plated on sucrose plus tetracycline gave only small colonies at a 70-fold-reduced frequency. The much lower frequency of tetracycline-resistant colonies indicates that dnaQ::tet segregants suffered a very serious growth disadvantage prior to plating. Furthermore, the small colonies showed varied colony morphology, and many but not all were dark blue (SOS constitutive) on a plate containing X-Gal. Some of the colonies were clearly sectored, either for colony morphology or for SOS levels. The variation in colony morphology and SOS levels and the sectoring presumably arise because dnaQ::tet cells are highly mutable (see reference 49). Indeed, many or all of the small colonies that were obtained might have already acquired a suppressor mutation to allow growth. We conclude that a dnaQ knockout mutation is lethal or nearly lethal in the JH39 background, similar to results obtained in other genetic backgrounds previously (49). Because our dnaQ insertion mutants disrupt a large fraction of the coding region, this result implies that our insertion strains do indeed carry a suppressor mutation(s) that restores robust growth, but this must be in some gene(s) other than dnaE or recQ.
Rescue of the dnaQ mutant phenotype.
Based on the above data, we could not rule out the possibility that the SOS phenotype is caused by a common suppressor mutation that occurs frequently in dnaQ strains, particularly if that mutation is cotransducible with dnaQ. We therefore tested whether the SOS phenotype is caused by the dnaQ insertion by asking whether the phenotype could be rescued by introducing a plasmid containing wild-type dnaQ (19, 27). We performed a control experiment in parallel, transforming the wild-type dnaQ-containing plasmid into the wild-type JH39 strain before introducing the dnaQ insertion allele into the chromosome by P1 transduction (selecting for kanamycin resistance). Rescue strains created by the latter procedure were never without a wild-type dnaQ gene and therefore should never have experienced pressure to accumulate a suppressor mutation.
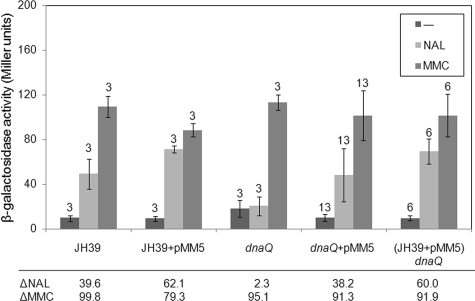
The rescue strains described above were tested in the β-galactosidase assay in the presence of nalidixic acid or mitomycin C and compared to wild-type positive controls and dnaQ mutant negative controls. All of the strains created by adding wild-type dnaQ to JH39 followed by transduction of the dnaQ allele [(JH39 + pMM5) dnaQ] showed an SOS profile similar to those of JH39 and the JH39 plus plasmid pMM5 controls (Fig. 2). Importantly, a large majority of the strains created by direct transformation of the wild-type dnaQ plasmid into the dnaQ mutant strain behaved much like wild-type JH39 or the JH39 plus pMM5 controls (Fig. 2). Two of 13 transformants behaved more like the original dnaQ insertion mutation, arguing that they had sustained an additional mutation (due to the mutator background) that alters their SOS phenotype (see Fig. S2 in the supplemental material). All of the tested strains retained both the antibiotic resistance of the plasmid and the transposon after growth under β-galactosidase assay conditions, arguing that recombination had not occurred to generate dnaQ+/dnaQ+ partial diploids (data not shown). In conclusion, complementation of a large majority of the dnaQ transformants by the wild-type dnaQ gene strongly argues that the ɛ subunit of DNA polymerase III is involved in the nalidixic acid-induced SOS response in wild-type cells.
FIG. 2.
Wild-type dnaQ complements the dnaQ mutant phenotype. β-Galactosidase activity levels (Miller units) are shown for uninduced control samples (−), nalidixic acid-induced samples (NAL), and mitomycin C-induced samples (MMC) for each strain. Error bars indicate ±1 standard deviation, with the number of trials indicated above the error bar. The differential responses to nalidixic acid (ΔNAL) and mitomycin C (ΔMMC) are listed at the bottom.
Analysis of additional SOS genes.
