Abstract
Among the 67 predicted TonB-dependent outer membrane transporters of Caulobacter crescentus, NagA was found to be essential for growth on N-acetyl-β-d-glucosamine (GlcNAc) and larger chitin oligosaccharides. NagA (93 kDa) has a predicted typical domain structure of an outer membrane transport protein: a signal sequence, the TonB box EQVVIT, a hatch domain of 147 residues, and a β-barrel composed of 22 antiparallel β-strands linked by large surface loops and very short periplasmic turns. Mutations in tonB1 and exbBD, known to be required for maltose transport via MalA in C. crescentus, and in two additional predicted tonB genes (open reading frames cc2327 and cc3508) did not affect NagA-mediated GlcNAc uptake. nagA is located in a gene cluster that encodes a predicted PTS sugar transport system and two enzymes that convert GlcNAc-6-P to fructose-6-P. Since a nagA insertion mutant did not grow on and transport GlcNAc, diffusion of GlcNAc through unspecific porins in the outer membrane is excluded. Uptake of GlcNAc into tonB and exbBD mutants and reduction but not abolishment of GlcNAc transport by agents which dissipate the electrochemical potential of the cytoplasmic membrane (0.1 mM carbonyl cyanide 3-chlorophenylhydrazone and 1 mM 2,4-dinitrophenol) suggest diffusion of GlcNAc through a permanently open pore of NagA. Growth on (GlcNAc)3 and (GlcNAc)5 requires ExbB and ExbD, indicating energy-coupled transport by NagA. We propose that NagA forms a small pore through which GlcNAc specifically diffuses into the periplasm and functions as an energy-coupled transporter for the larger chitin oligosaccharides.
TonB-dependent outer membrane proteins actively transport Fe3+ siderophores, heme, Fe3+, and vitamin B12 across the outer membrane of gram-negative bacteria (4, 6, 39, 40, 52, 54). It is thought that energy is required for two steps: (i) release of the tightly bound substrates from the transporters and (ii) movement of the hatch to open a pore in the β-barrel (10, 53). The energy is provided by the proton motive force of the cytoplasmic membrane. A protein complex composed of TonB, ExbB, and ExbD is involved in the energy transfer from the cytoplasmic membrane to the outer membrane. TonB interacts with the TonB box in the N-terminal region of the transporters (9, 36, 37, 48). In Escherichia coli, eight such transporters have been related to the transport of specific substrates.
Analysis of the genome sequence of Caulobacter crescentus predicts 67 TonB-dependent outer membrane proteins (32, 33). We have identified the tonB, exbB, and exbD genes, required for the transport of maltose and larger maltodextrins (29, 32). Transport is mediated by the outer membrane protein MalA (32). malA, tonB, exbB, and exbD gene insertion mutants transport maltose at 2% of the wild-type rate. This was the first description of TonB-dependent energy-coupled transport across the outer membrane for substrates other than Fe3+, Fe3+ chelates, and vitamin B12.
In nature, maltodextrins are derived from abundant starch. It is reasonable to assume that C. crescentus takes advantage of this carbon source, because it thrives in nutrient-poor freshwater lakes. Since C. crescentus encodes such a large number of predicted transporters, we hypothesized that many substrates in low abundance are actively transported across the outer membrane. In this study, we examined whether C. crescentus can grow on chitin, the second-largest carbon source in nature after cellulose, and its degradation products N-acetyl-β-d-glucosamine (GlcNAc) and higher homologs N-acetyl-chitobiose, -triose, -tetraose, and -pentaose. We determined growth on chitin and its degradation products, identified the gene cluster responsible for utilization of GlcNAc, identified the outer membrane protein that allows growth on GlcNAc, and tested whether transport depends on TonB, ExbB, and ExbD. It was found that GlcNAc uptake differs from maltose transport across the outer membrane in that in contrast to the case with maltose uptake, TonB and ExbB ExbD were not required, and uptake of GlcNAc across the cytoplasmic membrane occurs via the sugar transferase system (phosphotransferase transport system [PTS]) and maltose uptake via proton (ion) coupling.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
The C. crescentus and E. coli strains and the plasmids used in this study are listed in Table 1. The C. crescentus strains are derived from the sequenced CB15 strain. E. coli was grown at 37°C on tryptone-yeast extract medium (32), on which C. crescentus does not multiply. C. crescentus was grown at 30°C on PYE medium or on M2 minimal medium (32) supplemented with 0.3% of the carbon source to be tested. Growth on chitin was tested with chitin from crab shells purchased from Sigma-Aldrich (Seelze, Germany) or from Carl Roth (Karlsruhe, Germany). Chitin from Roth was hydrolyzed to determine contaminating protein. The sum of the amino acids was 0.7% of the value of GlcNAc. Small amounts of pure (GlcNAc)2, (GlcNAc)3, (GlcNAc)4, (GlcNAc)5, and (GlcNAc)6 were obtained from Hans Jürgen Plattner (Microbiology, University of Hannover, Hanover, Germany); (GlcNAc)3 and (GlcNAc)5 were purchased from Seikagaku America through Medac (Wedel, Germany). Antibiotics were used at the following concentrations (in μg per ml) for C. crescentus and in parentheses for E. coli: ampicillin, 50 on plates, 7.5 (50) in liquid culture; chloramphenicol, 2 (25); tetracycline, 2 on plates, 1 in liquid culture; gentamicin, 50 on plates, 25 (50) in liquid culture; spectinomycin, 50 on plates, 25 (50) in liquid culture; streptomycin, 5 (30); kanamycin (30); and nalidixic acid (20). C. crescentus JS1003 is resistant to nalidixic acid, ampicillin, and kanamycin.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype and/or phenotype | Reference or source |
|---|---|---|
| Caulobacter crescentus | ||
| JS1003 | NA1000 rsaA::KSAC Kmr cassette, Ampr | 15 |
| HB2004 | JS1003 tonB2::Ω cassette, Kmr Ampr Spcr Strr | 32 |
| HB2007 | JS1003 exbBD1::Gmr cassette, Kmr Ampr Gmr | 32 |
| UJ2602 | CB15 ΔrsaA | 23 |
| SE0446 | UJ2602 nagA::Ω cassette, Spcr Strr | This study |
| SE0446(pSE0446) | SE0446, pSE0446, Kmr | This study |
| SL2334a | UJ2602 tonB1::Ω cassette, Spcr Strr | This study |
| SL3508 | UJ2602 tonB3::Ω cassette, Spcr Strr | This study |
| SL8 | UJ2602 tonB2::Ω cassette, tonB3::Ω cassette, Spcr Strr | This study |
| SL9 | UJ2602 tonB1::Ω cassette tonB2::Ω cassette, Spcr Strr | This study |
| Escherichia coli | ||
| BL21(DE3)omp8 | F−hds gal, T7 polymerase under lac control | 32 |
| BL21(DE3)omp8a | As BL21(DE3) but degP | D. Linke |
| S17-1 λ pir | hsdR hsdM+ RP4-2-Tc::Mu Km::Tn7 (Tpr Smr) Tra+ λ pir | 1 |
| JM109WΩ | endA1 recA1 gyrA96 thi hsdR17 (rk− mk+) relA1 supE44 Δ(lac-proAB) [F′traD36 proAB lacIqZΔM15] | Promega |
| Serratia marcescens CDC08:H3 | This institute | |
| Plasmids | ||
| pBBnagA | nagA in pBBR1MCS-2, Kmr | This study |
| pDrive cloning vector | Kmr Ampr cloning vector | Qiagen |
| pGEMTeasy | Ampr cloning vector | Promega |
| pBAD/Myc-HisB | T7 expression vector, Kmr | Novagen |
| pNPTS138Tet | Kmr derivative of pLITMUS with sacB and oriT, Tetr | 32 |
| pETextonB | pETcctonB2 exbBD1 | 32 |
| pHP45Ω | Ω cassette, Specr Strepr | 42 |
| pBBR1MCS-2 | Kmr, MCS, MOB | 26 |
| pSEXΩY | cc0445Ωcc0447 Specr Strepr Tetr | This study |
Growth promotion was tested on M2 salt agar plates on which 108 bacteria were seeded in 3 ml M2 soft salt agar. Ten microliters of a 5% solution of the nutrients to be tested was placed on sterile filter paper disks of 6 mm in diameter. Bacterial growth around the disks was scored after 16 and 24 h of incubation at 30°C.
