Abstract
The symbiotic nitrogen-fixing bacterium Sinorhizobium meliloti possesses the Sin quorum-sensing system based on N-acyl homoserine lactones (AHLs) as signal molecules. The Sin system consists of SinI, the AHL synthase, and SinR, the LuxR-type regulator. This system regulates the expression of a multitude of S. meliloti genes through ExpR, another LuxR-type regulator. Analysis of the activity of the sinI promoter showed that the expression of sinI is dependent on sinR and enhanced by a combination of expR and Sin AHLs. The characterization of the ExpR binding site upstream of sinI and the identification of binding sites upstream of the galactoglucan biosynthesis genes wgaA (expA1) and wgeA (expE1) allowed the definition of a consensus sequence for these binding sites. Based on this consensus, two additional ExpR binding sites in the promoter regions of exoI and exsH, two genes related to the production of succinoglycan, were found. The specific binding of ExpR to the wgaA and wgeA promoters was enhanced in the presence of oxo-C14-HL. Positive regulation of the galactoglucan biosynthesis genes by ExpR was shown to be dependent on WggR (ExpG) and influenced by MucR, both of which are previously characterized regulators of these genes. Based on these results, a reworked model of the Sin-ExpR quorum-sensing regulation scheme of galactoglucan production in S. meliloti is suggested.
The soil bacterium Sinorhizobium meliloti converts atmospheric dinitrogen to ammonia in symbiotic association with the host plant Medicago sativa (alfalfa). During the establishment of successful symbiosis, the extracellular polysaccharides produced by S. meliloti are crucial for this process. Mutants that are unable to produce at least one of the three symbiotically active polysaccharides, including succinoglycan, galactoglucan, or K antigen, are defective in nodule invasion and primarily induce the formation of symbiotically ineffective root nodules that are devoid of bacteria and bacteroids (18, 28, 41). The mechanisms by which each of these polysaccharides functions revolve around mediating infection thread initiation and extension on alfalfa (11, 37).
The galactoglucan biosynthesis genes are located in a 32-kb gene cluster on pSymB that is composed of 21 genes organized into five putative operons. The nomenclature of these genes, previously named the exp cluster (6), was recently revised (GenBank accession no. AL591985). The five putative operons are as follows: wga (expA) (nine genes), wgcA (expC) (one gene), wggR (expG) (one gene), wgd (expD) (two genes), and wge (expE) (eight genes) (6). The regulation of the galactoglucan biosynthesis genes is controlled by WggR, a transcriptional regulator that activates the expression of wgaA (expA1), wggR and/or wgdA (expG and expD1, respectively), and wgeA (expE1) by binding to sites consisting of a conserved palindromic region and two associated sequence motifs in the promoter regions of these genes (2, 4). Another major regulator of galactoglucan production in S. meliloti is MucR, a protein containing a C2H2 zinc finger DNA binding motif (26). In addition to activating the production of succinoglycan through binding sites in the promoter regions of the succinoglycan genes exoH and exoY (8), MucR inhibits the production of galactoglucan through additional sites in the promoter regions of the galactoglucan biosynthesis genes wgaA, wgeA, wgdA, and wggR (1, 43). Yet another protein, PhoB, is involved in regulating the galactoglucan biosynthesis genes under phosphate-limiting growth conditions, probably through direct interactions with the so-called PHO boxes in the promoter regions of these genes (13, 50). Recent work on the regulation of galactoglucan production revealed a very complex system involving two promoters (distal and proximal) for wggR and for each of the wga, wge, and wgd operons, and a model involving WggR, MucR, and PhoB has been proposed (1, 43).