One caveat to the conclusion that ɛ is involved in the SOS response is that the effects on dinD::lacZ expression could be related to some regulatory aspect of the dinD fusion construct other than the SOS response. Thus, we sought to confirm a general defect in SOS induction by nalidixic acid in the dnaQ mutant. We began by measuring the levels of the SOS-inducible RecA protein in the wild-type versus dnaQ mutant strains in the presence and absence of nalidixic acid. Surprisingly, RecA protein was induced by nalidixic acid in both strains (see Fig. S3 in the supplemental material). This result could mean that the results with the dinD::lacZ reporter are misleading or alternatively could reflect some temporal or regulatory complexity of the SOS response (see Discussion). Therefore, we analyzed the transcript levels for a collection of SOS genes.
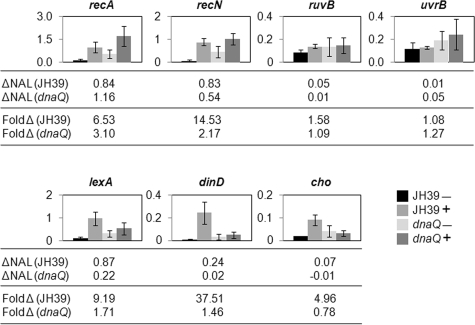
Quantitative real-time PCR was used to measure expression levels of SOS genes in the wild-type versus dnaQ mutant strains in the presence and absence of nalidixic acid (2-hour treatment). Total RNA was isolated from three separate experiments to account for possible variation stemming from the dnaQ mutator phenotype. To control for variation in reverse transcriptase efficiency, the constitutively expressed gapA (d-glyceraldehyde 3-phosphate dehydrogenase) gene was measured and gene expression values were normalized to gapA in each sample. In the JH39 wild-type strain, five of the seven genes analyzed showed strong induction after nalidixic acid treatment, with expression levels increasing from 4.96- to 38-fold over those of uninduced cells (Fig. 3; recA, recN, lexA, dinD, and cho). For the two genes with uncertain induction by nalidixic acid in the wild-type strain (ruvB and uvrB), the corresponding dnaQ induction results are not easily interpreted. Considering the five genes that were strongly induced in JH39, two showed modest induction by nalidixic acid in the dnaQ mutant strain (recA and recN), while three displayed reduced or ablated induction by nalidixic acid in the dnaQ mutant strain (lexA, dinD, and cho) (Fig. 3). These results indicate that the ɛ subunit of DNA polymerase III is involved in the regulation of a subset of SOS genes in response to nalidixic acid (see below).
FIG. 3.
Nalidixic acid-induced expression of selected SOS genes. Quantitative real-time PCR was used to measure expression levels of the genes indicated in the JH39 wild-type and dnaQ mutant strains in the presence (+) and absence (−) of nalidixic acid. Graphs represent the averages of three separate experiments where expression values are normalized to gapA in each sample, and error bars indicate ±1 standard deviation. The responses by each gene to nalidixic acid (nalidixic acid-induced samples minus uninduced samples; ΔNAL) are listed for both the JH39 wild-type strain and the dnaQ mutant, as are the increases (nalidixic acid-induced samples divided by uninduced samples; FoldΔ) within each strain.
DISCUSSION
Quinolones such as nalidixic acid and norfloxacin stabilize topoisomerase cleavage complexes, leading to induction of the SOS response and cell death. Various results argue that the pathway to cytotoxicity involves the conversion of stabilized cleavage complexes into frank DSBs. In order to search for proteins involved in this conversion event, we took advantage of the SOS response to isolate mutants that are specifically defective in the SOS induction to nalidixic acid treatment. Our initial attempt to isolate such mutants (37) uncovered 18 insertions into the recB and recC genes, which were already known to be involved in the SOS response to nalidixic acid (31).