Recombinant DNA techniques.
Isolation of plasmids, use of restriction enzymes, ligation, agarose gel electrophoresis, transformation, hybridization, and Southern blotting followed standard methods (45). PCR of E. coli DNA was started with 3 min at 94°C, followed by 1 min at 94°C for 35 cycles, 2 min of annealing at 54°C, 3 min of elongation at 72°C, and 10 min at 72°C. GC-rich DNA was amplified as described for E. coli, but the annealing temperature started at 64°C and was lowered at each cycle by 0.5°C. Information on the primers used for PCR will be provided upon request.
Cloning of nagA and construction of a nagA mutant.
Chromosomal DNA of C. crescentus UJ2602 was isolated using the Easy-DNA kit of Invitrogen (Groningen, The Netherlands). nagA was cloned by PCR with appropriately designed primers from the C. crescentus chromosome. Plasmid pBBnagA contains the nagA gene on a 3-kb XhoI-HindIII fragment cloned in the pBBR1MCS-2 vector. nagA was mutated by insertion of an Ω cassette (1, 16) into cc0466, resulting in pSEXΩY, as described previously for mutagenesis of malA (32). pnagAHis, encoding NagA with six C-terminal histidine residues, was constructed by cloning a 3-kb XhoI-HindIII fragment into pBAD/Myc-HisB.
[14C]GlcNAc transport assay.
Cultures were incubated at 30°C in M2 medium supplemented with either 0.3% GlcNAc or 0.3% glucose as a control to an optical density at 578 nm (OD578) of 0.5. Cells were harvested by centrifugation, washed with M2 medium, and then suspended in the transport assay medium (0.1 mM HEPES, 0.23 mM MgCl2, 0.15 mM CaCl2, 10 mM NaH2PO4, 5 mM KH2PO4, pH 7.2) to an OD578 of 0.4. Purchased [14C]GlcNAc (3.63 mM; specific radioactivity, 2.04 GBq/mmol) was diluted to 150 μM; 9 μl of the dilution was added to 0.9 ml cell suspension, and the mixture was then shaken at 30°C. Aliquots of 0.2 ml were withdrawn after 0.2, 1, 2, and 3 min, collected on cellulose nitrate filters, washed twice with 5 ml M2 salt medium, and dried. The radioactivity was determined in a liquid scintillation counter. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP), used to inhibit transport, was dissolved in 5% acetic acid and then adjusted with 1 M NaOH to pH 7.1. Cells were incubated with 0.1 mM CCCP or 1 mM 2,4-dinitrophenol (DNP) for 10 min prior to the addition of radiolabeled substrate.
Isolation of the outer membrane fraction of C. crescentus, two-dimensional gel electrophoresis, and mass spectrometry for protein identification.
Isolation of the outer membrane fraction, two-dimensional gel electrophoresis, and mass spectrometry were carried out as previously described (32), except that the carbon source consisted of chitin degradation products obtained by chitin hydrolysis instead of maltose.
Preparation of NagA.
NagA was solubilized from outer membranes in 2% dodecyl-N-N′-dimethylamine-N-oxide (LDAO), which did not solubilize MalA-His6 (32).
Chitinase test.
Chitinase (Sigma) was dissolved in 0.1 mM phosphate buffer, pH 7, to 0.1 mg/ml and added to a filter paper containing 0.1 ml of 20 μM of the chitinase substrate 4-methylumbelliferyl-β-d-N,N′-diacetyl chitobioside hydrate in 0.1 mM phosphate buffer (35). After 10 min of incubation, the filter paper fluoresced at 365 nm. C. crescentus JS1003 was grown in M2 medium supplemented with 0.3% GlcNAc to an OD578 of 0.5. A 1-ml aliquot was centrifuged, and the pelleted cells were resuspended in 0.1 ml of phosphate buffer. The supernatant (0.1 ml) and the cell suspension were added to a filter paper containing the chitinase substrate. As a control, the supernatant and the cell suspension of a culture of the chitinase-positive Serratia marcescens strain CDC08:H3 were prepared, following the procedures for the C. crescentus samples.
Preparation of chitin degradation products.
For the isolation of outer membranes of C. crescentus, larger amounts of N-acetyl-chitin oligosaccharides were needed for 1-liter cultures. Following a protocol obtained from H. J. Plattner, 50 g chitin (Roth) was suspended in 500 ml 37% HCl and stirred for 2 h at 4°C. It was then incubated for 2 h at 40°C, cooled to 0°C, and adjusted to pH 7 with 50% NaOH. The suspension was centrifuged for 15 min, and the supernatant was filtered through a glass filter and concentrated to 50 ml on a rotary evaporator. The degradation products were size fractionated on a Biogel P-2 acrylamide column (Bio-Rad Laboratories, München, Germany) with water as an eluent and analyzed by high-performance liquid chromatography on a Reprosil NH 2.5-μm column using an isocratic gradient of 30 to 60% acetonitrile in double-distilled water. Fractions containing predominantly GlcNAc, (GlcNAc)2, (GlcNAc)3, (GlcNAc)4, (GlcNAc)5, and (GlcNAc)6 were pooled and used as a carbon source. Fifty grams of chitin gave 4.8 g of structurally defined degradation products.
