Abstract
The plasmid-borne quinolone resistance gene qnrA1 is prevalent in multidrug-resistant Enterobacteriaceae. A chromosomally encoded homologue in Shewanella algae, qnrA3, has been described. We isolated two qnrA3-positive strains, one of Klebsiella pneumoniae (He96) and one of Kluyvera ascorbata (Kas96), from the feces of an immunocompromised outpatient. The qnrA3 allele was identical to that of S. algae except for 5 nucleotides and differed from qnrA1 by 29 nucleotides affecting three amino acids. The analysis of the qnrA3 genetic environment showed that qnrA3 was inserted downstream from an ISCR1 element at a recombination crossover site described for other resistance genes, including qnrA1, and immediately upstream from IS26, a situation not described before. IS26 preceded an incomplete class 1 integron which contained, among other genes, aac(6′)-Ib-cr, another transferable quinolone resistance gene, and the β-lactamase gene blaOXA-1/30. The 10-kb fragment encompassing qnrA3 was compared to previously described qnrA1-containing plasmids and multidrug-resistant plasmids; it shares identical sequences with pC15a, pHSH2, pQR1, pQKp311H, and pSAL-1 but with rearrangements, deletions, and mutations. Conjugal transfer of qnrA3 was highly efficient (10−2) from K. pneumoniae He96 or K. ascorbata Kas96 to Escherichia coli J53 but less so (10−5) from either donor to a clinical strain of Enterobacter cloacae. This first description of a plasmid-borne copy and of the in vitro transfer of qnrA3 is taken to illustrate its likely in vivo transfer from S. algae to the Enterobacteriaceae.
Two novel mechanisms of resistance to fluoroquinolones in Enterobacteriaceae were recently described: quinolone acetylation mediated by AAC-(6′)-Ib-cr, an altered form of the original aminoglycoside 6′-N-acetyltransferase, and Qnr-mediated topoisomerase protection (40, 46). They differ markedly from the classical mechanisms, and their genes are plasmid rather than chromosome borne. The classical mechanisms comprise alterations in DNA gyrase and topoisomerase IV (the quinolone targets), enhancement of drug efflux, a decrease in the permeability of the bacterial cell wall, or a combination thereof (14, 36).
The qnr genes known so far are qnrA, qnrS, and qnrB (38), with qnrA first described in Klebsiella pneumoniae, qnrS in Shigella flexneri, and qnrB in K. pneumoniae and Escherichia coli (20). Subsequent reports showed that these genes are also present in other species of Enterobacteriaceae, especially in multidrug-resistant isolates (6, 25, 32, 41). Variants were successively described for each gene, i.e., qnrA1 to -A6, qnrB1 to -B19, and qnrS1 and -S2 (2, 6, 20, 30, 37-39, 43). They have recently been reclassified, especially the qnrB alleles (17). While the amino acid identity among the proteins encoded by gene variants is between 91 and 99%, it is only 35 to 60% among QnrA, QnrB, and QnrS.
Transferable qnr genes are usually carried by large conjugative plasmids (50 to 180 kb) that often encode extended-spectrum β-lactamases (ESBLs) or AmpC-type β-lactamases (18, 30). The qnr genes were shown to be located in the vicinity of intact, antibiotic resistance determinant-containing class 1 integrons (20, 22, 26, 50). Transfer of plasmid-borne qnr was shown to occur by conjugation (19, 27, 50). Chromosome-borne qnr-type genes were discovered in environmental bacteria such as Photobacterium profundum (43), Vibrionaceae (9, 33, 43), and Shewanella algae (30, 34). In S. algae, which is hypothesized to be the origin of qnrA, the allele is highly homologous (90%) to qnrA1.
We recently screened strains of Enterobacteriaceae isolated in 2004 from the Hôpital Européen Georges Pompidou in Paris (France) for qnrA and found only two qnrA-positive isolates (one of K. pneumoniae and one of Kluyvera ascorbata). They were isolated from the feces of the same immunocompromised patient and possessed an original qnrA allele (GenBank accession no DQ435306) different from those in other strains isolated in Paris (6, 26). We found this allele to be homologous to qnrA3 of S. algae, raising the question of how it had been transferred to and among the clinical isolates. It was the purpose of this study to analyze the genomic environment of qnrA3 in both isolates and to evaluate its in vitro transferability.
MATERIALS AND METHODS
Bacterial strains.
K. pneumoniae He96 was isolated in 2004 at Hôpital Européen Georges Pompidou and was screened from 129 nonconsecutive enterobacterial isolates for the coincidence of resistance to third-generation cephalosporins and presence of qnr, as previously described (6). This strain was isolated from the feces of a 49-year-old outpatient with Hodgkin's disease. K. ascorbata Kas96 was isolated from the same specimen of feces at 106 CFU/mg. Both strains were identified using 16S rRNA and rpoB gene sequencing, with K. ascorbata ATCC 33433 and Kluyvera cryocrescens ATCC 33435 as the reference strains.
