Abstract
The plant cell wall, which consists of a highly complex array of interconnecting polysaccharides, is the most abundant source of organic carbon in the biosphere. Microorganisms that degrade the plant cell wall synthesize an extensive portfolio of hydrolytic enzymes that display highly complex molecular architectures. To unravel the intricate repertoire of plant cell wall-degrading enzymes synthesized by the saprophytic soil bacterium Cellvibrio japonicus, we sequenced and analyzed its genome, which predicts that the bacterium contains the complete repertoire of enzymes required to degrade plant cell wall and storage polysaccharides. Approximately one-third of these putative proteins (57) are predicted to contain carbohydrate binding modules derived from 13 of the 49 known families. Sequence analysis reveals approximately 130 predicted glycoside hydrolases that target the major structural and storage plant polysaccharides. In common with that of the colonic prokaryote Bacteroides thetaiotaomicron, the genome of C. japonicus is predicted to encode a large number of GH43 enzymes, suggesting that the extensive arabinose decorations appended to pectins and xylans may represent a major nutrient source, not just for intestinal bacteria but also for microorganisms that occupy terrestrial ecosystems. The results presented here predict that C. japonicus possesses an extensive range of glycoside hydrolases, lyases, and esterases. Most importantly, the genome of C. japonicus is remarkably similar to that of the gram-negative marine bacterium, Saccharophagus degradans 2-40T. Approximately 50% of the predicted C. japonicus plant-degradative apparatus appears to be shared with S. degradans, consistent with the utilization of plant-derived complex carbohydrates as a major substrate by both organisms.
An estimated 1011 tons of plant cell wall material is recycled annually, making this composite structure the most abundant source of organic carbon in the biosphere (11). Sugars released from the degradative process represent important nutrients for microorganisms, plants, and herbivores. Currently, microbial enzymes that attack plant cell walls are widely used in the biotechnology sector for the production of detergents, paper, textiles, and animal and human foods. However, the most important application of these biocatalysts is in the production of renewable biofuels (3, 28, 43).
The complex chemical structure of the plant cell wall, as well as the extensive interactions between polysaccharides, presents a significant challenge to microorganisms, as it greatly restricts enzymatic attack. As a result, the microbial species that rely on the plant cell wall as a growth substrate produce an extensive repertoire of glycoside hydrolases, lyases, and esterases to target the numerous linkages that are present in this composite structure (see references 40 and 43 for reviews). These enzymes often contain noncatalytic carbohydrate binding modules (CBMs) which, by bringing the appended catalytic module into intimate contact with its substrate, reduce the “accessibility problem” (6). In contrast to fungal plant cell wall-degrading hydrolases, which generally contain a single CBM, the corresponding bacterial enzymes often contain multiple CBMs belonging to the same or different families. Of the 49 CBM families identified to date, 47 contain bacterial proteins (http://afmb.cnrs-mrs.fr/CAZY/). The screening of gene libraries has provided insight into the portfolio of microbial plant cell wall hydrolases, but the current data are still fragmentary as they rely upon the expression of these enzymes in a heterologous host and an appropriate screen to identify clones displaying the appropriate hydrolytic activity. Therefore, genome sequencing and comparative analyses present a unique opportunity to obtain a complete picture of the microbial plant cell wall-degrading apparatus.
The terrestrial saprophytic bacterium Cellvibrio japonicus was first isolated from a Japanese soil in 1952 (41) and named “Pseudomonas fluorescens subsp. cellulosa.” Recent studies, however, demonstrated that C. japonicus is not a member of the genus Pseudomonas but is closely related to Cellvibrio mixtus, and hence the bacterium was renamed (31). C. japonicus has been experimentally shown to degrade all of the major plant cell wall polysaccharides, including crystalline cellulose, mannan, and xylan (7, 22, 23, 25, 26), and is a able to grow on media in which these polysaccharides are the sole carbon and energy source. Unlike the case for anaerobic plant cell wall-degrading organisms, exemplified by Clostridium thermocellum, the C. japonicus enzymes that target polysaccharides, which are integral to the plant cell wall, are fully secreted into the culture media and do not assemble into large multienzyme cellulosome-like complexes (7, 22, 23, 26). Since the first cellulase genes from C. japonicus were cloned ∼20 years (22) ago, numerous plant cell wall-degrading glycoside hydrolases, esterases, and lyases have been biochemically characterized (for a review, see reference 25). Furthermore, a genetic system has recently been developed (2), and thus C. japonicus represents an excellent system for studying the mechanism of plant cell wall degradation in a gram-negative, noncellulosomic saprophyte. We report in this communication the genome sequence of C. japonicus, which reveals a remarkable similarity between the plant cell wall-degrading apparatus of C. japonicus and that of the marine bacterium Saccharophagus degradans (38). Interestingly enough, the uniqueness of S. degradans comes from an ability to degrade many complex polysaccharides from algal, plant, and invertebrate sources, and this organism presented the first functional system from a marine bacterium (38). The genome sequences of the two organisms provide insight into the selection pressures imposed by marine and terrestrial ecosystems on the plant cell wall-degrading apparatus of prokaryotes.
MATERIALS AND METHODS
Genome sequencing, assembly, and gap closure.