Wild-type laboratory S. meliloti strain Rm1021, whose genome has been sequenced (16), produces a non-symbiotically-active galactoglucan under low-phosphate conditions (21, 34, 51) or upon the disruption of the mucR gene (26). However, the production of symbiotically active galactoglucan requires a functional ExpR, which is not present in Rm1021 (38). Characterization of the expR locus, located on the 3.4-Mb main chromosome (18), revealed an insertion sequence (ISRm2011-1) within the coding region of the gene (38). A spontaneous mutation involving the precise, reading frame-restoring excision of the insertion sequence from the coding region of expR resulted in a functional ExpR and mucoid colony morphology, which is indicative of high levels of galactoglucan production, including symbiotically active galactoglucan (38). ExpR is a member of the LuxR family of proteins, many of which are receptors for N-acyl homoserine lactones (AHLs) and are transcription regulators involved in the control of gene expression in response to changes in population density, a process known as quorum sensing (14, 38). Quorum-sensing systems have been discovered in S. meliloti (31, 33). One of these, the Sin system, consists of the autoinducer synthase SinI and its LuxR-type regulator, SinR. SinI is responsible for the production of a series of long-chain AHLs, including C12-HL, oxo-C14-HL, oxo-C16:1-HL, C16:1-HL, and C18-HL. A disruption of sinI was found to abolish galactoglucan production as well as the expression of several genes in the galactoglucan biosynthesis operons. This phenotype was complemented by the addition of AHL extracts from the wild-type strain, but not from a sinI mutant, and by the addition of synthetic C16:1-HL (32). The absence of symbiotically active galactoglucan in a sinI mutant was confirmed in plant nodulation assays, emphasizing the role of quorum sensing in symbiosis (32). In addition to regulating the galactoglucan biosynthesis genes, the Sin-ExpR combination also regulates a multitude of S. meliloti genes, including genes that participate in low-molecular-weight succinoglycan production, motility, and chemotaxis as well as other cellular processes (17, 23).
Given that ExpR is significant in the quorum-sensing-based regulation of so many S. meliloti genes, the question of its exact mechanism of regulation is rather interesting. We therefore sought to unravel the pathway by which quorum sensing and ExpR regulate the genes responsible for the production of galactoglucan. LuxR-type regulators, of which ExpR is a member, usually bind to a DNA consensus sequence known as a lux box, which is typically located upstream of the promoters of its target genes. These regulators bind to the lux box upon activation by binding to an AHL (12, 46). A putative lux box 70 bp upstream of sinI has been identified, but ExpR does not bind to it. Rather, as we previously reported, ExpR binds to a site approximately 100 bp upstream of sinI and results in approximately four-times-higher sinI mRNA levels (3), indicating positive regulation of sinI by ExpR, presumably upon AHL activation. However, this cannot be the only function of ExpR, because the upregulation of sinI occurs both in expR+ and expR strains (3), but only the expR+ strain produces detectable levels of galactoglucan.
In this study, we characterized the effect of ExpR on the Sin quorum-sensing system and identified additional ExpR DNA binding sites in the promoter regions of the galactoglucan and succinoglycan biosynthesis gene clusters. Furthermore, the effect of ExpR-stimulated galactoglucan biosynthesis gene transcription was studied in relation to those of the transcriptional regulators WggR and MucR. Cooperative and competitive interactions between these regulators are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains used in this study are listed in Table 1. S. meliloti strains were incubated at 30°C in Luria-Bertani (LB) medium supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LBmc) or tryptone-yeast medium (7). Escherichia coli strains were incubated at 37°C in LB medium. Antibiotics were added at the following concentrations: 120 μg ml−1 neomycin (Nm), 40 μg ml−1 gentamicin (Gm), 10 μg ml−1 nalidixic acid (Nx), 600 μg ml−1 streptomycin (Sm), and 10 μg ml−1 tetracycline (Tc) for S. meliloti and 50 μg ml−1 kanamycin (Kan), 100 μg ml−1 ampicillin (Ap), and 10 μg ml−1 Gm for E. coli.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference(s) or source |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| Rm2011 | Wild type; Nxr Smr | 10 |