Sequence alignments.
C. crescentus sequences of the genome (33) were compared with sequences deposited in the NCBI data bank (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genom&cmd=Retrieve&dopt=overview&list_uids=117) using http://www.expasy.org/tools/BLAST/?CAUCR to identify the nag genes by an ExPaSy BLAST analysis. The domain structure of NagA was analyzed by Andrei Lupas using the HHpred program (49). Transmembrane segments were determined using the “DAS” transmembrane prediction server (13).
RESULTS
Growth of C. crescentus on chitin and chitin products.
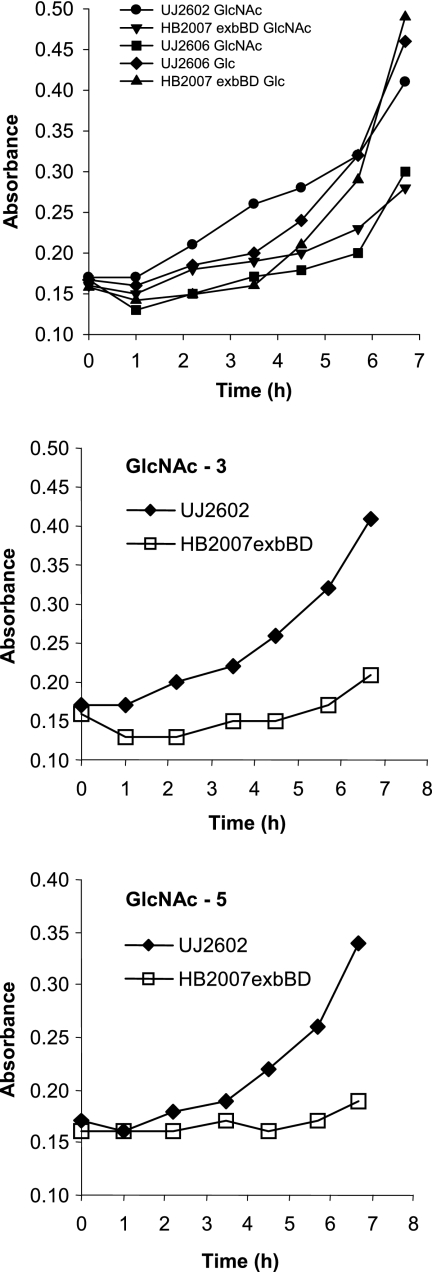
C. crescentus JS1003 incubated for several days with chitin did not multiply. The spent medium and a cell suspension did not contain chitinase activity, in contrast to the cell suspension of chitinase-positive S. marcescens, which yielded strong fluorescence with the chitinase substrate 4-methylumbelliferyl-β-d-N,N′-diacetyl chitobioside hydrate. The lack of chitinase activity agreed with the lack of a predicted chitinase gene in the genome of C. crescentus CB15 (33). If C. crescentus grows on chitin products, it must rely on other organisms to degrade chitin. Wild-type C. crescentus UJ2602 multiplied at similar rates on GlcNAc, (GlcNAc)3, and (GlcNAc)5. Growth on the chitin products was slower than growth on glucose (Fig. 1).
FIG. 1.
Growth of the C. crescentus wild type and exbBD mutant on M2 medium supplemented with GlcNAc, (GlcNAc)3, or (GlcNAc)5 as a carbon source at 30°C in liquid culture. For comparison, growth on glucose (Glc) was determined.
Identification of the NagA outer membrane protein as a GlcNAc transporter.
Initially we attempted to find a GlcNAc-specific transporter in the outer membrane of C. crescentus, since we had identified the MalA transporter for maltodextrins (32). An rsaA mutant of strain NA1000 (17) was used which lacks the quantitatively dominant RsaA S-layer surface protein (15). Since the (GlcNAc)2 transport system of E. coli is induced by (GlcNAc)2 and not by GlcNAc (24), C. crescentus JS1003 was grown on a mixture of chitin oligosaccharides obtained by chitin hydrolysis to induce the synthesis of the predicted outer membrane protein. Two-dimensional gel electrophoresis of isolated outer membrane fractions initially did not reveal a spot that was present in cells grown on GlcNAc and absent in cells grown on glucose.
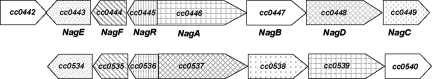
We then searched for genes homologous to those known to be involved in (GlcNAc)2 utilization in E. coli and chitin utilization in Vibrio cholerae. The level of identity of the well-characterized chitin utilization genes of V. cholerae, including those encoding an outer membrane chitoporin and PTS for unacetylated (GlcN)2 (30), to putative C. crescentus transport genes was not high enough (<30% identity). A PTS system in C. crescentus was not identified in searches using the E. coli chbABC genes encoding a PTS for (GlcNAc)2 (24), However, an open reading frame (cc0446) specifying an outer membrane transporter with a molecular mass of 95,775 Da was located in a cluster of genes (Fig. 2) which are predicted to encode a phosphoenolpyruvate:carbohydrate phosphotransferase system (8, 14, 41). This includes an EIIA protein and an EIIBC protein, which specifically phosphorylate and translocate sugars across the cytoplasmic membrane. EIIA is part of a fusion protein that includes EI and HPr, which phosphorylate EIIA with phosphoenolpyruvate. In addition, a deacetylase and a deaminase for the degradation of GlcNAc, a hexosaminidase for the cleavage of chitin oligosaccharides, and a transcriptional regulator are encoded in this gene cluster.
FIG. 2.
Arrangement of the genes in the nag locus of C. crescentus (top). The numbers with the prefix “cc” indicate open reading frames annotated in the genome (33). We designated the encoded proteins NagABCDEF. cc0442 encodes a predicted outer membrane transporter that does not belong to the nag locus. The lower line illustrates another locus in C. crescentus, with high sequence similarity to the nag locus. The same type of shading indicates predicted similar functions of the proteins. No shading means no sequence identity.
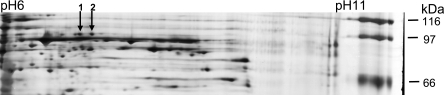
Based on this result, we searched the two-dimensional gels of the outer membrane proteins with matrix-assisted laser desorption-ionization time-of-flight (MALDI-TOF) mass spectrometry for 96-kDa protein spots. In some of the gels, the resolution of the spots was such that a double spot was discernible that was present in GlcNAc-grown cells (Fig. 3) but not in glucose-grown cells (not shown). Tryptic peptides generated from both spots were identical to predicted tryptic peptides encoded by the open reading frame cc0446. The double spot probably arose from protein modifications such as deamidation during the preparation of the samples for two-dimensional electrophoresis. The protein is designated NagA and the encoding gene nagA.
FIG. 3.