C. japonicus sp. nov. strain Ueda107 was obtained from the National Collections of Industrial, Marine and Food Bacteria (www.ncimb.co.uk/index.php; accession number 10462) and was cultured at 37°C to mid-log phase in minimal medium (M9 salts) containing 0.2% wheat arabinoxylan. Genomic DNA was purified using the Qiagen Genomic-tip 100/G kit, and cloning, sequencing, and genome assembly were performed as described previously (21). Two plasmid libraries containing small (2- to 3-kb) and medium (8- to 10-kb) inserts were generated, and ∼8-fold sequence coverage was achieved from both libraries. All sequencing and physical gaps were closed by editing the ends of sequence traces, primer walking or transposon-primed sequencing on plasmid clones, and combinatorial PCR followed by sequencing of the PCR products.
ORF prediction and annotation.
A set of open reading frames (ORFs) likely to encode proteins was identified by GLIMMER, and those shorter than 30 codons were eliminated. All ORFs were searched against a nonredundant protein database and annotated using two sets of hidden Markov models (HMMs): 7,459 HMMs from PFAM version 14.0 (www.sanger.ac.uk/Software/Pfam/index.shtml) and 2,250 HMMs from the TIGR ortholog resource version 4.0 (www.tigr.org/TIGRFAMs/index.shtml). Membrane-spanning regions were predicted with TMHMM, and signal peptides were predicted with SignalP. ORFs thought to be involved in plant cell wall degradation and CBM-containing ORFs were annotated manually. ORFs that overlapped were inspected visually and, in some cases, removed.
Phylogenetic analysis.
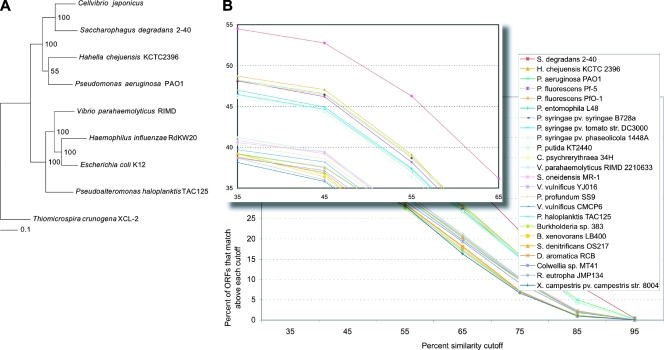
Concatenated protein maximum-likelihood trees were generated as described previously (21), with the following protein markers: initiation factor 2 (InfB); ribosomal proteins L2 (RplB), S5 (RpsE), S8 (RpsH), and S11 (RpsK); DNA topoisomerase I (TopA); signal recognition particle protein (Ffh); DNA gyrase B subunit (GyrB); GTP binding protein LepA; CTP synthase (PyrG); DNA recombinase A (RecA); and DNA-directed RNA polymerase β chain (RpoB). RecA and RpoB were substituted for elongation factors G (FusA) and Tu (Tuf), which were in multiple copies in C. japonicus. The protein markers were chosen because they are conserved across bacterial taxa (9, 15, 36) and produce monophyletic domains similar to rRNA trees (9, 15). The organisms were chosen based on top BLAST matches and their positions on 16S rRNA trees (data not shown). The concatenated protein trees contained sequences from completely sequenced genomes and were rooted to the gammaproteobacterium Thiomicrospira crunogena strain XCL-2 sequences (Fig. 1). Bootstrapped data sets were generated using the SEQBOOT program, and consensus trees were determined using CONSENSE. The final trees with preserved branch lengths were computed using DNAML and PROML (21).
FIG. 1.
Phylogenetic analysis and summary of BLAST results. (A) Consensus maximum-likelihood tree using 12 concatenated protein data sets. The numbers along the branches denote the percent occurrence of nodes among 100 bootstrap replicates. The scale bar represents the number of amino acid substitutions. (B) Distribution of protein percent similarities for the top 25 matching organisms in BLAST searches of the predicted C. japonicus proteome against a database of 382 completely sequenced bacterial genomes.
Comparative genomics.
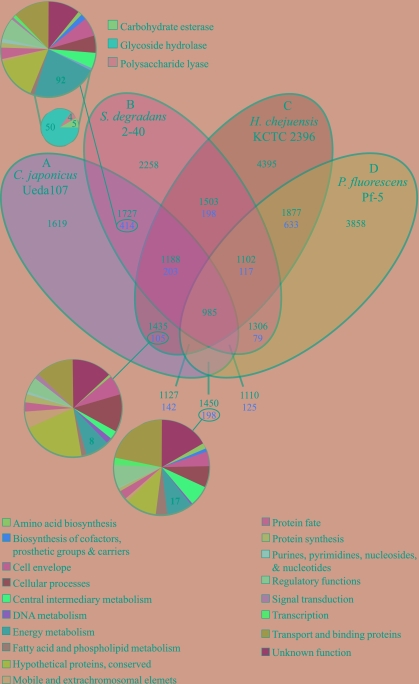
All predicted proteins from the C. japonicus genome were compared using BLAST to those from all other completed genomes (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi). Significant BLAST matches were scored for each ORF using cutoffs of 10−5 for the P value and 70% for the length of the alignment compared to both query and database proteins. ORFs with a top match to another C. japonicus protein rather than to a protein in another species were collected as recent duplication candidates; transposases were excluded from this list. The Venn diagram in Fig. 2 was constructed as previously described (21), except that local synteny (±5 ORFs) was examined prior to bidirectional best matches.
FIG. 2.