| Rm101 | Rm2011 mucRΩ; Spcr cassette from pHP45Ω inserted into the PmacI site within mucR | 6 |
| SmSRΔG | Rm2011 ΔwggR; deletion of the wggR gene comprising 490 nucleotides of the 3′ terminus of the wggR coding region and 17 nucleotides downstream of wggR | 43 |
| SmBBΔG101 | Rm2011 ΔwggR mucRΩ; double mutant | 1 |
| 2011mTn5STM.2.01.B05 | Rm2011 sinR::mini-Tn5 | 39 |
| Rm1021 | Wild type; Nxr Smr | 27 |
| Rm8530 | Rm1021 expR+ | 18 |
| Rm11511 | Rm1021 sinI::Km | 33 |
| Rm11527 | Rm8530 sinI::Km | 32 |
| Rm1021 sinR | Rm1021 sinR::mini-Tn5 | This work |
| Rm8530 sinR | Rm8530 sinR::mini-Tn5 | This work |
| E. coli | ||
| DH5α | F−endA1 supE44 thi-1 λ-recA1 gyrA96 relA1 deoRΔ(lacZYA-argF)U169 | 22 |
| S17-1 | E. coli 294 Thi RP4-2-Tc::Mu-Km::Tn7 integrated into the chromosome | 44 |
| M15pREP4 | Nxs Strs Rifs Thi− Lac− Ara+ Gal+ Mtl− F− RecA+ Uvr+ Lon+ | Qiagen, Hilden, Germany |
| Plasmids | ||
| pSRPP18 | Promoter probe vector, pUC derivative; promoterless lacZ gene; integrates between the exoP terminator and the thiD gene | 1 |
| pCR-TOPO | TOPO cloning kit | Invitrogen, Karlsruhe, Germany |
| pJN105 | araC-PBAD cassette cloned in pBBR1MCS5; Gmr | 36 |
| pJNexpR | pJN105 containing expR | 3 |
| pHU231 | Tcr; pRK290 with a 388-bp HaeII insert containing pUC18 polylinker | 25, 42 |
| pLK64 | pPHU231 containing sinI-EGFP translational fusion; Tcr | This work |
Plasmids.
The construction of plasmids pJNexpR (3) and pSRPP18 (1), used in this work, was described previously. For the fusion of the sinI promoter with the gene coding for enhanced green fluorescent protein (EGFP), the sinI promoter region along with the region containing the first 9 codons of SinI were amplified using primers sinI_forward_HindIII (5′-CCTAAAGCTTCAACGATTCTCGGCATATCC) and sinI_reverse_XbaI (5′-TCCTTCTAGAACCGTTTCCGTTCACTATCCT). The EGFP coding sequence was amplified using primers EGFP_forward_XbaI (5′-AAGATCTAGAGTGAGCAAGGGCGAGGAGCT) and GFP_reverse_EcoRI (5′-GTACGAATTCTTACTTGTACAGCTCGTCCATG). The PCR-amplified DNA fragments were cloned into EcoRI-HindIII sites of pPHU231, yielding plasmid pLK64.
Expression and purification of His6-ExpR.
The expression and purification of recombinant His6-ExpR were performed essentially as described previously (3) except that the purified protein was mixed with glycerol (1:1 volume) and stored at −20°C, which resulted in prolonged stability.
Binding site cloning and DNA labeling.
The DNA probes from the promoter regions of sinI, wgaA, wgeA, and wggR and/or wgdA used in the electrophoretic mobility shift assays (EMSAs) were prepared as described previously (3). Specific primers labeled with 5′ Cy3 were used in a PCR with genomic DNA as the template to produce the Cy3-labeled fragments. The sinI promoter region was derived from a 216-bp region that included 31 bp of the 3′ end of sinR, the 156-bp intergenic region between sinR and sinI, and 29 bp of the 5′ end of sinI using primers 5′-TGTTCGACATGCTCTGATCC and 5′-CGACCGTTTCCGTTCACTAT. The wgaA promoter was derived from a 301-bp region that included 115 bp of the 3′ end of wgcA, the 183-bp intergenic region between wgcA and wgaA, and 3 bp of the 5′ end of wgaA using primers 5′-CAGAACGGTCGAACAGAGGT and 5′-CATCAACTCTTGCACGCAGC. The wgeA promoter was derived from a 301-bp region that included 41 bp of the 3′ end of wgdB (expD2), the 274-bp intergenic region between wgdB and wgeA, and 11 bp of the 5′ end of wgeA by using primers 5′-CAGTCTCCGACAGTTTCAAC and 5′-CATCAACTCTTGCACGCAGC. The wgdA and wggR promoters were derived from a 334-bp region that included 131 bp of the 5′ end of wgdA, 7 bp of the 5′ end of wggR, and the 196-bp intergenic region between wggR and wgdA by using primers 5′-ATAAAGAAGCGTCACGACGA and 5′-TCTCCATTGGGAACGTACTT. The exsH promoter was derived from a 238-bp region that included 230 bp upstream of exsH plus 8 bp of the 5′ end of exsH by using primers 5′-CGCGGTACCAAGTCGTGACATCGTCAATC and 5′-CGCGGATCCACCACGGCGTTCAATACGGTT. The exoI promoter was derived from a 273-bp region that included 270 bp upstream of exoI plus 3 bp of the 5′ end of exoI by using primers 5′-CGCGGTACCACGGCAACAACATGGATGTTC and 5′-CGCGGATCCCATTCCCCATCCCCGTTCAG. For the cloning of the ExpR binding sites, oligonucleotides (indicated by the gray boxes in Fig. 3) were annealed and cloned into vector pCR using the TOPO cloning kit (Invitrogen) according to the manufacturer's instructions. DNA fragments from the pCR-oligonucleotide constructs were amplified and Cy3 labeled using pUC18 universal sequencing primers 5′-AGCGGATAACAATTTCACACAGGA and 5′-GTTTTCCCAGTCACGAC.