E. coli J53 Azir (50) and Enterobacter cloacae Ecl115, a clinical strain isolated from the same ward and during the same period as K. pneumoniae He96 and K. ascorbata Kas96, served as recipient strains for transfer experiments.
MICs of quinolones were determined using the agar dilution method, and susceptibility to antibiotics other than quinolones was tested using the disk diffusion method (disks from Bio-Rad, Marnes La Coquette, France) on Mueller-Hinton agar as specified elsewhere (http://www.sfm.asso.fr).
Transfer experiments.
Conjugal quinolone resistance transfer between K. pneumoniae He96 or K. ascorbata Kas96 and E. coli J53 was tested. Strains were grown to logarithmic phase in brain heart infusion broth, and 2 ml of the donor and the recipient strain suspensions were mixed in a 50-ml flask and incubated at 37°C for 40 min without shaking. Transconjugant selection was performed on Mueller-Hinton plates containing sodium azide (100 μg/ml) and either ampicillin (100 μg/ml) or tetracycline (20 μg/ml). Plates were incubated at 37°C and inspected at 24 and 48 h. Conjugal transfer between K. pneumoniae He96 or K. ascorbata Kas96 and an in vitro-selected sodium azide-resistant mutant of the clinical strain E. cloacae Ecl115 was similarly tested.
Plasmid analysis.
Plasmids were extracted with the High Speed Plasmid Midi kit (Qiagen, Courtaboeuf, France). PCR-based replicon typing was performed on the transconjugants of E. coli J53 and of E. cloacae Ecl115 after mating with the two donor strains K. pneumoniae He96 and K. ascorbata Kas96 (strains E. coli Tc He96/J53, E. coli Tc Kas96/J53, E. cloacae Tc He96/Ecl115, and E. cloacae Tc Kas96/Ecl115). Primers for PCRs were chosen for the identification of the most frequent replicons, using sets 1 (HI1, HI2, and I1), 2 (X, L/M, and N), 3 (FIA, FIB, and W), and 4 (Y, P, and FIC) in multiplex reactions and primer pairs A/C and OR1/CA1 in simplex reactions. Primer sequences and assay conditions were those described previously (7), except for OR1/CA1 (31).
Amplification and sequencing of qnrA and analysis of its genetic environment.
The sequence of qnrA was determined after amplification with intragenic primers qnrA5s and qnrA6as, using plasmid DNA as the template (6), and that of its environment after amplification with primers specific for genes usually surrounding qnrA (PCR sets A, B, and C [Table 1]). PCR was carried out with Long Expand polymerase (Roche Diagnostics, Meylan, France) in an iCycler (Bio-Rad) as follows: 2 min at 94°C; 10 cycles of 10 s at 94°C, 30 s at 55°C, and 2 min at 68°C; 25 cycles of 15 s at 94°C and 30 s at 55°C; and cycle elongation at 68°C starting from 2 min with a further 20 s for each successive cycle. PCR-amplified fragments were sequenced after purification with the Montage PCR Millipore purification kit (Millipore, Saint Quentin-en-Yvelines, France). Sequencing was performed using the ABI Prism BigDye Terminator v3.1 cycle sequencing kit and the ABI Prism 3100 sequencer (Applied Biosystems, Courtaboeuf, France). Nucleotide sequences were analyzed with SeqScape (Applied Biosystems) and compared with each other and with related sequences in the data banks.
TABLE 1.
Oligonucleotides primers used for PCR and sequencing of the ca. 10-kb DNA fragment encompassing the qnrA3 gene in pHe96, pKas96, and Tc He96/Ecl115
| DNA fragment (primer set) | Primera | Primer sequence (5′→3′) |
|---|---|---|
| qacΔE1-qnrA3 (A) | qacΔE1 s | GGCTTTTTCTTGTTATCGCA |
| qnrA7 as | GCGAAAACGGCTGTCACTC | |
| orf513 s | CCATGTCGCTGGCAAGGAA | |
| orf513 s2 | TCGCATCGCTCGCTGCATG | |
| orf513 as | GCTGCCACCAGAACGAGCGCC | |
| sul1 s | GAGGCGGACTGCAGGCTGGT | |
| qnrA3 (B) | qnrA5 s | GGGTATGGATATTATTGATAAAG |
| qnrA6 as | CTAATCCGGCAGCACTATTA | |
| qnrA9 s | GCCATAAGATGTACTTCTGCT | |
| qnrA10 as | AGCAGAAGTACATCTTATGGC | |
| qnrA-qacΔE1 (C) | qnrA8 s | GTCAAGATCTGTGCCCTGGCA |
| qacΔE1 as | CAAGCTTTTGCCCATGAAGC | |
| tnpA3 s | AGCTGCATACCGGTTTCTGGG | |
| 51pb s | CGCTAACTTTGCAACAGTGC | |
| int2 s | GCGAACCACTCATCCGGGGT | |
| aac(6′)-Ib-cr s | CGATTGGGTATGCCCAGTCG | |
| blaOXA1/30 s | CGATGCATCCACAACGCT | |
| blaOXA1/30 s2 | CCGCACTTACAGGAAACT | |
| catB3 s | GGGCATCGGTACGACTGG | |
| arr-3 s | GCGGCTACATATACATAG | |
| dfr s | AGTTGTCAGCCGCTCAGG | |
| ant(3′)-Ij-aac(6′)-Ib as | GCAGCGCAAGGACATTCTTG |
Primers used for PCR are in bold; the other primers were used for sequencing. s, sense primer; as, antisense primer.