Whole-genome comparison of C. japonicus (A) and three related organisms (B to D). The Venn diagram shows the number of proteins shared (black) or unique (red) within a particular relationship. The large pie chart plots the number of gene sequences by main functional role category for C. japonicus. The small pie chart plots the distribution of predicted hydrolytic enzymes that target the plant cell wall and are shared by C. japonicus and S. degradans.
Nucleotide sequence accession number.
The nucleotide sequence of the whole genome of C. japonicus was submitted to GenBank under accession number c_japonicus_305 CP000934.
RESULTS
Main genomic features of C. japonicus.
The genome of C. japonicus is a single circular chromosome of 4,576,573 bp with an average G+C content of 52%. A total of 3,790 proteins were predicted in the genome, and a function could be assigned for 2,485 proteins (65.57%) (see Table S1 in the supplemental material). In addition to novel insertion sequence (IS) elements in the chromosome, we identified a 36-kb Tn3-related element (transposase CJA_1851/CJA_1852 and cointegrate resolution proteins CJA_1822 and CJA_1849) that likely confers resistance to heavy metals. A single copy of a transposon, Tn7-Cj, was found downstream of glmS similar, to Escherichia coli Tn7 (12). This ∼20 kb element encodes five proteins similar to the Tn7 transposition proteins TnsABCDE (CJA_3801 to CJA_3797). In addition, there are at least eight nontransposition genes, three of which potentially encode enzymes involved in purine metabolism (CJA_3790 to CJA_3787) and another similar to a helicase (CJA_3794). Tn7-Cj carries two insertions of an IS element, ISCja1, so that Tn7-Cj might be the originating source of the other three copies of ISCja1 in the chromosome. At both ends of Tn7-Cj there are three 20-bp repeats that likely bind transposase and 3′-terminal nucleotides (ACA) that resemble those of Tn7.
The genome of C. japonicus contains a single 4.7-kb array of clustered regularly interspaced short palindromic repeats (CRISPRs). CRISPRs are a newly identified class of repeats and consist of repeat elements interspersed with a unique sequence (the spacers) of approximately the same length. CRISPR elements are associated with a group of putative protein-encoding genes (CRISPR-associated sequences [cas genes]) and have been found in the genomes of a broad range of microbial species. The C. japonicus CRISPR array carries 72 spacer sequences, which range in length from 33 to 36 bp. It has previously been shown in a different bacterial system that CRISPR spacer sequences derive from phage genomic DNA and that they are added after viral challenge (1). The presence of the spacer sequences in the host genome provides immunity against infection by the phages from which the spacer sequences are derived. Thus, the CRISPR locus in C. japonicus may protect against phage attack. Indeed, no prophage could be detected in the C. japonicus genome using the Phage_Finder program (20). One such phage that C. japonicus might have encountered previously is bacteriophage PA73 (Pseudomonas phage 73) or a close relative, because 2 of the 72 CRISPR spacer sequences in C. japonicus match different regions of PA73 (accession number DQ163913.1). One spacer (AATGTTAACAGAACAACAAGTATCGCATTGTTCG) matches 33 of 34 bp between positions 30288 and 30321 of PA73. The second spacer (ATGCGATTATCGGCAAAGCGACCGAAGCGGGCGAA) matches 34 of 35 bp between positions 32805 and 32771 of PA73. The other 70 of the 72 spacers did not have any match in the NCBI BLAST databases. This suggests that a significant cache of horizontally transferred genetic elements, which C. japonicus has encountered previously, awaits isolation and study.
C. japonicus was previously described as having a flagellum and being motile (31). Two of the three major flagellar operons that are present in the E. coli genome could be found in the C. japonicus genome (33). Specifically, operons I and IIIb have synteny with the E. coli system, while operons II and IIIa differ in gene content and order from E. coli. The genome also encodes two type IV pili, which have been shown to be involved in twitching motility as well as adherence to plant cells (14). One of these type IV pili (CJA_2401-CJA_2385) displays similarity to the mannose-sensitive hemagglutinin pilus from Pseudoalteromonas tunicata, an antifouling marine bacterium (30), which mediates attachment to the cellulose-containing surface of the green alga Ulva australis (13, 37). C. japonicus is also predicted to produce a type I fimbria (CJA_1284-CJA_1290) (39). Despite the presence of three different pili, no filamentous phage region could be identified in the genome.
Comparative genomics.
Phylogenetic and genome-wide analyses agree with the conclusions of recent studies that resulted in the movement of C. japonicus from its original classification as a pseudomonad, P. fluorescens subsp. cellulosa. Fewer than 13% of the C. japonicus genes have a top BLASTP match to predicted proteins from the genomes of P. aeruginosa, P. fluorescens (Pf-5, PfO-1), P. putida, P. entomophila, or P. syringae (1448A, B728a, and DC3000). By contrast, 35% of the C. japonicus genes have a top BLASTP match with predicted proteins from the genome of the gram-negative bacterium S. degradans 2-40T. As mentioned previously, S. degradans was originally isolated from decaying salt marsh cord grass (16) and is closely related to the marine bacterial genera Microbulbifer and Teridinibacter spp. The organisms belonging to these three genera have the ability to degrade complex marine carbohydrates. The high degree of overlap between the C. japonicus and S. degradans genomes is also evident in a graph of all significant BLAST matches of predicted C. japonicus proteins (Fig. 1): over 54% of the C. japonicus predicted proteome displays similarity to S. degradans, whereas the next closest organism has less than 49% overlap.