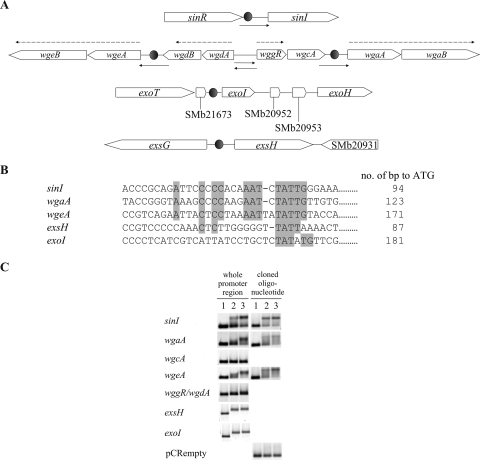
FIG. 3.
Promoter regions containing ExpR binding sites. Empty boxes indicate WggR binding sites (2), dark gray boxes indicate ExpR binding sites (oligonucleotides cloned into vector pCR), and the light gray box in the sinI promoter region indicates the lux box. Transcriptional start sites for wgaA, wgeA (1), and sinI are indicated by arrows with a “+1.” Translational starts are underlined.
Rapid amplification of 5′ cDNA ends.
The rapid amplification of 5′ cDNA ends was performed essentially according to the kit manufacturer's instructions (Roche). Cells were grown in LBmc medium to late logarithmic phase, centrifuged, and frozen in liquid nitrogen. RNA was extracted and purified using the RNeasy purification kit (Qiagen). RNA was then reverse transcribed at 55°C for 1 h using primer 5′-ATCGGTGACCGTGACGATATGG. A homopolymeric A tail was added to the 3′ end of the cDNA, and the cDNA was then amplified by PCR using a poly(T) primer and primer 5′-ATGGTGACCTGGTTCGATGC. The resulting PCR product was cloned using the TOPO cloning kit (Invitrogen) according to the manufacturer's instructions. Multiple positive clones were sequenced, and the transcription start was mapped to the promoter region of sinI.
PCR-based mutation of the ExpR binding site upstream of sinI.
For the mutation of each nucleotide within the ExpR binding site upstream of sinI, a series of complementary primers based on the binding site were designed. Within each set of complementary primers, a single nucleotide was replaced. From each set of complementary primers, the forward primer was used together with the universal reverse primer and the reverse primer was used with the universal forward primer using the UpsinI fragment cloned into pUC18 as the template (3). The resulting PCR products were gel purified, combined to allow annealing, and used as a template for a second PCR together with the universal primers to produce a Cy3-labeled fragment containing a mutated nucleotide.
EGFP fluorescence assay.
S. meliloti strains were first cultivated in liquid LBmc medium. Cell suspensions (20 μl) with an optical density at 600 nm (OD600) of 0.5 were spotted onto LBmc plates supplemented with streptomycin and tetracycline, with or without added oxo-C14-HL, and grown at 30°C for 20 to 22 h. The cells were collected and resuspended in 0.9% NaCl to an OD600 of 1.0. The cell density (OD600) and EGFP fluorescence (excitation at 485 nm and emission at 538 nm with a 97% scanning rate) were measured using a Tecan Infinite M200 reader (Tecan Trading AG, Switzerland) using 200 μl of the cell suspension in 96-well microtiter plates.
Analysis of promoter expression using the promoter probe vector.