Amplification and sequencing of the quinolone resistance-determining regions of gyrA, gyrB, parC, parE, and β-lactamase genes.
Total DNA was extracted, and the quinolone resistance-determining regions of gyrA, gyrB, parC, and parE were amplified as described previously (24). bla genes of the TEM, SHV, CTX-M, and OXA types were screened for by PCR as described previously (12, 29).
Nucleotide sequence accession number.
The qnrA sequence determined in this study was submitted to GenBank under accession number DQ435306. The nucleotide sequences of the encompassing fragment of 10,776 bp from pHe96 and pKas96 were submitted to GenBank/EMBL/DDJB under accession numbers EU495237 and EU495238.
RESULTS AND DISCUSSION
The plasmid-borne qnrA3 allele and its genetic environment.
K. pneumoniae He96 and K. ascorbata Kas96 were identified on the basis of their 16S rRNA sequences and particularly their rpoB sequences, which are more discriminatory in the identification of Enterobacteriaceae (28). Each strain contained a plasmid of ca.70 kb (pHe96 and pKas96, respectively) (data not shown).
Since this is the first observation of qnrA3 as a plasmid-borne gene, we analyzed in detail the sequences of qnrA3 and its environment and compared them to published sequences of plasmids containing other qnrA genes, such as pHSH2 from qnrA1-positive E. coli strains isolated in Hong-Kong (50), pQR1 from a qnrA1-positive E. coli strain isolated in France (Paris) (26), and pQKp311H from a qnrA1-positive K. pneumoniae strain isolated in Spain (Barcelona) (25).
The qnrA sequence was the same for K. pneumoniae He96 and K. ascorbata Kas96 and was 95.6% identical to that of qnrA1 (46), with 29 nucleotide differences accounting for three amino acid substitutions, i.e., Arg39Gln, Ile108Val, and Ala127Thr. The deduced amino acid sequence was identical to that of the chromosome-borne qnrA3 gene of S. algae (30), but there were five nucleotide differences (99.2% identity). No qnrB or qnrS gene was detected.
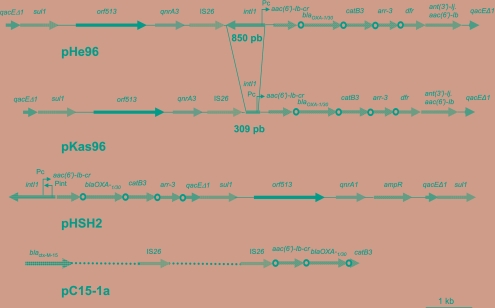
The sequences up- and downstream from qnrA3 are schematically shown in Fig. 1 and are detailed in Fig. 2. They were obtained from two fragments amplified on one hand with a sense primer in qacEΔ1 and an antisense primer in qnrA3 and on the other hand with a sense primer in qnrA3 and an antisense primer in qacEΔ1, with sequences being obtained using primers within the fragments (Table 1). Nucleotide sequences of the encompassing fragment of 10,776 bp from pHe96 and pKas96 were identical in He96 and Kas96, except for 541 bp which were absent in Kas96.
FIG. 1.
Genetic environment of qnrA3 in K. pneumoniae He96 (pHe96) and in K. ascorbata Kas96 (pKas96). The 59-bp elements are indicated by circles. The sequences of pHe96 are compared with those described for pHSH2 (50) (GenBank accession number AY259086) and pC15-1a (5) (GenBank accession number NC005327).
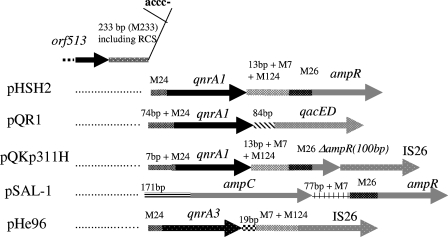
FIG. 2.