These results are also supported by phylogenetic analyses using a subset of 12 putative proteins from the genomes of C. japonicus and eight other completely sequenced gammaproteobacteria. A maximum-likelihood tree built from a concatenated alignment of these 12 predicted proteins shows that C. japonicus and S. degradans are more closely related to each other than they are to other species in the tree (Fig. 1). The 12 proteins used in the tree were carefully chosen and have been shown to be stable phylogenetic markers (see Materials and Methods). However, it is surprising that C. japonicus appears to be more closely related to S. degradans than to P. aeruginosa, since S. degradans and Pseudoalteromonas haloplanktis belong to the order Alteromonadales, and C. japonicus and P. aeruginosa belong to the order Pseudomonadales. Also, this finding of relatedness is limited by the number of completely sequenced genomes available at the time of this study. Ideally, a more robust phylogenetic analysis would include other relatives of C. japonicus and S. degradans. The genomes which have not yet been sequenced, but might be included in the future, are known members of the genus Teredinibacter, the genus Microbulbifer, and other members of the genus Cellvibrio. Regardless of this shortcoming, the current maximum-likelihood tree of 12 proteins and the graph of all significant BLAST matches both show that there are many genome features that are similar in C. japonicus and S. degradans.
The C. japonicus genes displaying a top match to S. degradans have diverse functions and are predicted to be in diverse role categories. Of the putative plant-degradative C. japonicus enzymes, approximately 50% (n = 74) display closest similarity to predicted proteins from S. degradans, and 59 of these are shared exclusively with S. degradans (Fig. 2). This is consistent with the utilization of plant-derived complex carbohydrates as a major growth substrate by both organisms (7, 22, 23, 26, 38) and the retention of this hydrolytic apparatus by both species during the course of evolution. The other 50% (n = 77) of these predicted degradative enzymes are found among the 1,619 “unique” proteins that are predicted in the C. japonicus genome and absent from the other three related organisms (Fig. 2). At least 19 of these “unique” putative enzymes have a top match with another C. japonicus potential protein rather than a protein from another species and likely represent recently duplicated genes in C. japonicus. Thus, the majority of the hydrolytic enzymes in C. japonicus either are shared with S. degradans or have arisen by apparent gene duplication/expansion events since C. japonicus and S. degradans diverged from a common ancestor. Alternatively, the genes encoding these 19 “unique” enzymes might have been lost from the S. degradans lineage after the split from a common ancestor.
The plant cell wall-degrading apparatus of C. japonicus.
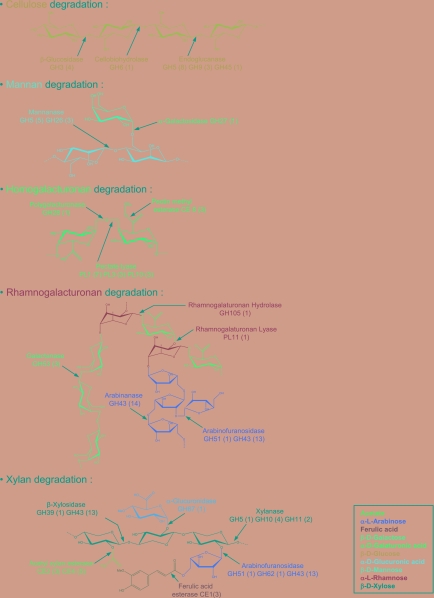
Consistent with its ability to degrade an extensive range of plant structural polysaccharides (25), C. japonicus possesses 123 glycoside hydrolases, 14 carbohydrate lyases, and 17 CBM-containing proteins with no known catalytic module (Fig. 3; see Table S2 in the supplemental material). In general, the genes encoding these enzymes are not clustered, although exceptions to this rule include the following loci; note that the names of the genes and enzymes indicate their activity and CAZy family (e.g., cellulases [Cel], mannanases [Man], arabinofuranosidase [Abf], and xylanase [Xyn]). One cluster encodes genes for an exo-1,4-β mannosidase, Man5D, and an α-galactosidase, Aga27A, (24), which remove terminal mannose residues from the mannan backbone and galactose side chains, respectively. Thus, genes encoding enzymes that remove the terminal sugars from galactomannan are tightly linked. Indeed, this cluster is identical in arrangement to a genetic locus found in the related bacterium Cellvibrio mixtus (10). The cluster also encodes an epimerase which in C. mixtus has been shown to convert mannose into glucose, thus providing a mechanism for the integration of mannose into the metabolic pathways of Cellvibrio (10). There is also a cluster of genes encoding Abf62A, Xyn10B, and two esterases, Fae1A and Fae1B, all of which have similar (or identical in Abf62A, Xyn10B, and Fae1A [19, 32]) N-terminal regions containing a CBM2 and a CBM35, which catalyze the degradation of xylan. An endo-β-galactosidase, Ebg98A, which also has an N-terminal CBM2 and a C-terminal CBM35, is also closely linked to this locus. The expansion of CBM2 and CBM35 in these enzymes appears to be an example of gene duplication in C. japonicus. Other examples of clusters (genes within 0.4 Mb of each other) include man26A, which is near abf51, a general-acting arabinofuranosidase (2); man5A, which is physically linked to cae1, cel5A (esterase genes), and a putative transcriptional regulator; and man5B, which is part of a large cluster containing man26B, cel74, cel6A, cel9A, and cbp2F.