S. meliloti strains carrying the promoter probe constructs were grown at 30°C either to late log phase in LBmc liquid medium or overnight on LBmc agar containing 120 μg. ml−1 Nm. Cells harvested from LBmc agar were resuspended in 0.9% NaCl. The cell density (OD600) and relative β-galactosidase activity were measured using a Tecan Infinite M200 reader (Tecan Trading AG, Switzerland) using 100 μl of the cell suspension in 96-well microtiter plates. The β-galactosidase activity assay was performed, and relative units (Miller units) were calculated as described previously (35).
EMSA.
The EMSA protocol was described previously (3). The Cy3-labeled DNA fragments were mixed with purified His6-ExpR in a reaction buffer containing approximately 0.1 mg ml−1 of sonicated herring testes DNA and 1.0 mg ml−1 bovine serum albumin (Sigma) in a final volume of 20 μl of DNA binding buffer (20 mM Tris-HCl [pH 8.0], 5 mM KCl). In the 20-μl reaction mixture, the Cy3-labeled DNA was included at 0.05 pmol; His6-ExpR was included at 0.1 pmol for the sinI-derived fragment and 3.0 pmol for fragments derived from the galactoglucan biosynthesis gene cluster as well as exsH- and exoI-derived fragments. Oxo-C14-HL was included at 20 pmol. The reaction mixtures were incubated at 24°C for 20 min. Loading buffer (5 μl; 78% glycerol) was added, and the reaction mixtures were loaded onto a 2% agarose gel. Following electrophoresis at 5 V/cm for 2 h, gel images were acquired using a Typhoon 8600 variable-mode imager (Amersham Bioscience, Freiburg, Germany).
RESULTS
Characterizing the UpsinI ExpR binding site.
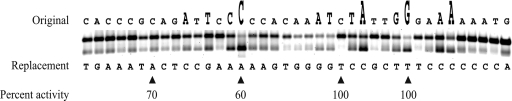
As established in our previous report (3), plasmid pJN105, carrying the His6-expR gene, complemented the nonfunctional expR locus in Rm1021, demonstrating that the His6-ExpR fusion protein functions in vivo. In that study, purified His6-ExpR was shown to bind to a site within the promoter region of sinI based on DNA footprinting. However, the specific nucleotides that are important for the binding of ExpR were unknown. In this study, a total of 35 bp was systematically modified within the sinI promoter using a “gene SOEing-type” PCR approach (24). The effect on the relative strength of binding of His6-ExpR to each DNA fragment that included a modified nucleotide was observed using gel shift assays in the presence of oxo-C14-HL, one of the identified Sin AHLs demonstrated to activate binding between His6-ExpR and the sinI promoter region (3) and previously shown to cause a maximal induction of sinI expression in S. meliloti (29). Figure 1 shows the resulting shift after mixing His6-ExpR, the modified Cy3-labeled sinI promoter fragment, and oxo-C14-HL. Of the 35 nucleotides that were modified, 12 were found to be important for His6-ExpR binding. Of these 12 nucleotides, 4 nucleotides, indicated by an increase in size in Fig. 1, were found to be critical for binding, although residual binding remained observable in each case. Also, as indicated in Fig. 1, the effects of four of the mutations on the expression of sinI were individually measured using the promoter probe vector containing a fusion of the sinI promoter region to lacZ in order to compare the in vitro and in vivo results. The selection of mutations was based on the EMSA result, where the sinI promoter-carrying mutations at both critical and insignificant locations were included. The first of the mutations (from left to right) corresponds to an insignificant reduction of binding in the EMSA and a reduction in expression to 70% compared to that of the wild type. The second mutation corresponds to a significant reduction in binding in the EMSA and to a reduction in expression to 60%. The third mutation appears to have resulted in an improved binding in the EMSA, although the level of expression was not significantly altered. The remaining mutation corresponds to a significant reduction in binding but not in expression. Under identical conditions, using both the promoter probe vector (results not shown) and plasmid pLK64, which contains a fusion of the sinI promoter region to the EGFP gene (see Fig. 4), the absence of expR resulted in a reduction of sinI expression to approximately 30% of the levels of a strain carrying a functional expR. Thus, it appears that although a high level of sinI expression is dependent on the presence of ExpR, a reduction in binding strength between ExpR and the sinI promoter, as observed in vitro, does not necessarily correspond to a reduction in the activation of sinI expression by ExpR in vivo.