Schematic map (not to scale) of the regions upstream and downstream from qnrA3 and comparison with published qnrA1-containing plasmids (pHSH2 [50]; pQR1 [26]; pQKp331H [25]) and pSAL-1, containing DHA-1, the AmpC-type β-lactamase gene originating from chromosomal DNA of Morganella morganii (48). RCS, recombination crossover site (1). Sequences that were found in at least two plasmids are shown as motifs (M) with the number of base pairs: M233 (sequence under GenBank accession no. EU495237), M7 (5′-ACAAGAG-3′ in all plasmids, except 5′-ACAAGAG-3′ in pHe96), M24 (5′-CCCTCCCTGATTAAAGGAAGCCGT-3′), M26 (5′-CCTAAAGAAAAACTTACAGGTGGATT-3′), and M124 (sequence under GenBank accession no. EU495237). Specific features of pHe96 are the following, from 5′ to 3′: the recombinase gene orf513; a 233-bp sequence (M233) common to all plasmids; the 24-bp (M24) sequence common to all qnrA plasmids with an additive sequence of 7-bp (GTTAGCA) and 74 bp (GCAAAGGTTGTTGGGAAGGCGCGAACCAACCCCATGTTTGCCTGCCTAGGCAAAGCTCGCCGAAAGAGTTAGCA) upstream in pQKp331H and pQR1, respectively; the qnrA gene; 19 bp homologous to the S. algae chromosome; M7 (detailed above); M124, common to pHSH2 and pQKp311H; and IS26, composed of a complete tnpA gene (717 bp) and the specific 51-bp inverted repeat.
The qnrA3 gene was observed downstream from an ISCR1 element similar to that described previously (45). The 233-bp sequence downstream from orf513 (M233 in Fig. 2) was identical to those in most of the other qnrA1-containing plasmids described so far (19, 21, 25, 26, 46, 50) and to that in pSAL-1 upstream from the ampC-ampR operon (48) (Fig. 2). In several qnrA1-containing plasmids, such as pHSH2 and pQKp311H, ampR was observed immediately downstream from qnrA1, but this was not the case in our plasmids. This suggests that the insertion of qnrA3 downstream from ISCR1 may have followed the same mechanism as that of qnrA1 and also as that of the insertion of the chromosome-borne region encoding ampC-ampR from Morganella morganii. Indeed, the insertion probably occurred at the recombination crossover site (ACCC-) at the 3′ end of ISCR1 (Fig. 2), as described previously (1, 48).
The sequence immediately downstream from qnrA3 was identical to that found downstream from qnrA3 in the S. algae chromosome (34), which confirms that qnrA3 has been excised from chromosomal DNA of S. algae or similar organisms, and was ended by a 7-bp element (M7) found in the other qnr-containing plasmids and also in pSAL-1. After 124 bp (M124) that were 92% identical to pHSH2 and other qnrA1-containing plasmids but of unknown origin, there was an unusual insertion of an IS26 element (11). Proximity of IS26 and qnrA1 was recently reported for pQKp311H (25).
Downstream from IS26, 52 bp were identical with those found in pC15-1a, a multiresistance plasmid containing blaCTX-M-15 and described from an E. coli outbreak in Canada (5) (Fig. 3). Sequence identity with pC15-1a ended at the site of insertion of an incomplete intlI gene and resumed with the succession of resistance gene cassettes [aac(6′)-Ib-cr to catB3] (Fig. 1 and 3). The same sequence organization was also found in In37, present in pHSH2 (Fig. 1) (50). Compared to In37, a 217-bp deletion was observed in pHe96 upstream from aac(6′)-Ib-cr, which included the 83-bp 5′ terminus of intI1 (Fig. 3). The intI1 gene also lacked 81 bp at its 3′ end. The 217-bp deletion accounted for the absence of start and stop codons in intlI and of the promoter Pint, while the promoter Pc was complete (Fig. 3). However, the −10 box of the promoter Pc differed from In37 by two mutations and was identical to that found in pCTX-M-3, isolated from a multidrug-resistant strain of Citrobacter freundii (42). What distinguished the sequence in K. ascorbata Kas96 from that in K. pneumoniae He96 was a further 541-bp deletion in the integrase gene at the 3′ end (Fig. 1 and 3).
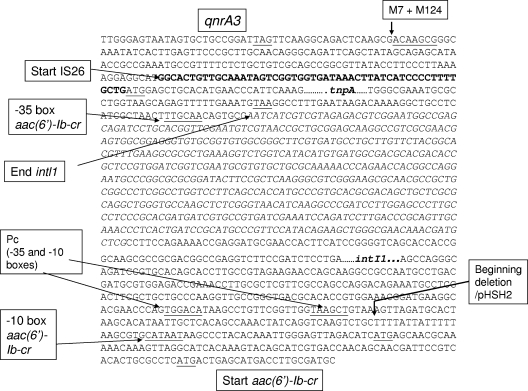
FIG. 3.
Detailed sequence downstream from qnrA3 for pHe96 and pKas96. Italics indicate the part which is deleted in pKas96. M7 and M124 are defined in Fig. 2. Underlined sequences correspond to the following features: stop codon of qnrA3, 7-bp element, start codon tnpA, stop codon tnpA, −35 promoter box of aac(6′)-Ib-cr, −35 and −10 promoter boxes of Pc in intI1, −10 promoter box of aac(6′)-Ib-cr, start codon of aac(6′)-Ib-cr as described for pC15-a, and start codon of aac(6′)-Ib-cr as described for pHSH2.