Two-dimensional gel electrophoresis of outer membrane proteins of C. crescentus grown in M2 minimal medium with GlcNAc as a carbon source. Arrows 1 and 2 indicate NagA identified by MALDI-TOF. The gel was stained with Coomassie blue.
To further identify the NagA protein, nagA was expressed in E. coli BL21 omp8. A protein of 93 kDa was detected after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, but the yield was low (data not shown). The amount of NagA was greatly increased when synthesized in a degP derivative of E. coli BL21 omp8, indicating that most of NagA was degraded by the periplasmic DegP protease before it was incorporated into the E. coli outer membrane.
NagA is required for growth on GlcNAc, (GlcNAc)3, and (GlcNAc)5.
The open reading frame cc0446 of the C. crescentus genome was inactivated by insertion of an Ω cassette. In contrast to UJ2602, the nagA mutant SE0446 did not grow on M2 minimal medium around filter paper discs containing 10 μl of a 5% solution of GlcNAc, (GlcNAc)3, or (GlcNAc)5. The mutant also failed to grow on M2 liquid medium with 0.3% GlcNAc as a carbon source. A polar effect of the nagA mutation on the transcription of essential downstream genes can be excluded, since nagA is the last of four genes with the same transcription polarity in the nag gene cluster and complementation of the mutant by the wild-type nagA gene restored growth on the chitin oligosaccharides and transport of GlcNAc (see below).
NagA is required for GlcNAc uptake.
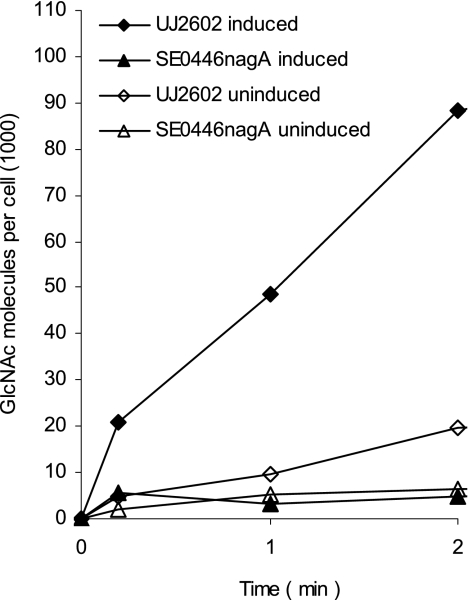
The rates of transport of [14C]GlcNAc into wild-type UJ2602 and the nagA mutant SE0446 were determined quantitatively. UJ2602 transported [14C]GlcNAc, and transport was strongly enhanced by growing cells on GlcNAc. The mutant SE0446 did not transport GlcNAc (Fig. 4; Table 2), which explains its failure to grow on GlcNAc. In competition experiments with unlabeled GlcNAc, 1 μM GlcNAc reduced the transport rate of the wild type to half of the rate for the untreated culture, for which a dissociation constant (Kd) of approximately 1 μM was deduced. Since binding of sugar substrates to the EII enzymes of E. coli PTS transport systems is also in the 1 to 10 μM range (14, 41), it is unclear whether the Kd value reflects uptake across the outer membrane or the cytoplasmic membrane or both.
FIG. 4.
Transport of [14C]GlcNAc into wild-type C. crescentus UJ2602 and the nagA mutant SE0446 grown in M2 medium supplemented with GlcNAc (induced) or glucose (uninduced).
TABLE 2.
Comparison of rates of GlcNAc and maltose transport into wild-type and mutant cells of C. crescentusa
| Strain (treatment) | % Transport
|
|
|---|---|---|
| [14C]GlcNAc | [14C]maltose | |
| UJ2602 (induced) | 100 | 100 |
| UJ2602 (uninduced) | 8 ± 1 | 35 ± 3 |
| SE0446 nagA | 2 ± 1 | 96 ± 6 |
| SL2334a tonB1 | 99 ± 8 | 7 ± 1 |
| HB2004 tonB2 | 101 ± 5 | 100 ± 7 |
| SL3508 tonB3 | 115 ± 12 | 113 ± 11 |
| HB2007 exbBD | 102 ± 10 | 8 ± 2 |
| SL8 tonB2 tonB3 | 88 ± 9 | 97 ± 6 |
| SL9 tonB1 tonB2 | 100 ± 5 | ND |
| UJ2602 (0.1 mM CCCP) | 23 ± 4 | 2 ± 0 |
| UJ2602 (1 mM DNP) | 24 ± 5 | 1 ± 0 |
Percent transport is relative to that for induced wild-type cells. Transport was calculated by subtracting the counts taken up into the cells at 0.2 min from the counts at 3 min. Cultures were incubated in M2 salt medium under inducing conditions (0.3% GlcNAc or 0.3% maltose) unless otherwise indicated; uninduced cultures contained 0.3% glucose. ND, not determined. The values are the means from three experiments ± standard errors of the means.
Energy requirement for GlcNAC uptake.
The essential requirement of NagA for uptake of and growth on GlcNAc suggested an energy-dependent uptake across the outer membrane, for which TonB and ExbBD should be required. To examine this prediction, we determined the uptake of [14C]GlcNAc into the mutant SL2334a tonB1, which no longer takes up maltose (Table 2). Unexpectedly, transport of [14C]GlcNAc into the tonB1 mutant was as high as transport into the wild type (Table 2). In parallel transport assays with [14C]maltose, these mutants displayed strongly reduced transport rates (Table 2). Inactivation of a second tonB gene (cc2327), named tonB2, which was not required for maltose uptake (32), also did not affect GlcNAc uptake (Table 2). Inactivation of a third, distantly related tonB gene (cc3508), named tonB3, which is larger (401 residues, compared to 243 for tonB1 and 240 for tonB2) and contains five predicted transmembrane regions instead of one as found in TonB1, TonB2, and E. coli TonB, also did not reduce GlcNAc uptake (Table 2). To exclude functional replacement of the tonB genes, two double mutants were constructed. Strains SL8 tonB2 tonB3 and SL9 tonB1 tonB2 both showed GlcNAc uptake similar to that of the parent strain (Table 2). Sequence analysis of the C. crescentus genome did not reveal another tonB gene.
In E. coli, TonB forms a functional complex with ExbB and ExbD. Mutations in any of these genes abolish outer membrane transport (6, 40). Transport of [14C]GlcNAc into the HB2007 exbBD mutant was as high as transport into the wild-type strain, in contrast to [14C]maltose transport, determined in parallel, which amounted to 8% of the wild-type rate (Table 2). Independence of [14C]GlcNAc transport of ExbBD agrees with independence of TonB. However, growth of the exbBD mutant HB2007 was slower than that of the wild-type on GlcNAc and very slow on (GlcNAc)3 and (GlcNAc)5 (Fig. 1). The larger the substrates become, the more their uptake is reduced in the exbBD mutant.