FIG. 1.
Mutation-based characterization of the ExpR binding site upstream of sinI. Purified His6-ExpR was mixed with 10 μM oxo-C14-HL and Cy3-labeled UpsinI fragments, each carrying a single point mutation, and the effect of the mutation was determined using gel shift assays. Each shift is a representation of the effect caused by a single modifying nucleotide on ExpR-DNA binding strength. A weak shift indicates a significant disruptive effect caused by the modification, and a strong shift indicates a moderate or negligible effect. The letter above each shift indicates the original nucleotide, and the letter below each shift indicates the modifying nucleotide. The nucleotides very important for ExpR binding are indicated by a large increase in the size of the letter above the shift; nucleotides of lesser importance are indicated by an intermediate increase in size. The resulting shift using the native Cy3-labeled UpsinI fragment (not shown) showed no observable difference from the first five shifts on the left side of the figure. The gel shift experiment was repeated three times. Four UpsinI fragments, each with a single point mutation (indicated by the arrows below), were cloned into the promoter probe vector, in addition to the native UpsinI fragment. Percent activity indicates the β-galactosidase activity that was measured from the mutated fragment in comparison to that from the native fragment.
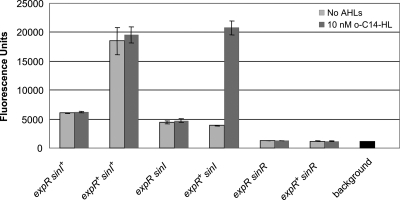
FIG. 4.
Expression of sinI is dependent on sinR and is induced by expR and Sin AHLs. sinI expression was measured as EGFP fluorescence in strains carrying plasmid pLK64 in the absence (dark gray bars) and presence (light bars) of synthetic oxo-C14-HL (abbreviated as o-C14-HL). Background fluorescence was determined in Rm1021 devoid of plasmid pLK64 and is shown at the far right of the graph. Strains are included as follows: lane 1, Rm1021 (sinI+ expR); lane 2, Rm8530 (sinI+ expR+); lane 3, Rm11511 (sinI expR); lane 4, Rm11527 (sinI expR+); lane 5, Rm1021 sinR (sinR sinI+ expR); lane 6, Rm8530 sinR (sinR sinI+ expR+).
ExpR binds to several promoter regions in the galactoglucan and succinoglycan biosynthesis gene clusters.
DNA fragments directly upstream of the galactoglucan biosynthesis genes wgaA, wgdA, wgcA, wgeA, and wggR (Fig. 2A), representing the promoter regions of the five putative galactoglucan biosynthesis operons, were included in gel shift assays with purified His6-ExpR. Of these regions, only those from wgaA and wgeA shared homology with the ExpR binding site upstream of sinI (Fig. 2B) and bound to His6-ExpR in an oxo-C14-HL-enhanced manner (Fig. 2C). Covering the homologous regions within the promoter regions of wgaA and wgeA, 31 and 34 nucleotides (indicated by the dark gray boxes in Fig. 3), respectively, were cloned into vector pCR. Universal sequencing primers based on the vector were used to amplify and Cy3 label DNA fragments containing the cloned regions and the surrounding vector sequence. In addition, a 33-nucleotide sequence (indicated by a dark gray box in Fig. 3) from the promoter region upstream of sinI, covering the ExpR binding site, was also cloned in the same manner and included as a positive control. A Cy3-labeled PCR product from the empty vector was included as a negative control. Figure 2C shows the resulting mobility after mixing His6-ExpR, the Cy3-labeled fragment, and oxo-C14-HL and confirms that the ExpR binding sites are indeed located within the cloned regions consisting of <35 bp (Fig. 2B) derived from the promoter regions of sinI, wgaA, and wgeA. The identification of a general ExpR binding site consensus allowed the location of other potential ExpR binding sites in the promoter regions of two more genes, exoI and exsH, both shown to exhibit ExpR/Sin quorum-sensing-dependent expression (19). Unlike the promoter regions of sinI, wgaA, and wgeA, where the His6-ExpR-induced shifts are maximal in the presence of oxo-C14-HL, the promoter regions of both exoI and exsH showed a shift in the presence of His6-ExpR independent of the presence of oxo-C14-HL. Labeled DNA derived from several other gene promoters, including SMc00690, SMc02032, SMc04171, SMa1067, and SMc01116, were included on the basis of homology to the ExpR binding site consensus but did not show a shift under the same conditions (results not shown).