The sequence of the aac(6′)-Ib gene present in pHe96 and pKas96, was identical to that of the alleles found in In37 (pHSH2) and pC15-1a (5, 50), with two substitutions, Thr104Arg and Asp181Tyr with respect to the initially sequenced AAC(6′)-Ib (47), which designated it a cr1 variant (25, 40). The following gene, blaOXA-30 (44) (recently found to be identical with blaOXA-1 [4]), and the sequence between aac(6′)-Ib-cr and blaOXA-1/30 are identical to those found in In37 and pC15-1a, although in pC15-1a neither gene is present as an integron-borne cassette (5). The chloramphenicol acetyltransferase gene catB3, and the rifampin ADP-ribosylating transferase gene arr-3 are in the same order downstream from blaOXA-1/30 in In37, at variance with the gene organization in pC15-1a (Fig. 1). Since the DNA segments comprising IS26-aac(6′)-Ib-cr-blaOXA-1/30-catB3 are identical in In37, pC15-1a, and pHe96, these elements may be derived from the same ancestral strain or plasmid or have been mobilized similarly. Whether pC15-1a contains a reduced form of In37 with deletion of intl1 and rearrangements due to IS26 or whether In37 has gained the antibiotic resistance cassettes from pC15-1a cannot be said. However, the fact that the −35 and −10 promoter boxes of aac(6′)-Ib-cr were found adequately upstream from aac(6′)-Ib-cr in pC15-1a but that the −35 box is absent in In37 favors the second hypothesis. It seems that in pHe96 the integrase gene was inserted between tnpA and aac(6′)-Ib-cr, since the −35 and the −10 boxes were separated by 940 bp (mostly the intl1 gene) in pHe96 (Fig. 3), with the sequence found at the 5′end of aac(6′)-Ib-cr being identical to that in pC15-1a (5) (Fig. 1). Consequently, pHe96 cannot be derived from In37 but may have been derived from pC15-1a.
Characterization of antibiotic resistance determinants.
K. pneumoniae He96 exhibited higher levels of resistance to quinolones than K. ascorbata Kas96 (Table 2). Since qnrA3 was reported to confer only decreased susceptibility to quinolones (34) and the MICs of ciprofloxacin and of levofloxacin were high in K. pneumoniae He96, we sought an additional quinolone resistance mechanism such as topoisomerase mutation. K. pneumoniae He96 harbored one gyrA mutation (Ser83Phe), with no mutation in gyrB or in the topoisomerase IV genes. This gyrA mutation added to the quinolone resistance phenotype conferred by the two plasmid-borne quinolone resistance genes, qnrA3 and aac(6′)-Ib-cr.
TABLE 2.
MICs of quinolones and aminoglycosides, and susceptibility phenotypes for other antibiotics, for K. pneumoniae He96, K. ascorbata Kas96, E. coli J53, E. cloacae Ecl115, and their respective qnrA3-positive transconjugants
| Species and strain | MIC (μg/ml)a
|
Susceptibility phenotypeb
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Quinolones
|
Aminoglycosides
|
|||||||||||||
| NAL | NOR | CIP | LVX | MXF | GAT | GEN | KAN | TOB | AMK | TMP | TET | PIP | TZP | |
| K. pneumoniae He96 | 128 | 32 | 8 | 4 | 4 | 2 | 0.25 | 48 | 12 | 6 | R | R | R | I |
| K. ascorbata Kas96 | 64 | 8 | 2 | 1 | 4 | 2 | <0.06 | 6 | 2 | 1.5 | R | R | R | S |
| E. coli | ||||||||||||||
| J53 | 4 | 0.03 | 0.008 | 0.01 | 0.06 | 0.01 | <0.06 | 0.75 | 0.19 | 0.75 | S | S | S | S |
| Tc He96/J53 | 16 | 1 | 0.25 | 0.12 | 0.5 | 0.25 | 0.09 | 16 | 4 | 2 | R | R | I | S |
| Tc Kas96/J53 | 8 | 0.5 | 0.12 | 0.12 | 0.5 | 0.25 | 0.09 | 8 | 3 | 1.5 | R | R | I | S |
| E. cloacae | ||||||||||||||
| Ecl115 | 4 | 0.06 | 0.01 | 0.06 | 0.12 | 0.06 | 32 | 8 | 4 | 1.5 | S | S | R | S |
| Tc He96/Ecl115 | 8 | 0.5 | 0.25 | 0.12 | 0.5 | 0.25 | 24 | 16 | 6 | 2 | R | R | R | I |
| Tc Kas96/Ecl115 | 8 | 0.5 | 0.25 | 0.12 | 0.5 | 0.25 | 24 | 24 | 8 | 2 | R | R | R | I |
NAL, nalidixic acid; NOR, norfloxacin; CIP, ciprofloxacin; LVX, levofloxacin; MXF, moxifloxacin; GAT, gatifloxacin; GEN, gentamicin; KAN, kanamycin; TOB, tobramycin; AMK, amikacin.