In E. coli, energy-coupled transport across the outer membrane is inhibited by energy poisons that dissipate the electrochemical potential of the cytoplasmic membrane (6, 39, 53). If this applies to C. crescentus, transport of GlcNAc across the outer membrane is inhibited by CCCP and DNP without affecting transport across the cytoplasmic membrane, since the latter is most likely energized by phosphoenolpyruvate. Addition of 0.1 mM CCCP or 1 mM DNP to the assay reduced transport to 24% but did not abolish transport of GlcNAc (Table 2). Transport of maltose was completely inhibited (Table 2). In a control experiment, 0.01 mM and 0.1 mM CCCP inhibited PTS-mediated [14C]GlcNAc transport into E. coli W3110 to 82 and 58%, respectively. CCCP at a concentration of 0.01 mM is sufficient to dissipate the electrochemical potential of the cytoplasmic membrane of E. coli, but 0.1 mM CCCP is required to completely inhibit maltose transport across the outer membrane and the cytoplasmic membrane of C. crescentus (32). CCCP might exert secondary effects on GlcNAc uptake into E. coli, but the stronger reduction of GlcNAc transport into C. crescentus may arise from direct effects on the outer membrane transport.
Prediction of the NagA structure.
NagA is among the 67 outer membrane transporters assigned in silico (32, 33). Its molecular mass of 93 kDa is, as with other C. crescentus outer membrane transporters, larger than masses of those of E. coli, which lie around 80 kDa. For example, the mature maltose transporter MalA of C. crescentus is 96 kDa in mass (32). NagA contains a typical signal sequence of 26 residues (Fig. 5). The sequence EQVVIT is similar to TonB box sequences of energy-coupled outer membrane transporters. The TonB box of MalA reads EEVVIT. The NagA TonB box starts with residue 8 of the mature protein; this is where TonB box sequences are usually located, except in cases in which an N-terminal extension of the transporters is involved in gene transcription regulation, such as in FecA of E. coli (7, 25). Computer-assisted sequence analysis with the HHpred program predicts a 147-residue N-terminal sequence that resembles the folding of the hatch domains of the five known crystal structures of outer membrane transporters (four-stranded β-sheet and two amphipathic α-helices). The β-barrel consists of 22 antiparallel β-strands connected by short turns in the periplasm and large loops above the cell surface. The large surface loops of the C. crescentus transporters are the reason for the large size of these proteins. The four largest loops of NagA comprise 89, 65, 62, and 54 residues, similar in size to the MalA loops, the largest of which consists of 79 residues (32). In contrast, the largest loop of an E. coli outer membrane transporter identified in a crystal structure is that of FepA, with 37 residues. The sequence identity of NagA and MalA is only 25%.
FIG. 5.
Genome-derived amino acid sequence of the NagA protein. The predicted signal sequence is shown in italics, the TonB box is shown in bold, and the 22 antiparallel β-strands are indicated by numbered lines above the sequence. The hatch domain is located between the TonB box and the four residues before β-strand 1.
Analysis of the nag gene cluster.
The nagA gene is contained in a gene cluster that can be assigned to GlcNAc transport and metabolism (Fig. 2). cc0447 encodes a β-N-acetylhexosaminidase synthesized with a signal sequence which may degrade chitin oligosaccharides in the periplasm. cc0448 encodes an 882-residue-long polypeptide, which is predicted to consist of three domains. Domain 1 encodes an EIIA protein, domain 2 a Hpr protein, and domain 3 an E1 protein. The location of a gene with an EI and HPr domain in a GlcNAc gene cluster is unusual, since EI and Hpr are usually not sugar specific and form separate proteins (8, 14, 41). EI uses phosphoenolpyruvate to phosphorylate HPr, and HPr phosphorylates EIIA. EIIA phosphorylates EIIB, which phosphorylates at the inner side of the cytoplasmic membrane GlcNAc to GlcNAc-6-P. The open reading frame cc0449 encodes a fusion protein with EIIB and EIIC domains that transports GlcNAc across the cytoplasmic membrane. Fusions between EIIB and EIIC occur frequently. The nag locus contains three other genes of opposite transcriptional polarity; cc0443 encodes an N-acetylglucosamine-6-phosphate deacetylase which converts GlcNAc-6-P into GlcN-6-P, and cc0444 encodes a GlcN-6-P deaminase which converts GlcN-6-P into fructose-6-P, which can be further metabolized by glycolysis. This protein contains a SIS motif found in sugar phosphate isomerases and sugar phosphate binding proteins. cc0445 encodes a transcriptional regulator of the GntR family. In E. coli and Serratia marcescens, GntR represses transcription of genes involved in gluconate metabolism (18, 22). In Streptomyces coelicolor, the GntR homolog DasR represses transcription of the GlcNAc regulon, needed for uptake of GlcNAc. Binding of DasR to its target genes is abolished by GlcNAc-6-P (44). These observations suggest that GntR of C. crescentus most likely functions as a transcriptional repressor that is inactivated by GlcNAc-6-P. In the nag gene cluster of C. crescentus, promoters are expected to be located upstream of nagA and nagR, which are the first genes in the two operons with opposite transcription polarity. Comparative genomics suggests a promoter binding site for repressors of the GntR type that reads TGGTATTxxxTGGTATT (56). The 211-nucleotide intercistronic sequence between nagA and nagR contains the sequence TGGTA only once, upstream of nagA at the −63 position, which makes it unlikely that this sequence serves as a binding site for NagR. The nag gene cluster contains all the genes required for GlcNAc transport and metabolism.
Another predicted TonB-dependent transporter is located adjacent to the nag locus (cc0442). It plays no role in GlcNAc transport since the nagA insertion mutant is transport deficient.
The genome sequence of C. crescentus predicts a locus, cc0534 to cc0540 (Fig. 2), that encodes proteins with 68 to 83% sequence identity to the proteins encoded by the nag locus, except for cc0539, which encodes an outer membrane transporter with 27% sequence identity to NagA, and cc0540, which displays no sequence similarity to the N-acetylglucosaminidase (cc0447). Since cc0539 cannot replace NagA and the sequence of cc0540 is very different from that of cc0447, it is unlikely that the predicted PTS transporter encoded by cc0448 and cc0449, the deacetylase encoded by cc0534, and the deaminase encoded by cc0535 transport and metabolize GlcNAc.