FIG. 2.
(A) The galactoglucan biosynthesis gene region from wgaA to wgeB (expE2) (10,330 bp) from the galactoglucan biosynthesis gene cluster of S. meliloti (16). Promoter regions used for gel shift assays and determination of expression levels are indicated by solid arrows. ExpR binding sites are indicated by filled circles. Transcriptional units are marked by broken arrows. (B) ExpR binding-site consensus. The identified ExpR binding sites from the promoter regions of sinI, wgaA, and wgeA were aligned in the 5′-to-3′ orientation to show homology. Sequences from the promoter regions of exsH and exoI exhibiting weak homology to the consensus are included. (C) AHL-enhanced DNA binding of purified His6-ExpR to the galactoglucan biosynthesis gene promoter regions. (Left) Cy3-labeled promoter regions of wgaA, wgeA, and wgdA and/or wggR were used in the gel shift assay. (Right) Oligonucleotides (indicated by the dark gray boxes in Fig. 3) covering the ExpR binding sites within the sinI, wgaA, and wgeA promoters cloned into vector pCR. DNA fragments from the pCR-oligonucleotide constructs were amplified and Cy3 labeled and are included in the gel shift assays, along with a Cy3-labeled fragment from the empty vector as a negative control. For each panel, lane 1 is the Cy3-labeled DNA only, lane 2 is Cy3-labeled DNA mixed with His6-ExpR, and lane 3 is a mixture of Cy3-labeled DNA, His6-ExpR, and oxo-C14-HL.
The transcription start for sinI was determined by rapid amplification of 5′ cDNA ends and is located 28 bp upstream of the translation start, 37 bp downstream from the middle of the lux box, and approximately 80 bp downstream of the ExpR binding site (Fig. 3). The ExpR binding site upstream of wgaA is centered approximately 35 bp downstream of the WggR binding site (2), either overlapping or immediately downstream of the distal transcription start, 47 bp upstream of the proximal transcription start (1), and 138 bp upstream of the ATG (Fig. 3). Interestingly, MucR was shown to bind between the WggR and ExpR binding sites or possibly overlap the ExpR binding site (1). The ExpR binding site upstream of wgeA is centered approximately 44 bp downstream of the WggR binding site (2), overlapping the distal transcription start, 132 bp upstream of the proximal transcription start (1), and 188 bp upstream of the ATG (Fig. 3). The MucR binding site was once again shown to be located between the WggR and ExpR binding sites (1).
Expression of sinI is dependent on sinR and is activated by expR in the presence of Sin AHLs.
In order to characterize the regulation of sinI expression, we measured fluorescence in S. meliloti strains Rm1021, Rm8530 (Rm1021 expR+), Rm11511 (Rm1021 sinI), Rm11527 (Rm8530 sinI), Rm1021 sinR, and Rm8530 sinR, carrying plasmid pLK64, in which the sinI promoter region is fused to the EGFP gene. During growth on LBmc agar, the expression of sinI-EGFP was activated in the presence of ExpR, since an approximately three-times-higher level of sinI expression was measured in Rm8530 (expR+ sinI+ sinR+) than in Rm1021 (expR sinI+ sinR+) (Fig. 4). The activation by ExpR is dependent on the Sin AHLs, since sinI expression was reduced in strain Rm11527 (expR+ sinI) but restored by the addition of 10 nM oxo-C14-HL to the medium. Activation by ExpR is also completely dependent on SinR, since only background levels of fluorescence could be detected in both Rm8530 sinR and Rm1021 sinR strains (expR+ sinI+ sinR and expR sinI+ sinR, respectively). The Sin AHLs may be required for the full activity of SinR, although this is unclear, since the positive effect of Sin AHLs on sinI expression is only weak in the absence of ExpR (in strain Rm1021 [expR sinI+ sinR+]), and sinI expression was not activated by the addition of oxo-C14-HL to strain Rm11511 (expR sinI sinR+). However, SinR was able to promote sinI expression to moderate levels even in the absence of ExpR, SinI, or synthetic oxo-C14-HL, since fluorescence levels were higher in Rm11511 (expR sinI sinR+) and Rm11527 (expR+ sinI sinR+) than in Rm1021 sinR (expR sinI+ sinR) and Rm8530 sinR (expR+ sinI+ sinR).