TMP, trimethoprim; TET, tetracycline; PIP, piperacillin; TZP, piperacillin-tazobactam; R, resistant; I, intermediate; S, susceptible.
K. pneumoniae He96 exhibited a β-lactam resistance phenotype including resistance to piperacillin as well as combinations of clavulanic acid with amoxicillin or ticarcillin and a decrease in susceptibility to piperacillin combined with tazobactam, cefotaxime, and cefepime, as previously described for OXA-1/30 producers (10), whereas K. ascorbata appeared to be susceptible to amoxicillin-clavulanic acid, piperacillin-tazobactam, cefotaxime, and cefepime. However, both strains harbored blaOXA-1 and produced an enzyme compatible with an OXA-type β-lactamase (data not shown). In addition, in He96 we found an SHV-1 variant (Leu35Gln, Thr149Ser) not previously described and different from the chromosomal gene blaLEN-1, but no TEM-type or CTX-M-type genes were found. Both strains were resistant to tetracycline and co-trimoxazole but susceptible to chloramphenicol.
Plasmids of ca. 70 kb containing qnrA3 were transferred at a frequency of 10−2 from K. pneumoniae He96 or K. ascorbata Kas96 to E. coli J53. This frequency is among the highest reported for other plasmid-borne qnr alleles (19, 49). The blaOXA-1/30 gene was cotransferred along with tetracycline and trimethoprim resistance determinants. Transfer of aac(6′)-Ib-cr was first assumed on the basis of increased MICs of kanamycin and tobramycin for the E. coli transconjugants (Table 2). Since similar MICs were observed in both transconjugants (Table 2), the expression of the aac(6′)-Ib-cr gene from the two plasmids is probably similar, despite the differences in its surrounding sequences mentioned above. Furthermore, the presence of this gene, which has the peculiar property of conferring selective resistance to quinolones with a nonsubstituted piperazinyl group at C-7, may explain why the transconjugants of E. coli J53 showed a greater increase in the MICs of ciprofloxacin and norfloxacin (16- and 32-fold increases) than in those of levofloxacin, moxifloxacin, and gatifloxacin (2- to 16-fold) in comparison to parental strains. The second aminoglycoside resistance gene in the 10-kb fragment, ant(3′)-Ij-aac(6′)-Ib, was truncated at its 3′ end for the last 585 bp and thus was not assumed to confer additional aminoglycoside resistance.
For the E. coli J53 transconjugants obtained from the parental strains He96 and Kas96, PCR-based replicon typing was positive for N-type replicons and negative for the other types, which suggests that qnrA3 is harbored on an IncN plasmid. qnrA1 genes have been associated so far with the IncA/C-type plasmids but not with IncN-type plasmids (35). Conversely, IncN-type plasmids were previously associated with the β-lactamase genes blaVIM-1 (8) and blaCTX-M-3 (15) but not with qnr genes, although replicon typing was seldom done in qnr-positive strains.
Interspecies transfer of qnrA3.
In light of the observation of the qnrA3 variant, so far described only for Shewanella (30), on two close-to-identical ca. 10-kb plasmid-borne fragments in isolates of different bacterial species from the same patient, we suspected the possibility of an in vivo interspecies transfer of qnrA3. We therefore tried to reproduce the presumptive in vivo transfer from K. ascorbata Kas96 or K. pneumoniae He96 to other clinical isolates of Enterobacteriaceae. A qnrA-negative E. cloacae strain that was susceptible to tetracycline and trimethoprim, thereby allowing for the selection of the qnrA3 plasmid-containing transconjugants, was successfully conjugated. However, the transfer was observed at a frequency of 10−5, i.e., 1,000-fold lower than that between K. ascorbata and E. coli J53. The low frequency of transfer may be due to inefficient conjugation or to the occurrence of recombination within the host plasmid (the strain was an ESBL producer) mediated by integron-like structure (3).
To check this hypothesis, we first compared the plasmid contents in E. cloacae Ecl115 and in the two transconjugants E. cloacae Tc He96/Ecl115 and E. cloacae Tc Kas96/Ecl115. In E. cloacae Ecl115, the presence of two plasmids of the IncHI2 and IncL/M types, but none of the IncN type, was suspected on the basis of replicon typing results. The IncHI2 type has previously been associated with blaCTX-M-9-containing plasmids (15), while to our knowledge an association of the L/M group with antibiotic resistance genes has not been reported. The E. cloacae transconjugants were indeed positive for both HI2 and L/M replicons and also for the N-type replicon corresponding to the plasmid from strains He96 and Kas96.