In this study, we screened for mutants that are defective in the SOS response to nalidixic acid among insertions in 47 different genes that cause a partially constitutive SOS response; their constitutive phenotype would have prevented them from being uncovered in the Newmark et al. (37) screen. We found that insertions in one gene, dnaQ, cause a specific defect in SOS induction by nalidixic acid but not by mitomycin C (Table 1; Fig. 1). Two independent transposon insertions in the dnaQ gene were shown to cause this phenotype, the phenotype could be cotransduced with the transposon insertion, and complementation experiments supported the conclusion that the phenotype is caused by the lack of dnaQ function (Fig. 2; see also Fig. S2 in the supplemental material).
Mutations in dnaQ were previously shown to cause hypersensitivity to nalidixic acid (27), and we also detected a modest hypersensitivity of our insertion strain using qualitative disk diffusion tests such as those in the work of Newmark et al. (37) (data not shown). The hypersensitivity raises a possible trivial explanation for the lack of SOS induction, namely, that dnaQ mutants fail to induce simply because they are rapidly ceasing metabolism upon exposure to the drug. This explanation seems very unlikely for the following reasons: (i) other nalidixic acid-hypersensitive mutants still induce the nalidixic acid SOS response (e.g., uvrD, ruvA, ruvB, ruvC, polA, acrA, acrB, tolC, ftsX, ftsE, and tdcE [Table 1]); (ii) the dnaQ mutant increased 3.4-fold in mass during the 2-hour incubation after addition of nalidixic acid (data not shown); (iii) dnaQ mutants are also hypersensitive to mitomycin C (27) but are still mitomycin C inducible (Fig. 1); and (iv) while a subset of SOS genes show a deficiency in response to nalidixic acid, two others show some induction (Fig. 3).
The two dnaQ insertion strains are likely completely null for function of the ɛ subunit of DNA polymerase III. The transposons are located just after codons 99 and 158 in the ɛ subunit, flanking the site of the previously studied truncation mutant (truncated just after residue 128) that behaves as a complete null (49). However, dnaQ knockout strains are essentially inviable without an additional suppressor mutation(s). A suppressor mutation in dnaE has been shown to reduce the constitutive SOS phenotype and allow cell growth but does not relieve the mutator phenotype (21, 22, 24, 49). These results argue for at least two distinct functions of the ɛ subunit, one in proofreading and one in some aspect of replication fork stability that normally prevents SOS induction. Furthermore, both in vivo and in vitro experiments indicate that ɛ-deficient replication forks are somehow feeble, and the suppressor mutations in the dnaE-encoded α subunit may relieve this polymerization/elongation defect (24, 35, 44, 49, 50).
By analyzing loss of dnaQ function from a duplication construct in the JH39 genetic background (see Results), we conclude that our dnaQ insertion strains likely contain one or more suppressor mutations. In search of these suppressor mutations, we sequenced both the dnaE and recQ genes of our dnaQ insertion mutants but found no mutations (except for a single point mutation in the dnaE gene of only one out of five dnaQ strains). The dnaE and recQ genes are the only strong candidates for possible suppressor mutations for dnaQ deficiency, based on prior studies (21, 22, 25, 49). Despite our inability to identify a suppressor mutation(s) in our dnaQ strains, we are confident that any suppressor mutation that might exist cannot be the major/sole cause of the SOS induction phenotype. Indeed, the plasmid rescue experiment (Fig. 2) demonstrates that the SOS phenotype can be rescued by provision of wild-type ɛ protein. The only remaining caveat is that the SOS phenotype might conceivably require both a deficiency of DnaQ and a mutation at a common suppressor locus, but even in this case, deficiency of the ɛ subunit of DNA polymerase III would still be involved in the SOS phenotype.
A variety of models could explain the involvement of the ɛ subunit in the SOS response to nalidixic acid, particularly since ɛ is at the heart of the replisome. One model involves the behavior of the replisome upon encountering a block. dnaQ mutants display large increases in direct repeat recombination, which has been attributed to excessive single-stranded regions and/or broken replication forks (35, 44). Some (but not all) biochemical studies have revealed decreased polymerase III processivity in the absence of ɛ (29, 50), and so the biochemical basis of the aberrant replication is not entirely clear. However, if ɛ is important in triggering the disassembly of a blocked replisome, an ɛ-free replisome could have a longer half-life when the fork reaches a cleavage complex. A reversal of quinolone-induced topoisomerase cleavage complex at the blocked fork could then allow the original replisome to continue after a transient stall (rather than dissociating and exposing the fork to processing enzymes).