FIG. 3.
The plant cell wall-degrading apparatus of C. japonicus. The major enzymes that attack the plant cell wall and their sites of action are depicted with arrows. The values in parentheses refer to the predicted number of members that were identified for the corresponding enzyme family in the C. japonicus genome.
All but 28 of these proteins contain N-terminal signal peptides and are therefore extracytoplasmic. The catalytic modules of the C. japonicus glycoside hydrolases belong to 31 different families, with the largest families being GH5, GH13, and GH43 (15, 17, and 14 enzymes, respectively) (see Table S2 in the supplemental material). GH5 contains an extensive repertoire of endo-acting enzymes that hydrolyze plant structural polysaccharides, which is consistent with the observation that C. japonicus can target these polymers as major nutrients (7, 22, 23, 26). GH13 comprises starch-degrading enzymes, and this storage polymer likely represents an important carbon and energy source for the bacterium. GH43 enzymes generally display specificity for arabinose-containing substrates, which are particularly prevalent in the side chains appended to the rhamnogalacturonan backbone of pectins. These gene expansions reflect the capacity of these organisms to utilize starch and the complex array of arabinose-based side chains of pectin as important nutrients.
The complete hydrolysis of cellulose requires endoglucanases, which are believed to target noncrystalline regions of the polysaccharide, and exo-acting cellobiohydrolases that attack the crystalline structures (40, 43). These enzymes progressively release the disaccharide cellobiose from the reducing (GH7 and GH48) and nonreducing (GH6) ends of the β-glucan chains, which are then hydrolyzed to glucose by β-glucosidases. The C. japonicus genome encodes 12 endoglucanases and 4 β-glucosidases but only a single cellobiohydrolase (CJA_2473), located in GH6 (see Table S2 in the supplemental material). As a result, this bacterium lacks a “reducing-end” cellobiohydrolase, which is considered an important component of fungal (GH7) and clostridial (GH48) cellulose-degradative systems.
The extensive portfolio of C. japonicus enzymes that degrade noncellulosic plant structural polysaccharides includes numerous biocatalysts that can attack the backbone and side chains of the hemicellulose polysaccharides. These include four GH10 and two GH11 xylanases along with five GH5 and three GH26 mannanases (see Table S2 in the supplemental material). The bacterium also contains an extensive array of enzymes that hydrolyze pectins which, in addition to GH43 hydrolases, includes an extensive range of pectate lyases and carbohydrate esterases.
Complement of C. japonicus CBMs.
Many of the C. japonicus enzymes that attack the major plant structural polysaccharides contain CBMs that increase catalytic activity by reducing the “substrate accessibility problem” (5). C. japonicus possesses 57 proteins, primarily glycoside hydrolases, that contain a total of 66 CBMs derived from 13 different families (families 2, 4, 5, 6, 10, 13, 15, 26, 32, 33, 35, 41, and X14), with many enzymes containing two CBMs (see Fig. S1 and S2 in the supplemental material). The variation in the portfolio of CBMs appended to these hydrolytic enzymes may affect the carbohydrate targeting of these biocatalysts and thus influence their substrate specificity. For example, in the two xylanase families, GH10 and GH11, the catalytic modules are appended to a range of different CBMs that target crystalline cellulose (CBM10 and CBM2) and xylan (CBM15, CBMX14, and CBM35). The complexity of CBM combinations in these enzymes is consistent with recent studies that show that the specificity of crystalline cellulose and xylan binding CBMs in vivo is influenced by the context of their target ligands within the plant cell wall (4, 34). To summarize, a key feature of the architecture of the C. japonicus hydrolases, which degrade polysaccharides that are integral components of the plant cell wall, is that they are tethered to these composite structures by a crystalline cellulose binding CBM and are then directed to their specific substrate by a second CBM which binds to the target polysaccharide. As this is one of the few reports on the genome of an aerobic plant cell wall-degrading saprophytic bacterium, it is unclear whether the molecular architecture of the C. japonicus enzymes represents a generic model for the plant cell wall-degrading apparatuses of other aerobic prokaryotes. It is clear, however, that the molecular organization of the bacterium's hydrolases is more complex than that of the corresponding aerobic fungal enzymes, which generally contain only a single CBM.
Although CBMs enable catalytic activity by bringing the cognate enzyme into close contact with its insoluble substrate, a noncatalytic protein containing a CBM33 was shown to disrupt the crystalline structure of chitin, greatly enhancing the activity of chitinases against this recalcitrant substrate (42). Thus, the C. japonicus CBM33 (CJA_3139) and CBM33-CBM5 (CJA_2191) proteins may also disrupt crystalline chitin and therefore potentiate the action of the bacterium's chitinases. One or more of the other CBMs that are not appended to known catalytic entities may also play a role in the degradative process by disrupting crystalline polysaccharides such as cellulose.
Cellular location of the plant cell wall-degrading apparatus.