We also studied E. cloacae Ecl115 for genes similar to those included in pHe96 and the two transconjugants for additional genes in the same order as in the 10-kb fragment containing qnrA3. In E. cloacae Ecl115, we detected an IS26 element, similar to that of pHe96 and pKas96, with a partial aac(3)-II gene upstream from IS26, in the same genetic context as described for pC15-1a (5) but downstream from an ISCR1 element in the case of E. cloacae Ecl115. The ESBL gene was blaCTX-M-3, a gene shown to originate from the K. ascorbata chromosome and from which blaCTX-M-15, contained in pC15-1a, was derived by a point mutation leading to Asp240Gly (42). Sequencing of the 10-kb fragment amplified from the transconjugant Tc He96/Ecl115 revealed sequence identity with pHe96, including qnrA3. Overall, this favors the hypothesis of a conjugative transfer between two strains of clinical origin, strains which usually are more difficult to conjugate than laboratory strains (50).
K. ascorbata is an environmental, waterborne bacterium that may cause food-borne infections in humans (13). Infections due to K. ascorbata have rarely been described but may occur in immunocompromised patients such as the patient with Hodgkin's disease from whom strain Kas96 was isolated. In this patient, the strain did not cause infection; however, it was found to be dominant in his gut flora, together with K. pneumoniae. The patient had received β-lactams and vancomycin but not quinolones during the month before the isolation of K. pneumoniae He96 and K. ascorbata Kas96. The role of K. ascorbata as a reservoir of resistance genes has been recognized with the discovery, in their chromosomes, of a variety of CTX-M genes (16, 23, 42). Since K. ascorbata may live for long periods as a commensal in the human gut, it may well contribute to resistance gene transfer by conjugation to other inhabitants of this ecosystem, transient or not, such as K. pneumoniae and E. cloacae (13).
Acknowledgments
We thank Guillaume Arlet for helpful discussion.
This work was supported by a grant from the Chancellerie de l'Université de Paris.
The authors have no conflicts of interest.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Bae, I. K., Y. N. Lee, W. G. Lee, S. H. Lee, and S. H. Jeong. 2007. Novel complex class 1 integron bearing an ISCR1 element in an Escherichia coli isolate carrying the blaCTX-M-14 gene. Antimicrob. Agents Chemother. 513017-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonemann, G., M. Stiens, A. Puhler, and A. Schluter. 2006. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents Chemother. 503075-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvier, M., G. Demarre, and D. Mazel. 2005. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 244356-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D. A., and M. R. Mulvey. 2006. OXA-1 is OXA-30 is OXA-1. J. Antimicrob. Chemother. 58224-225. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 483758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cambau, E., C. Lascols, W. Sougakoff, C. Bebear, R. Bonnet, J. D. Cavallo, L. Gutmann, M. C. Ploy, V. Jarlier, C. J. Soussy, and J. Robert. 2006. Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002-2005. Clin. Microbiol. Infect. 121013-1020. [DOI] [PubMed] [Google Scholar]
- 7.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63219-228. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli, A., V. Miriagou, A. Bertini, A. Loli, C. Colinon, L. Villa, J. M. Whichard, and G. M. Rossolini. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg. Infect. Dis. 121145-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattoir, V., L. Poirel, D. Mazel, C. J. Soussy, and P. Nordmann. 2007. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 512650-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubois, V., C. Arpin, C. Quentin, J. Texier-Maugein, L. Poirel, and P. Nordmann. 2003. Decreased susceptibility to cefepime in a clinical strain of Escherichia coli related to plasmid- and integron-encoded OXA-30 beta-lactamase. Antimicrob. Agents Chemother. 472380-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 5714-23. [DOI] [PubMed] [Google Scholar]
- 12.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 481249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farmer, J. J., III, G. R. Fanning, G. P. Huntley-Carter, B. Holmes, F. W. Hickman, C. Richard, and D. J. Brenner. 1981. Kluyvera, a new (redefined) genus in the family Enterobacteriaceae: identification of Kluyvera ascorbata sp. nov. and Kluyvera cryocrescens sp. nov. in clinical specimens. J. Clin. Microbiol. 13919-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooper, D. 2003. Mechanisms of quinolone resistance, p. 41-67. In D. Hooper and E. Rubinstein (ed.), Quinolone antimicrobial agents. ASM Press, Washington, DC.