Another general model posits that ɛ has a previously unrecognized activity that acts directly on the cleavage complex or blocked fork. For example, an endonuclease activity may cleave the fork when it is blocked for a prolonged period of time. Alternatively, the ɛ subunit might recruit another protein that acts directly on the blocked fork, although we did not uncover any strong candidates for such a protein in the transposon screens. It is even possible that ɛ interacts directly with the topoisomerase cleavage complexes in some manner—suppressors of the mutator phenotype of dnaQ mutants have been found in the gyrA gene (which encodes DNA gyrase subunit A), although the mechanistic basis of this suppression is unclear (6).
It is interesting that ɛ has been implicated in a bacterial replication checkpoint response dependent on the UmuD2C complex (53). When replication forks encounter DNA damage, the SOS response is induced, and as a result, levels of UmuD and UmuC increase. Although the molecular mechanism of the checkpoint response is not clear, the UmuD2C complex further inhibits DNA replication, presumably to allow more time for conventional repair reactions. Interactions of UmuD2C with both ɛ and the β processivity clamp are implicated in the checkpoint response (51, 52). We see no obvious model in which the lack of a checkpoint response could relate to an inability of dnaQ mutants to induce particular SOS genes after nalidixic acid treatment; furthermore, neither umuD nor umuC mutants were uncovered in these genetic screens.
We focused on the dnaQ insertion mutant because of its defective response to nalidixic acid, as measured by dinD-driven expression. However, the dnaQ mutant exhibited an apparently normal induction of RecA protein levels in response to nalidixic acid, arguing that the SOS defect is not complete (see Fig. S3 in the supplemental material). To further characterize the SOS defect, we measured the possible induction of a variety of SOS transcripts in the wild type versus the dnaQ insertion mutant. A subset of SOS genes, including lexA, dinD, and cho, showed little or no induction in the dnaQ mutant, similar to the behavior of the dinD-lacZ reporter construct (Fig. 3). On the other hand, transcripts for the recA and recN genes were upregulated by nalidixic acid in both the wild type and the dnaQ mutant, correlating with the induction of RecA protein levels that we detected by Western blotting. We conclude that the dnaQ mutation blocks only a subset of the nalidixic acid-induced SOS response.
There is ample evidence in the literature for SOS genes responding differentially and for the SOS response being much more complex than an on/off switch (for a review, see reference 13). In particular, the induction ratios for various SOS genes differ dramatically, different SOS genes are both induced and turned off at different times during the SOS response, a subset of SOS promoters are induced only with relatively high levels of DNA damage, proteins including DinI and RecX can affect LexA autoproteolysis, and the levels of certain SOS proteins are modulated by cellular proteases and chaperones. We infer that the partial blockage of SOS induction in the dnaQ mutant reflects some aspect of this complexity, although the collection of genes that are inducible versus noninducible does not suggest a simple model to us. The main conclusion that seems clear at this point is that the ɛ subunit of DNA polymerase III plays an important role in modulating the SOS response to nalidixic acid, implicating the replisome in at least some aspect of the processing of drug-stabilized cleavage complexes. Given that the SOS defect is incomplete, the dnaQ mutations may affect only one of two or more redundant pathways for DSB formation.