As expected, all the predicted plant cell wall-degrading enzymes contain signal peptides and are thus extracytoplasmic. The signal peptides on the CBM-containing enzymes are generally cleaved by type I signal peptidases and are likely secreted into the extracellular milieu by a type II secretion system (CJA_3333-CJA_3323), similar to the secretion of pectate lyases by the plant pathogen Erwinia chrysanthemi (27). While it is possible that some of these proteins may interact with the cell membrane through hydrophobic and/or ionic interactions, where analyzed in vivo, the majority of the CBM-containing enzymes appear to be present in the culture medium and thus do not associate with the outer envelope of C. japonicus (7, 17, 22, 23, 26). Interestingly, ∼33% of the exported C. japonicus plant cell wall-degrading enzymes contain signal peptides that are cleaved by type II signal peptidases and are predicted to be lipoproteins. The cellular location of these extracytoplasmic enzymes is consistent with a survival strategy for C. japonicus that maximizes its capacity to utilize the nutrients released from the plant cell wall. This is exemplified by the bacterium's hemicellulose-degrading systems. The two GH10 and two GH11 xylanases and three GH5 endo-acting mannanases (see Table S2 in the supplemental material), which contain CBMs that target crystalline polysaccharides (6), are secreted into the culture medium and release soluble polysaccharides and oligosaccharides. These small polymers are then hydrolyzed by enzymes on the bacterial cell surface that target small saccharide polymers (29, 35) to generate mono- and disaccharides. This model of temporal and spatial regulation has been shown for enzymes that catalyze mannan and xylan degradation in C. japonicus (17) and for cellulose degradation in S. degradans (38).
In addition to the Sec secretion system to translocate proteins across the inner/outer membranes, C. japonicus encodes the Sec-independent twin-arginine translocation “TAT” secretion system: tatA (CJA_0743), tatB (CJA_0742, CJA_0489), and tatC (CJA_0741, CJA_0488). At least two cell wall-degrading enzymes were predicted to encode a TAT export signal in addition to the Sec export signal: polygalacturonase (pga28A) (CJA_0172) and endo-1,4-beta-xylanase (xyn10D) (CJA_2888). Except for the flagellar system, no other type III secretion system components were found. There were three putative type I secretion systems identified, typified by the presence of adjacent inner membrane and membrane fusion protein pairs (CJA_1515/CJA_1516, CJA_1393/CJA_1394, and CJA_3604/CJA_3605). There were no type V or type VI secretion systems found.
DISCUSSION
Plant cell wall material is the most abundant source of organic carbon in the biosphere, and its enzymatic degradation represents an important source of nutrients for microorganisms, plants, and herbivores. Plant cell wall-degrading enzymes are widely used in the biotechnology sector for the production of detergents, paper, textiles, and animal and human foods, as well as for the production of renewable biofuels. Therefore, the discovery of new and more efficient plant cell wall-degrading enzymes can potentially have numerous and important biotechnology applications.
Because of its complex chemical structure, enzymatic attack of the plant cell wall presents a significant challenge, and microorganisms usually rely on an extensive repertoire of enzymes to efficiently degrade complex plant cell wall materials. In order to obtain a complete picture of the plant cell wall-degrading apparatus, we completely sequenced, annotated, and analyzed the genome of one of the most efficient plant cell wall degraders, C. japonicus. The results presented here show that the genome of C. japonicus is remarkably similar to that of the gram-negative marine bacterium S. degradans 2-40T. Both C. japonicus and S. degradans produce a repertoire of glycoside hydrolases, lyases, and esterases that can degrade cellulose, xylan, mannan, chitin, pectin, starch, and laminarin, with GH13 and GH43 being two of the largest enzyme families.
The large number of GH43 glycoside hydrolases is an intriguing characteristic of C. japonicus (n = 14), a feature shared with the colonic bacterium Bacteroides thetaiotaomicron (n = 33) (44), as well as S. degradans (n = 13) and some aerobic fungi, including Aspergillus. B. thetaiotaomicron utilizes complex carbohydrates as major nutrients. However, rather than attacking the internal regions of integral plant structural polysaccharides (such as cellulose and hemicellulose), the vast majority of glycoside hydrolases are predicted to be exo-acting, e.g., removing monosaccharides from the complex glycans presented on the surface of mammalian cells and the easily accessible plant polysaccharides such as the pectins. Thus, these extensive, and highly accessible, pectin decorations are likely to represent an important source of nutrients for both colonic bacteria and soil saprophytic microorganisms that use plant biomass as an important nutrient source.
The complement of cellulases encoded by the genome of C. japonicus is similar to that of S. degradans, which also contains a single cellobiohydrolase from GH6. However, both bacteria lack a reducing-end cellobiohydrolase, which is considered an important component of fungal (GH7) and clostridial (GH48) cellulose degradative systems. As both S. degradans and C. japonicus efficiently degrade crystalline cellulose, it would appear that not all enzyme systems which hydrolyze this important polysaccharide require a GH7 or GH48 exo-acting glycoside hydrolase.
Examination of the C. japonicus CBM repertoire shows a dominance of CBM2 and CBM10 modules. This observation likely reflects specificity for crystalline cellulose, which, because of its prevalence in all plant cell walls, functions as a universal receptor for these degradative enzymes, as suggested previously (18). As described above, the plant cell wall hydrolases of C. japonicus contain CBMs from numerous families. In sharp contrast, the corresponding fungal enzymes generally contain only a family 1 CBM, suggesting that targeting these glycoside hydrolases to polysaccharides other than crystalline cellulose does not confer an obvious evolutionary advantage. In the case of C. japonicus, it is apparent that the different CBMs appended to the catalytic modules of the glycoside hydrolases will target these enzymes to diverse plant cell wall components, providing an explanation for the complex arrangement of these modules in the degradative enzymes.