- 15.Hopkins, K. L., E. Liebana, L. Villa, M. Batchelor, E. J. Threlfall, and A. Carattoli. 2006. Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 503203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. Beta-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 463045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby, G., V. Cattoir, D. Hooper, L. Martinez-Martinez, P. Nordmann, A. Pascual, L. Poirel, and M. Wang. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 522297-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2)S120-S126. [DOI] [PubMed] [Google Scholar]
- 19.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 501178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong, J. Y., H. J. Yoon, E. S. Kim, Y. Lee, S. H. Choi, N. J. Kim, J. H. Woo, and Y. S. Kim. 2005. Detection of qnr in clinical isolates of Escherichia coli from Korea. Antimicrob. Agents Chemother. 492522-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kehrenberg, C., S. Friederichs, A. de Jong, G. B. Michael, and S. Schwarz. 2006. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J. Antimicrob. Chemother. 5818-22. [DOI] [PubMed] [Google Scholar]
- 23.Lartigue, M. F., L. Poirel, D. Aubert, and P. Nordmann. 2006. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 501282-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lascols, C., J. Robert, V. Cattoir, C. Bebear, J. D. Cavallo, I. Podglajen, M. C. Ploy, R. Bonnet, C. J. Soussy, and E. Cambau. 2007. Type II topoisomerase mutations in clinical isolates of Enterobacter cloacae and other enterobacterial species harbouring the qnrA gene. Int J. Antimicrob Agents 29402-409. [DOI] [PubMed] [Google Scholar]
- 25.Lavilla, S., J. J. Gonzalez-Lopez, M. Sabate, A. Garcia-Fernandez, M. N. Larrosa, R. M. Bartolome, A. Carattoli, and G. Prats. 2008. Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J. Antimicrob. Chemother. 61291-295. [DOI] [PubMed] [Google Scholar]
- 26.Mammeri, H., M. Van De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 4971-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351797-799. [DOI] [PubMed] [Google Scholar]
- 28.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 261005-1011. [DOI] [PubMed] [Google Scholar]
- 29.Mugnier, P., I. Casin, A. T. Bouthors, and E. Collatz. 1998. Novel OXA-10-derived extended-spectrum beta-lactamases selected in vivo or in vitro. Antimicrob. Agents Chemother. 423113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56463-469. [DOI] [PubMed] [Google Scholar]
- 31.Osborn, A. M., R. W. Pickup, and J. R. Saunders. 2000. Development and application of molecular tools in the study of IncN-related plasmids from lakewater sediments. FEMS Microbiol. Lett. 186203-208. [DOI] [PubMed] [Google Scholar]
- 32.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 503953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel, L., A. Liard, J. M. Rodriguez-Martinez, and P. Nordmann. 2005. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 561118-1121. [DOI] [PubMed] [Google Scholar]
- 34.Poirel, L., J. M. Rodriguez-Martinez, H. Mammeri, A. Liard, and P. Nordmann. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 493523-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirel, L., L. Villa, A. Bertini, J. D. Pitout, P. Nordmann, and A. Carattoli. 2007. Expanded-spectrum beta-lactamase and plasmid-mediated quinolone resistance. Emerg. Infect. Dis. 13803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole, K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 5620-51. [DOI] [PubMed] [Google Scholar]
- 37.Quiroga, M. P., P. Andres, A. Petroni, A. J. Soler Bistue, L. Guerriero, L. J. Vargas, A. Zorreguieta, M. Tokumoto, C. Quiroga, M. E. Tolmasky, M. Galas, and D. Centron. 2007. Complex class 1 integrons with diverse variable regions, including aac(6′)-Ib-cr, and a novel allele, qnrB10, associated with ISCR1 in clinical enterobacterial isolates from Argentina. Antimicrob. Agents Chemother. 514466-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6629-640. [DOI] [PubMed] [Google Scholar]
- 39.Robicsek, A., D. F. Sahm, J. Strahilevitz, G. A. Jacoby, and D. C. Hooper. 2005. Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob. Agents Chemother. 493001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 1283-88. [DOI] [PubMed] [Google Scholar]
- 41.Robicsek, A., J. Strahilevitz, D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 502872-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez, M. M., P. Power, M. Radice, C. Vay, A. Famiglietti, M. Galleni, J. A. Ayala, and G. Gutkind. 2004. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob. Agents Chemother. 484895-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saga, T., M. Kaku, Y. Onodera, S. Yamachika, K. Sato, and H. Takase. 2005. Vibrio parahaemolyticus chromosomal qnr homologue VPA0095: demonstration by transformation with a mutated gene of its potential to reduce quinolone susceptibility in Escherichia coli. Antimicrob. Agents Chemother. 492144-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siu, L. K., J. Y. Lo, K. Y. Yuen, P. Y. Chau, M. H. Ng, and P. L. Ho. 2000. Beta-lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like beta-lactamase, OXA-30. Antimicrob. Agents Chemother. 442034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 995638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran van Nhieu, G., and E. Collatz. 1987. Primary structure of an aminoglycoside 6′-N-acetyltransferase AAC(6′)-4, fused in vivo with the signal peptide of the Tn3-encoded beta-lactamase. J. Bacteriol. 1695708-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verdet, C., G. Arlet, G. Barnaud, P. H. Lagrange, and A. Philippon. 2000. A novel integron in Salmonella enterica serovar Enteritidis, carrying the bla(DHA-1) gene and its regulator gene ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, M., D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 481295-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 472242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]