Results from the phage T4 system argue that topoisomerase inhibitors can lead to “collateral” DNA damage, in which blocked replication forks are cleaved by recombination nucleases that are branch specific (18). Furthermore, we previously found that quinolone-stabilized gyrase cleavage complexes block replication forks in vivo, and these blocked forks are susceptible to breakage in vivo by an unknown mechanism (41). We had therefore anticipated that a nuclease or resolvase might create the DSBs after nalidixic acid treatment in E. coli. The ɛ subunit could be indirectly involved in DSB formation, for example by altering the dynamics of fork stalling or by recruiting an endonuclease. The branch resolution system RuvABC could conceivably be involved in SOS induction by nalidixic acid by cleaving the blocked replication fork (after fork regression; see references 33 and 46). In the screen for SOS constitutive mutants (39), insertions were found in ruvA, ruvB, and ruvC that show a modest (approximately 30%) defect in induction by nalidixic acid (Table 1). However, these mutants are about equally defective in induction by mitomycin C (Mary Rogers and Kenneth Kreuzer, unpublished data), and so RuvABC is not specifically required for induction of the SOS response after nalidixic acid. It remains possible that RuvABC and an unknown redundant activity are involved in generating the SOS-inducing signal from stabilized gyrase cleavage complexes and/or blocked forks. Candidates for redundant activities include YqgF, an uncharacterized protein that is related to RuvC by both sequence (1) and structure (25), and SbcCD, which can cleave branched DNA including cruciform structures and protein-DNA complexes (4, 5). Neither yqfF nor sbcCD was found in our various screens for SOS induction defects, but we did find a number of other insertion mutants with partial reductions (Table 1). Future studies involving multiple mutants are necessary to address the possibility of redundant pathways.
In summary, the results presented here implicate the ɛ subunit of DNA polymerase III holoenzyme in particular, and the replisome in general, in the SOS response to nalidixic acid. Perhaps the most likely model is that the presence or absence of ɛ in the replisome alters the dynamics of replication fork stalling, which indirectly affects generation of DSBs at the stalled forks. Elucidation of the detailed mechanism of DSB formation may reveal important aspects of replication fork dynamics and could also have implications for antibacterial and anticancer chemotherapy with topoisomerase inhibitors.
Supplementary Material
Acknowledgments
We are very grateful to Roel Schaaper (NIEHS) for strains and for excellent advice.
This research was supported by NIH grants GM072089 and its predecessor GM065206.
Footnotes
Published ahead of print on 6 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Survey and summary: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 283417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breña-Valle, M., and J. Serment-Guerrero. 1998. SOS induction by gamma-radiation in Escherichia coli strains defective in repair and/or recombination mechanisms. Mutagenesis 13637-641. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. R., M. Malik, M. Snyder, and K. Drlica. 1996. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J. Mol. Biol. 258627-637. [DOI] [PubMed] [Google Scholar]
- 4.Connelly, J. C., E. S. de Leau, and D. R. Leach. 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair 2795-807. [DOI] [PubMed] [Google Scholar]
- 5.Connelly, J. C., L. A. Kirkham, and D. R. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 957969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, E. C. 1976. Bacterial mutator genes and the control of spontaneous mutation. Annu. Rev. Genet. 10135-156. [DOI] [PubMed] [Google Scholar]
- 7.Cozzarelli, N. R. 1980. DNA gyrase and the supercoiling of DNA. Science 207953-960. [DOI] [PubMed] [Google Scholar]
- 8.D'Arpa, P., C. Beardmore, and L. F. Liu. 1990. Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 506919-6924. [PubMed] [Google Scholar]
- 9.D'Arpa, P., and L. Liu. 1989. Topoisomerase-targeting antitumor drugs. Biochim. Biophys. Acta 989163-177. [DOI] [PubMed] [Google Scholar]
- 10.Deitz, W. H., T. M. Cook, and W. A. Goss. 1966. Mechanism of action of nalidixic acid on Escherichia coli. III. Conditions required for lethality. J. Bacteriol. 91768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fath, M., H. Mahanty, and R. Kolter. 1989. Characterization of a purF operon mutation which affects colicin V production. J. Bacteriol. 1713158-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedberg, E., G. Walker, W. Siede, R. Wood, R. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC.