Approximately 50% of the C. japonicus plant-degradative apparatus appears to be shared with S. degradans, consistent with the observation that the cell walls of terrestrial and marine plants have many polysaccharides in common. Although these two species share this high level of similarity in their substrate utilization machinery, it is still not clear whether the common ancestor of C. japonicus and S. degradans was a marine or a terrestrial bacterium, and additional analyses of their genome sequences might reveal what adaptations underlie these different lifestyles. For example, consistent with the marine environment of S. degradans, the bacterium produces an extensive array of agarases and carageenases, while the terrestrial C. japonicus lacks these enzymes. Agarases are found mostly in marine habitats. This is consistent with the fact that agar, being a product of marine algae, is available to and utilized by marine organisms, such as S. degradans, as a convenient carbon and energy source. Mannan, however, is likely to be more common in terrestrial plants than in salt marsh cord grass (8), and consistent with this, C. japonicus has a more extended mannan-degrading system than S. degradans. It is also apparent that while both organisms contain a large number of CBMs, there has been an extensive expansion of CBM6 and CBM32 modules in S. degradans, which likely reflects the diversity of sugars present in the marine-specific polysaccharides. These differences highlight how the selection pressures imposed by terrestrial and marine environments have influenced the evolution of the plant cell wall-degrading apparatus of saprophytic prokaryotes.
Supplementary Material
Acknowledgments
We thank Sean C. Daugherty for help with gene annotations, database management, and GenBank submission.
This project was supported by U.S. Department of Energy Office of Biological Energy Research Co-Operative Agreement DE-FC02-95ER61962 and the Biotechnology and Biological Sciences Research Council.
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. [DOI] [PubMed] [Google Scholar]
- 2.Beylot, M. H., K. Emami, V. A. McKie, H. J. Gilbert, and G. Pell. 2001. Pseudomonas cellulosa expresses a single membrane-bound glycoside hydrolase family 51 arabinofuranosidase. Biochem. J. 358599-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat, M. K. 2000. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 18355-383. [DOI] [PubMed] [Google Scholar]
- 4.Blake, A. W., L. McCartney, J. E. Flint, D. N. Bolam, A. B. Boraston, H. J. Gilbert, and J. P. Knox. 2006. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J. Biol. Chem. 28129321-29329. [DOI] [PubMed] [Google Scholar]
- 5.Bolam, D. N., A. Ciruela, S. McQueen-Mason, P. Simpson, M. P. Williamson, J. E. Rixon, A. Boraston, G. P. Hazlewood, and H. J. Gilbert. 1998. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 331775-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braithwaite, K. L., G. W. Black, G. P. Hazlewood, B. R. Ali, and H. J. Gilbert. 1995. A non-modular endo-beta-1,4-mannanase from Pseudomonas fluorescens subspecies cellulosa. Biochem. J. 3051005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brett, C. T. a. K. W. 1996. Physiology and biochemistry of plant cell walls, vol. 1. Chapman and Hall, London, United Kingdom.
- 9.Brown, J. R., C. J. Douady, M. J. Italia, W. E. Marshall, and M. J. Stanhope. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28281-285. [DOI] [PubMed] [Google Scholar]
- 10.Centeno, M. S., C. I. Guerreiro, F. M. Dias, C. Morland, L. E. Tailford, A. Goyal, J. A. Prates, L. M. Ferreira, R. M. Caldeira, E. F. Mongodin, K. E. Nelson, H. J. Gilbert, and C. M. Fontes. 2006. Galactomannan hydrolysis and mannose metabolism in Cellvibrio mixtus. FEMS Microbiol. Lett. 261123-132. [DOI] [PubMed] [Google Scholar]
- 11.Coughlan, M. P. 1985. The properties of fungal and bacterial cellulases with comments on their production and application, p. 39-109. In G. E. Russell (ed.), Biotechnology and genetic engineering reviews, vol. 3. Intercept, Paris, France. [Google Scholar]
- 12.Craig, N. L. 2002. Tn7, p. 423-456. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 13.Dalisay, D. S., J. S. Webb, A. Scheffel, C. Svenson, S. James, C. Holmstrom, S. Egan, and S. Kjelleberg. 2006. A mannose-sensitive haemagglutinin (MSHA)-like pilus promotes attachment of Pseudoalteromonas tunicata cells to the surface of the green alga Ulva australis. Microbiology 1522875-2883. [DOI] [PubMed] [Google Scholar]
- 14.Dorr, J., T. Hurek, and B. Reinhold-Hurek. 1998. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol. Microbiol. 307-17. [DOI] [PubMed] [Google Scholar]
- 15.Eisen, J. A. 1995. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J. Mol. Evol. 411105-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekborg, N. A., J. M. Gonzalez, M. B. Howard, L. E. Taylor, S. W. Hutcheson, and R. M. Weiner. 2005. Saccharophagus degradans gen. nov., sp. nov., a versatile marine degrader of complex polysaccharides. Int. J. Syst. Evol. Microbiol. 551545-1549. [DOI] [PubMed] [Google Scholar]