DISCUSSION
After MalA, NagA is the second example which demonstrates that the 67 predicted outer membrane transporters of C. crescentus function as substrate-specific transporters. They extend the hitherto-identified substrates beyond iron complexes and vitamin B12 to carbohydrates and support the notion that many substrates present in very low abundance are taken up by transporters across the outer membrane of gram-negative bacteria. Two-dimensional gel electrophoresis coupled to MALDI-TOF analysis of C. crescentus outer membrane proteins revealed a number of predicted TonB-dependent receptors which showed a distinct pattern after growth of the cells on minimal and rich medium, respectively (21, 38, 51). Genomic analysis and transcriptional profiling predicted many TonB-dependent receptors in C. crescentus (20) and most notably 106 TonB-dependent receptors of the SusC type in Bacteroides thetaiotaomicron (2, 55): SusC is essential for utilization of maltooligosaccharides (43). In Xanthomonas campestris pv. campestris, 72 TonB-dependent receptors were predicted, but only 48 have all the characteristics of TonB-dependent receptors, such as a signal peptide, a β-barrel, a hatch domain, a TonB box, and a C-terminal aromatic residue which is important for insertion into the outer membrane (3). One of the outer membrane proteins, SuxA, was experimentally studied. It is contained in a locus that encodes transport proteins, an amylosucrase and a transcriptional repressor. Transcription of suxA requires SuxA and is enhanced by low concentrations of sucrose. Up to now, four outer membrane transporters, MalA, NagA, SuxA, and SusC, have been related to the transport of specific carbohydrates, and many more substrate-specific transporters will be unraveled by genomic analyses and metabolic studies.
The nagA insertion mutant SE0446 did not grow on (GlcNAc)1 to (GlcNAc)5 and failed to transport GlcNAc. Since complementation of the mutant by plasmid-encoded wild-type nagA fully restored GlcNAc transport and growth, the transport defect of the mutant was confined to nagA. NagA is essential for entry of GlcNAc into the cells, and there is no alternative way for GlcNAc to cross the outer membrane—not by diffusion through unspecific porins and not through the predicted transporter encoded by cc0442 next to the nag locus or through other transporters, such as cc0539, encoded in a gene cluster highly similar to the nag gene cluster except for cc0539.
The lack of a NagA bypass for GlcNAc in the nagA mutant suggests that there are no unspecific porins of the E. coli OmpF, OmpC type in the outer membrane of C. crescentus. This conclusion is supported by the analysis of the C. crescentus genome, in which no genes encoding OmpF-type outer membrane porins were found (33). The finding that GlcNAc does not enter the periplasm through unspecific porins has consequences for the interpretation of the role of MalA in maltodextrin uptake into C. crescentus (32). Deletion of the malA gene diminishes maltose transport to 1% of the level for the malA wild-type strain and completely impairs transport of larger maltodextrins. However, in contrast to the failure of the nagA mutant to grow on GlcNAc and larger chitin oligosaccharides, the malA mutant can grow on maltodextrins up to the size of maltotetraose (32). MalA is essential for growth on maltopentaose and maltohexaose. The slow uptake by the malA mutant is sufficient to support growth on the smaller maltodextrins. Since the smaller GlcNAc (220 Da; maltose, 342 Da) does not diffuse through unspecific porins, it is unlikely that maltodextrins diffuse in the malA mutant through unspecific porins across the outer membrane. In E. coli, maltodextrins preferentially diffuse through LamB but the smaller maltodextrins also diffuse through the nonspecific porins OmpF and OmpC. Likewise, substrates other than maltodextrins diffuse through LamB (34). If the properties of LamB, OmpC, and OmpF apply to the properties of the presumptive C. crescentus porins, the specificity of the putative maltodextrin channel would not be so high as to exclude diffusion of GlcNAc. The slow uptake of maltodextrins across the outer membrane of the malA mutant may occur through a transporter that transports substrates other than maltodextrins but displays some specificity also for maltodextrins. However, it is not excluded that C. crescentus contains an outer membrane pore which is highly specific for maltodextrins through which the smaller maltodextrins but not the larger maltodextrins diffuse.
NagA has the typical structure of a TonB-dependent energy-coupled outer membrane transporter. Computer-assisted analyses of its structure predict a protein consisting of a β-barrel composed of 22 β-strands and a hatch domain that closes the pore of the β-barrel. It also contains a typical TonB box. In E. coli outer membrane transporters (FhuA, FecA, and BtuB), the TonB box serves as the binding site of TonB, as shown by genetic (19, 47), biochemical (9, 36), and crystallographic (37, 48) methods. It was therefore surprising to find that NagA-mediated GlcNAc transport was unaffected by insertion mutations in tonB1 (cc2334a) and exbBD. These genes are required for MalA-mediated maltodextrin transport (Table 2) (29, 32). Sequence analysis of the C. crescentus genome reveals two additional putative TonB open reading frames, designated TonB2 (cc2327) and TonB3 (cc3508). TonB1 (243 residues) displays 30% and TonB2 (240 residues) 26% sequence identity to TonB of E. coli (239 residues). TonB1 and TonB2 have 41% sequence identity. E. coli TonB, TonB1, and TonB2 are composed of three domains, a cytoplasmic domain, followed by a transmembrane domain, and a periplasmic domain, which is rich in proline. TonB1 and TonB2 contain a histidine residue in the transmembrane domain which is essential for the E. coli TonB activity (27, 50) and a highly conserved YP sequence (12) close to Q160, with which E. coli TonB interacts with the TonB box of outer membrane transporters (31, 32, 37, 39, 48). In contrast, TonB3 is much larger (401 residues) than TonB1 and TonB2 and does not contain a histidine in the five predicted transmembrane segments, ranging from residues 14 to 24, 42 to 51, 93 to 107, 187 to 192, and 280 to 289. The evidence for a TonB-like protein comes from the high sequence identity (53%) in the last 99 C-proximal residues, whereas the rest of the sequences are very dissimilar. It shares with TonB-like proteins of similar size, for example, those of X. campestris and B. thetaiotaomicron, a HExxH zinc-binding motif (residues 170 to 174) (12). It is therefore questionable whether the large TonB-like proteins function as TonB proteins, and if they do, the mechanism must be quite distinct from that of the TonB proteins of the E. coli type. Since TonB1 contains the E. coli TonB transmembrane motif SXXXH, which in E. coli interacts with ExbBD (40), it is highly unlikely that TonB3 can functionally replace TonB1 and TonB2. The lack of significant reduction in the initial transport rates of GlcNAc in the tonB1, tonB2, and tonB3 single mutants and in the tonB1 tonB2 and tonB2 tonB3 double mutants suggests a tonB-independent but highly GlcNAc-specific NagA-mediated uptake of GlcNAc across the outer membrane. More than one copy of a tonB gene has been identified in a number of bacterial species (3, 12, 28, 32, 57). In certain cases, the various tonB genes can functionally replace each other (52, 54, 57). Individual knockout of eight predicted tonB genes in Bacteroides thetaiotaomicron does not affect SuxA-mediated sucrose uptake, even though the structural analysis predicts a TonB-dependent function of SuxA (3). It is likely that more than one TonB protein functions in SuxA-mediated sucrose transport.