- 14.Gudas, L. J., and A. B. Pardee. 1976. DNA synthesis inhibition and the induction of protein X in Escherichia coli. J. Mol. Biol. 101459-477. [DOI] [PubMed] [Google Scholar]
- 15.Heitman, J., and P. Model. 1991. SOS induction as an in vivo assay of enzyme-DNA interactions. Gene 1031-9. [DOI] [PubMed] [Google Scholar]
- 16.Hishida, T., Y.-W. Han, T. Shibata, Y. Kubota, Y. Ishino, H. Iwasaki, and H. Shinagawa. 2004. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 181886-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holm, C., J. M. Covey, D. Kerrigan, and Y. Pommier. 1989. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 496365-6368. [PubMed] [Google Scholar]
- 18.Hong, G., and K. N. Kreuzer. 2003. Endonuclease cleavage of blocked replication forks: an indirect pathway of DNA damage from antitumor drug-topoisomerase complexes. Proc. Natl. Acad. Sci. USA 1005046-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horiuchi, T., H. Maki, M. Maruyama, and M. Sekiguchi. 1981. Identification of the dnaQ gene product and location of the structural gene for RNase H of Escherichia coli by cloning of the genes. Proc. Natl. Acad. Sci. USA 783770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horiuchi, T., H. Maki, and M. Sekiguchi. 1978. A new conditional lethal mutator (dnaQ49) in Escherichia coli K12. Mol. Gen. Genet. 163277-283. [DOI] [PubMed] [Google Scholar]
- 21.Lancy, E. D., M. R. Lifsics, D. G. Kehres, and R. Maurer. 1989. Isolation and characterization of mutants with deletions in dnaQ, the gene for the editing subunit of DNA polymerase III in Salmonella typhimurium. J. Bacteriol. 1715572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lancy, E. D., M. R. Lifsics, P. Munson, and R. Maurer. 1989. Nucleotide sequences of dnaE, the gene for the polymerase subunit of DNA polymerase III in Salmonella typhimurium, and a variant that facilitates growth in the absence of another polymerase subunit. J. Bacteriol. 1715581-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewin, C. S., B. M. Howard, N. T. Ratcliffe, and J. T. Smith. 1989. 4-Quinolones and the SOS response. J. Med. Microbiol. 29139-144. [DOI] [PubMed] [Google Scholar]
- 24.Lifsics, M. R., E. D. Lancy, Jr., and R. Maurer. 1992. DNA replication defect in Salmonella typhimurium mutants lacking the editing (epsilon) subunit of DNA polymerase III. J. Bacteriol. 1746965-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, D., Y. S. Wang, and D. F. Wyss. 2003. Solution structure of the hypothetical protein YqgF from Escherichia coli reveals an RNAse H fold. J. Biomol. NMR 27389-392. [DOI] [PubMed] [Google Scholar]
- 26.Liu, L. 1983. DNA topoisomerases—enzymes that catalyze the breaking and rejoining of DNA. Crit. Rev. Biochem. 151-24. [DOI] [PubMed] [Google Scholar]
- 27.Maki, H., T. Horiuchi, and M. Sekiguchi. 1983. Structure and expression of the dnaQ mutator and the RNase H genes of Escherichia coli: overlap of the promoter regions. Proc. Natl. Acad. Sci. USA 807137-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik, M., X. Zhao, and K. Drlica. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61810-825. [DOI] [PubMed] [Google Scholar]
- 29.Marians, K. J., H. Hiasa, D. R. Kim, and C. S. McHenry. 1998. Role of the core DNA polymerase III subunits at the replication fork. Alpha is the only subunit required for processive replication. J. Biol. Chem. 2732452-2457. [DOI] [PubMed] [Google Scholar]
- 30.McDaniel, L. S., L. H. Rogers, and W. E. Hill. 1978. Survival of recombination-deficient mutants of Escherichia coli during incubation with nalidixic acid. J. Bacteriol. 1341195-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McPartland, A., L. Green, and H. Echols. 1980. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell 20731-737. [DOI] [PubMed] [Google Scholar]
- 32.Meddows, T. R., A. P. Savory, J. I. Grove, T. Moore, and R. G. Lloyd. 2005. RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol. Microbiol. 5797-110. [DOI] [PubMed] [Google Scholar]
- 33.Michel, B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25173-178. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Morag, A. S., C. J. Saveson, and S. T. Lovett. 1999. Expansion of DNA repeats in Escherichia coli: effects of recombination and replication functions. J. Mol. Biol. 28921-27. [DOI] [PubMed] [Google Scholar]