- 17.Emami, K., T. Nagy, C. M. Fontes, L. M. Ferreira, and H. J. Gilbert. 2002. Evidence for temporal regulation of the two Pseudomonas cellulosa xylanases belonging to glycoside hydrolase family 11. J. Bacteriol. 1844124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira, L. M., A. J. Durrant, J. Hall, G. P. Hazlewood, and H. J. Gilbert. 1990. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem. J. 269261-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira, L. M., T. M. Wood, G. Williamson, C. Faulds, G. P. Hazlewood, G. W. Black, and H. J. Gilbert. 1993. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem. J. 294349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouts, D. E. 2006. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 345839-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 3e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert, H. J., G. Jenkins, D. A. Sullivan, and J. Hall. 1987. Evidence for multiple carboxymethylcellulase genes in Pseudomonas fluorescens subsp. cellulosa. Mol. Gen. Genet. 210551-556. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, H. J., D. A. Sullivan, G. Jenkins, L. E. Kellett, N. P. Minton, and J. Hall. 1988. Molecular cloning of multiple xylanase genes from Pseudomonas fluorescens subsp. cellulosa. J. Gen. Microbiol. 1343239-3247. [DOI] [PubMed] [Google Scholar]
- 24.Halstead, J. R., M. P. Fransen, R. Y. Eberhart, A. J. Park, H. J. Gilbert, and G. P. Hazlewood. 2000. Alpha-galactosidase A from Pseudomonas fluorescens subsp. cellulosa: cloning, high level expression and its role in galactomannan hydrolysis. FEMS Microbiol. Lett. 192197-203. [DOI] [PubMed] [Google Scholar]
- 25.Hazlewood, G. P., and H. J. Gilbert. 1998. Structure and function analysis of Pseudomonas plant cell wall hydrolases. Prog. Nucleic. Acids Res. Mol. Biol. 61211-241. [DOI] [PubMed] [Google Scholar]
- 26.Hazlewood, G. P., J. I. Laurie, L. M. Ferreira, and H. J. Gilbert. 1992. Pseudomonas fluorescens subsp. cellulosa: an alternative model for bacterial cellulase. J. Appl. Bacteriol. 72244-251. [DOI] [PubMed] [Google Scholar]
- 27.He, S. Y., C. Schoedel, A. K. Chatterjee, and A. Collmer. 1991. Extracellular secretion of pectate lyase by the Erwinia chrysanthemi out pathway is dependent upon Sec-mediated export across the inner membrane. J. Bacteriol. 1734310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Himmel, M. E., M. F. Ruth, and C. E. Wyman. 1999. Cellulase for commodity products from cellulosic biomass. Curr. Opin. Biotechnol. 10358-364. [DOI] [PubMed] [Google Scholar]
- 29.Hogg, D., G. Pell, P. Dupree, F. Goubet, S. M. Martin-Orue, S. Armand, and H. J. Gilbert. 2003. The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem. J. 3711027-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmstrom, C., S. James, B. A. Neilan, D. C. White, and S. Kjelleberg. 1998. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 481205-1212. [DOI] [PubMed] [Google Scholar]
- 31.Humphry, D. R., G. W. Black, and S. P. Cummings. 2003. Reclassification of “Pseudomonas fluorescens subsp. cellulosa” NCIMB 10462 (Ueda et al. 1952) as Cellvibrio japonicus sp. nov. and revival of Cellvibrio vulgaris sp. nov., nom. rev. and Cellvibrio fulvus sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 53393-400. [DOI] [PubMed] [Google Scholar]
- 32.Kellett, L. E., D. M. Poole, L. M. Ferreira, A. J. Durrant, G. P. Hazlewood, and H. J. Gilbert. 1990. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem. J. 272369-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 34.McCartney, L., A. W. Blake, J. Flint, D. N. Bolam, A. B. Boraston, H. J. Gilbert, and J. P. Knox. 2006. Differential recognition of plant cell walls by microbial xylan-specific carbohydrate-binding modules. Proc. Natl. Acad. Sci. USA 1034765-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy, T., K. Emami, C. M. Fontes, L. M. Ferreira, D. R. Humphry, and H. J. Gilbert. 2002. The membrane-bound alpha-glucuronidase from Pseudomonas cellulosa hydrolyzes 4-O-methyl-d-glucuronoxylooligosaccharides but not 4-O-methyl-d-glucuronoxylan. J. Bacteriol. 1844925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos, S. R., and H. Ochman. 2004. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 6754-759. [DOI] [PubMed] [Google Scholar]
- 37.Shime-Hattori, A., T. Iida, M. Arita, K. S. Park, T. Kodama, and T. Honda. 2006. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol. Lett. 26489-97. [DOI] [PubMed] [Google Scholar]
- 38.Taylor, L. E., II, B. Henrissat, P. M. Coutinho, N. A. Ekborg, S. W. Hutcheson, and R. M. Weiner. 2006. Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40T. J. Bacteriol. 1883849-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thanassi, D. G., E. T. Saulino, and S. J. Hultgren. 1998. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1223-231. [DOI] [PubMed] [Google Scholar]
- 40.Tomme, P., R. A. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 371-81. [DOI] [PubMed] [Google Scholar]
- 41.Ueda, K., S. Ishikawa, T. Itami, and T. Asai. 1952. Studies on the aerobic mesophilic cellulose-decomposing bacteria. 5-2. Taxonomical study on genus Pseudomonas. J. Agric. Chem. Soc. Jpn. 2635-41. [Google Scholar]
- 42.Vaaje-Kolstad, G., S. J. Horn, D. M. van Aalten, B. Synstad, and V. G. Eijsink. 2005. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 28028492-28497. [DOI] [PubMed] [Google Scholar]
- 43.Warren, R. A. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50183-212. [DOI] [PubMed] [Google Scholar]
- 44.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 2992074-2076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.