TonB-independent GlcNAc uptake is further supported by ExbBD independence. This finding excludes an unidentified TonB protein, because all TonB activities require ExbB and ExbD. Although we did not observe reduction of GlcNAc uptake in transport assays, we noticed reduction of growth of the exbBD mutant. The ExbBD proteins contribute to the uptake of GlcNAc, but they are not essential. The ExbBD proteins are increasingly important for the uptake of the larger chitin oligosaccharides from (GlcNAc)3 to (GlcNAc)5. Analysis of the C. crescentus genome for additional exbBD-like genes which could replace exbBD identified a typical tol gene cluster with the same gene arrangement as in E. coli: tolQ tolA tolB pal (32). In E. coli, tolQR can partially replace exbBD (5). We were unable to construct chromosomal insertion mutants of tolQR (32) and assume that the tol gene products are involved in the proper assembly of the outer membrane, as is discussed for E. coli (11), which may be essential for C. crescentus. Since TolQ and TolR do not replace ExbB and ExbD in maltose transport and there is no substrate specificity known for ExbBD function, it is virtually excluded that TolQ and TolR replace ExbB and ExbD in the uptake of the N-acetyl-chitin oligosaccharides.
To further test energy-driven GlcNAc transport across the outer membrane of C. crescentus, we dissipated the electrochemical potential across the cytoplasmic membrane by CCCP and DNP, which in E. coli impairs outer membrane transport (53). These energy poisons should inhibit GlcNAc transport across the outer membrane but not transport across the cytoplasmic membrane by the PTS system. CCCP and DNP reduced the transport rate to 24% of the wild-type rate but did not completely abolish transport. It is not certain whether the observed reduction of GlcNAc uptake reflects only an energy component of outer membrane translocation, since uptake of GlcNAc by the E. coli PTS system was also reduced by CCCP, although to a much lower degree. At the concentration used (0.1 mM), CCCP completely inhibited uptake of maltose into C. crescentus, for which uptake across the outer membrane and the cytoplasmic membrane should be CCCP sensitive.
Our data are consistent with the following model: GlcNAc uptake across the outer membrane requires NagA, which cannot be bypassed by the use of another transporter or porin. NagA functions as a GlcNAc-specific porin through which GlcNAc diffuses. GlcNAc can also be actively transported by NagA, as suggested by the reduction of uptake by CCCP and DNP and the reduction of growth in the exbBD mutant. Active transport becomes more important than diffusion for the uptake of the larger chitin oligosaccharides; their rate of diffusion becomes growth limiting in the exBD mutant. The proposed model for NagA function differs from the model of TonB-dependent transport across the outer membrane of gammaproteobacteria. In these organisms, the pore in the β-barrel of the outer membrane transporters is completely closed and does not allow diffusion of substrates unless energy input opens the pore. NagA functions as a highly specific porin for small substrates and presumably as an active transporter for larger substrates. The Kd values of about 1 μM for GlcNAc transport by NagA, 0.2 μM for the initial transport of maltose by MalA (32), and 0.03 μM for the transport of sucrose by SuxA (3) are much lower than the Kd value (10 mM) of maltose diffusion through LamB (34) but much higher than the Kd values of vitamin B12 binding to BtuB (5 nM) (53) and ferric enterobactin binding to FepA (1 nM) (10). The intermediary Kd value of C. crescentus NagA suggests that it functions as a highly specific porin for GlcNAc and as a transporter for the larger chitin oligosaccharides. The larger substrates may bind much more tightly to the transporters than the smaller substrates. Release of the larger substrates from NagA with rates sufficient to support growth may require energy input. In addition, the permanently open pore may be too small to allow diffusion of the larger substrates unless energy input moves the hatch. The specificity for GlcNAc comes from the tight binding of GlcNAc to NagA, presumably at the entrance of the pore, similar to the energy-coupled transporters of E. coli and Pseudomonas aeruginosa, where the crystal structures show the substrates bound close to the surfaces of the transporters, which are exposed to the medium (46). The open pore may allow diffusion of other small sugars which bind with low affinity to NagA, such as maltose, as long as NagA is not occupied by chitin oligosaccharides. Isolation of active NagA will be required to determine NagA conductance in liposomes and to determine its crystal structure. However, this requires further experiments, since the yield of NagA synthesized in E. coli is low and it is questionable whether it is properly incorporated into the outer membrane, as its degradation by the periplasmic DegP protease demonstrates. Isolation of NagA from C. crescentus faces the difficulty of many cosynthesized, very similar outer membrane proteins.
Acknowledgments
We thank A. Lupas for analysis of the NagA structure and generous support, H. J. Plattner for chitin oligosaccharides, K. Hantke and S. Patzer for valuable advice, and K. A. Brune for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (BR330-24/2) and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print on 6 June 2008.
REFERENCES
- 1.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. New mini-Tn5 derivatives for insertion mutagenesis and genetic engineering in gram-negative bacteria. Can. J. Microbiol. 411053-1055. [DOI] [PubMed] [Google Scholar]
- 2.Bjursell, M. K., E. C. Martens, and J. I. Gordon. 2006. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteriodes thetaiotaomicron, to the suckling period. J. Biol. Chem. 28136269-36279. [DOI] [PubMed] [Google Scholar]
- 3.Blanvillain, S., D. Meyer, A. Boulanger, M. Lautier, C. Guynet, N. Deancé, J. Vasse, E. Lauber, and M. Arlat. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE 2e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V., K. Hantke, and W. Köster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation, p. 67-145. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems. 35. Iron transport and storage in microorganisms. Marcel Dekker, New York, NY. [PubMed] [Google Scholar]
- 5.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between TonB-ExbB-ExbD and TolA-TolQ-TolR proteins. Mol. Microbiol. 8261-268. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., and S. Mahren. 2007. Transfer of energy and information across the periplasm in iron transport and regulation, p. 276-286. In M. Ehrmann (ed.), The periplasm. ASM Press, Washington, DC.