- 36.Myers, R. S., and F. W. Stahl. 1994. Chi and the RecBCD enzyme of Escherichia coli. Annu. Rev. Genet. 2849-70. [DOI] [PubMed] [Google Scholar]
- 37.Newmark, K. G., E. K. O'Reilly, J. R. Pohlhaus, and K. N. Kreuzer. 2005. Genetic analysis of the requirements for SOS induction by nalidixic acid in Escherichia coli. Gene 35669-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oller, A. R., I. J. Fijalkowska, and R. M. Schaaper. 1993. The Escherichia coli galK2 papillation assay: its specificity and application to seven newly isolated mutator strains. Mutat. Res. 292175-185. [DOI] [PubMed] [Google Scholar]
- 39.O'Reilly, E. K., and K. N. Kreuzer. 2004. Isolation of SOS constitutive mutants of Escherichia coli. J. Bacteriol. 1867149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picksley, S., P. Attfield, and R. Lloyd. 1984. Repair of DNA double-strand breaks in Escherichia coli K12 requires a functional recN product. Mol. Gen. Genet. 195267-274. [DOI] [PubMed] [Google Scholar]
- 41.Pohlhaus, J. R., and K. N. Kreuzer. 2005. Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo. Mol. Microbiol. 561416-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robbins-Manke, J. L., Z. Z. Zdraveski, M. Marinus, and J. M. Essigmann. 2005. Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J. Bacteriol. 1877027-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 21279-96. [DOI] [PubMed] [Google Scholar]
- 44.Saveson, C. J., and S. T. Lovett. 1997. Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics 146457-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheuermann, R. H., and H. Echols. 1984. A separate editing exonuclease for DNA replication: the epsilon subunit of Escherichia coli DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. USA 817747-7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seigneur, M., S. D. Ehrlich, and B. Michel. 2000. RuvABC-dependent double-strand breaks in dnaBts mutants require recA. Mol. Microbiol. 38565-574. [DOI] [PubMed] [Google Scholar]
- 47.Skaar, E. P., M. P. Lazio, and H. S. Seifert. 2002. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J. Bacteriol. 184919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slater, S., and R. Maurer. 1993. Simple phagemid-based system for generating allele replacements in Escherichia coli. J. Bacteriol. 1754260-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slater, S. C., M. R. Lifsics, M. O'Donnell, and R. Maurer. 1994. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J. Bacteriol. 176815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studwell, P. S., and M. O'Donnell. 1990. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J. Biol. Chem. 2651171-1178. [PubMed] [Google Scholar]
- 51.Sutton, M. D., M. F. Farrow, B. M. Burton, and G. C. Walker. 2001. Genetic interactions between the Escherichia coli umuDC gene products and the beta processivity clamp of the replicative DNA polymerase. J. Bacteriol. 1832897-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutton, M. D., S. Murli, T. Opperman, C. Klein, and G. C. Walker. 2001. umuDC-dnaQ interaction and its implications for cell cycle regulation and SOS mutagenesis in Escherichia coli. J. Bacteriol. 1831085-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34479-497. [DOI] [PubMed] [Google Scholar]
- 54.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 55.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3430-440. [DOI] [PubMed] [Google Scholar]
- 56.Whitby, M. C., and R. G. Lloyd. 1995. Altered SOS induction associated with mutations in recF, recO and recR. Mol. Gen. Genet. 246174-179. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, W. R., and G. F. Whitmore. 1981. Cell-cycle-stage specificity of 4′-(9-acridinylamino)methanesulfon-m-anisidide (m-AMSA) and interaction with ionizing radiation in mammalian cell cultures. Radiat. Res. 87121-136. [PubMed] [Google Scholar]
- 58.Zhao, X., M. Malik, N. Chan, A. Drlica-Wagner, J.-Y. Wang, X. Li, and K. Drlica. 2006. Lethal action of quinolones against a temperature-sensitive dnaB replication mutant of Escherichia coli. Antimicrob. Agents Chemother. 50362-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.