- 7.Breidenstein, E., S. Mahren, and V. Braun. 2006. Residues involved in FecR binding are localized on one side of the FecA signaling domain of Escherichia coli. J. Bacteriol. 1886440-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busch, W., and M. H. Saier. 2002. The transporter classification (TC) system, 2002. Crit. Rev. Biochem. Mol. Biol. 37287-337. [DOI] [PubMed] [Google Scholar]
- 9.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 9610673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, Z., P. Warfel, S. M. C. Newton, and P. E. Klebba. 2003. Spectroscopic observations of ferric enterobactin transport. J. Biol. Chem. 2781022-1028. [DOI] [PubMed] [Google Scholar]
- 11.Cascales, E., R. Lloubes, and J. N. Sturgis. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42795-807. [DOI] [PubMed] [Google Scholar]
- 12.Chu, B. C. H., R. S. Peacock, and H. J. Vogel. 2007. Bioinformatic analysis of the TonB protein family. Biometals 20467-483. [DOI] [PubMed] [Google Scholar]
- 13.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and E. Elofsson. 1997. Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10673-676. [DOI] [PubMed] [Google Scholar]
- 14.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, P., and J. Smith. 1991. A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface array. J. Gen. Microbiol. 133399-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204372-384. [DOI] [PubMed] [Google Scholar]
- 17.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fineran, P. C., L. Everson, H. Slater, and G. P. Salmond. 2005. A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology 1513833-3845. [DOI] [PubMed] [Google Scholar]
- 19.Heller, K. J., R. J. Kadner, and K. Günther. 1988. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene 64147-153. [DOI] [PubMed] [Google Scholar]
- 20.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 1861448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ireland, M. M., J. A. Karty, E. M. Quardokus, J. P. Reilly, and Y. V. Brun. 2002. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol. Microbiol. 451029-1041. [DOI] [PubMed] [Google Scholar]
- 22.Izu, H., O. Adachi, and M. Yamada. 1997. Gene organization and transcriptional regulation of the gntRKU operon involved in gluconate uptake and catabolism of Escherichia coli. J. Mol. Biol. 267778-793. [DOI] [PubMed] [Google Scholar]
- 23.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 175658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keyhani, N. O., and S. Roseman. 1997. Wild-type Escherichia coli grows on the chitin disaccharide N,N′-diacetylchitobiose, by expressing the cel operon. Proc. Natl. Acad. Sci. USA 9414367-14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, I., A. Stiefel, S. Plantör, A. Angerer, and V. Braun. 1997. Transcription induction of the ferric citrate transport system genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol. Microbiol. 23333-344. [DOI] [PubMed] [Google Scholar]
- 26.Kovach, M. E., K. J. Hughes, K. D. Everiss, and K. M. Peterson. 1994. Identification of a ToxR-activated gene, tagA, that lies within the accessory colonization factor gene cluster of Vibrio cholerae O395. Gene 14891-95. [DOI] [PubMed] [Google Scholar]
- 27.Larsen, R. A., G. E. Deckert, K. A. Kastead, S. Devanathan, K. L. Keller, and K. Postle. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol. 1892825-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Létoffée, S., P. Delepelaire, and C. Wandersman. 2004. Free and hemophore-bound heme acquisitions through the outer membrane receptor HasR have different requirements for the TonB-ExbB-ExbD complex. J. Bacteriol. 1864067-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmiller, S., K. Hantke, S. I. Patzer, and V. Braun. 2008. TonB-dependent maltose transport by Caulobacter crescentus. Microbiology 1541748-1754. [DOI] [PubMed] [Google Scholar]
- 30.Meibom, K. L., X. B. Li, A. T. Nielsen, C.-W. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 1012524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42835-849. [DOI] [PubMed] [Google Scholar]
- 32.Neugebauer, H., C. Herrmann, W. Kammer, G. Schwarz, A. Nordheim, and V. Braun. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 1878300-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, et al. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 984136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikaido, H. 2003. Molecular basis of outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Brien, M., and R. R. Colwell. 1987. A rapid test for chitinase activity that uses 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide. Appl. Environ. Microbiol. 531718-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogierman, M., and V. Braun. 2003. Interaction between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J. Bacteriol. 1851870-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawelek, P. D., N. Croteau, C. Ng-Thow-Hing, C. M. Khursigara, M. Moiseeva, M. Allaire, and J. W. Coulton. 2006. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 3121399-1402. [DOI] [PubMed] [Google Scholar]
- 38.Phadke, N. D., M. P. Molloy, S. A. Steinhoff, P. J. Ulintz, P. C. Andrews, and J. R. Maddock. 2001. Analysis of the outer membrane proteome of Caulobacter crescentus by two-dimensional electrophoresis and mass spectrometry. Proteomics 1705-720. [DOI] [PubMed] [Google Scholar]
- 39.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49869-882. [DOI] [PubMed] [Google Scholar]
- 40.Postle, K., and R. A. Larsen. 2004. The TonB, ExbB, and ExbD proteins, p. 96-112. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 41.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1996. Phosphoenolpyruvate:carbohydrate phosphotransferase systems, p. 1149-1174. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 42.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29303-313. [DOI] [PubMed] [Google Scholar]
- 43.Reeves, A. R., J. N. D'Elia, J. Frias, and A. A. Salyers. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigali, S., H. Nothaft, E. E. E. Noens, M. Schlicht, S. Colson, M. Müller, B. Joris, H. K. Koerten, D. A. Hopwood, F. Titgemeyer, and G. P. van Wezel. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GlntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 611237-1251. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, H., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schalk, I. J., W. W. Yue, and S. K. Buchanan. 2004. Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol. Microbiol. 5414-22. [DOI] [PubMed] [Google Scholar]
- 47.Schöffler, H., and V. Braun. 1989. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol. Gen. Genet. 217378-383. [DOI] [PubMed] [Google Scholar]
- 48.Shultis, D. E., M. D. Purdy, C. N. Branchs, and M. C. Wiener. 2006. Outer membrane active transport: structure of the BtuB:TonB complex. Science 3121396-1399. [DOI] [PubMed] [Google Scholar]
- 49.Söding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21951-960. [DOI] [PubMed] [Google Scholar]
- 50.Traub, I., S. Gaisser, and V. Braun. 1993. Activity domains of the TonB protein. Mol. Microbiol. 8409-423. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, J. K., S. Setayeshgar, L. A. Sharon, J. P. Reilly, and Y. V. Brun. 2006. A nutrient uptake role for bacterial cell envelope extensions. Proc. Natl. Acad. Sci. USA 10311772-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58611-647. [DOI] [PubMed] [Google Scholar]
- 53.White, J. C., P. M. Di Girolamo, M. L. Fu, Y. A. Preston, and C. Bradbeer. 1973. Transport of vitamin B12 in Escherichia coli. Location and properties of the initial B12-binding site. J. Biol. Chem. 2483978-3986. [PubMed] [Google Scholar]
- 54.Wyckoff, E. E., A. R. Mey, and S. M. Payne. 2007. Iron acqusisition in Vibrio cholerae. Biometals 20405-416. [DOI] [PubMed] [Google Scholar]
- 55.Xu, I., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human Bacteroides thetaiotaomicron symbiosis. Science 2992074-2076. [DOI] [PubMed] [Google Scholar]
- 56.Yang, C., D. A. Rodionov, X. Li, O. N. Laikova, M. C. Gelfand, O. G. Zagnitko, M. F. Romine, A. Y. Obraztsova, K. H. Nealson, and A. L. Osterman. 2005. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 28129872-29885. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, O., and K. Poole. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol. Lett. 184127-132. [DOI] [PubMed] [Google Scholar]