ExpR-activated expression of galactoglucan biosynthesis genes is dependent on WggR.
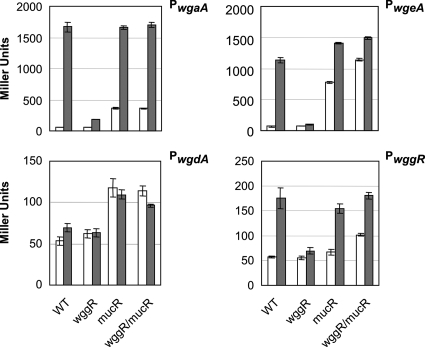
The promoter fragments of galactoglucan biosynthesis operons, including wga, wgd, wge, and wggR, were cloned into promoter probe vector pSRPP18, a mobilizable suicide vector carrying a promoterless lacZ gene (1). Following the integration of the plasmid into the genome by a single homologous recombination event downstream of exoP, the promoter strength, indicated by LacZ activity, was measured in four genetic backgrounds: wggR+ mucR+ expR (Rm2011), wggR mucR+ expR (SmSRΔG), wggR+ mucR expR (Rm101), and wggR mucR expR (SmBBΔG101) (Fig. 5). The effects of a functional expR locus on the promoter fragments were measured in each of the four genetic backgrounds using either pJNexpR, which was constructed as previously described (3), or empty vector pJN105 as a negative control. During growth on LBmc agar, the presence of a functional expR resulted in a mucoid phenotype that is easily distinguishable from the “dry” phenotype of strains without expR, as was previously reported (32). Consistent with this observation, a functional expR resulted in a dramatic increase in the activation of the wgaA promoter (28 times) and the wgeA promoter (17 times) but much lower activation of the wggR promoter (2.7 times). The presence of expR did not result in a significant increase in activity of the wgdA promoter. The putative promoter for wgcA was also measured but did not show any activity in any of our experiments (results not shown). In a wggR background, this ExpR-dependent activation is markedly reduced for the wgaA, wgeA, and wggR promoters, suggesting that ExpR is dependent on the presence of WggR for most of the activation of these promoters but that a low level of activation by ExpR is also present in the absence of WggR.
FIG. 5.
Effect of ExpR on expression of promoters of galactoglucan biosynthesis genes. The promoter regions of wgaA, wgdA, wgeA, and wggR were fused to lacZ via the promoter probe vector (1), and the resulting LacZ activity was measured in the absence (pJN105) (open boxes) and presence (pJNexpR) (gray boxes) of ExpR and various wggR and mucR backgrounds after growth in LBmc liquid medium to late log phase. LacZ activity was also measured in strains containing the empty promoter probe vector (without an inserted promoter) and was approximately 50 ± 5 Miller units (not shown).
Compared to the wild type in the absence of expR, the effect of a disruption of mucR was an increase in the levels of expression of wgaA (six times), wgdA (2.2 times), and wgeA (11 times), consistent with MucR as a repressor for the galactoglucan biosynthesis genes, as was reported previously (1, 26, 43). No significant change in wggR expression in the absence of mucR was observed under our conditions. In previous reports, however, the increase in expression of the galactoglucan biosynthesis genes due to the disruption of mucR was higher. This discrepancy is probably due to the use of different growth media. In strains carrying homogenote fusions of lacZ to these genes, growth in minimal medium supplemented with 2 mM phosphate showed a 10-times increase in the level of expression of wggR and a 40-times increase in the level of expression of wgdA due to the disruption of mucR (43). A disruption of mucR in strains carrying the identical promoter probe constructs used in this study resulted in a four-times increase in the levels of expression of both wgdA and wggR during growth in minimal medium supplemented with 2 mM phosphate (1).
In a mucR background, the effect of the presence of expR was again a very high level of expression of wgaA (28 times), similar to that in a mucR+ expR+ background, while that of wgeA was increased (21 times) by approximately 20% compared to that in the mucR+ expR+ background. In this case, it appears that MucR slightly negatively regulates the wgeA promoter in the presence of expR but not the wgaA promoter under the same conditions. The expression of wgdA was unchanged by the presence of expR in a mucR background, while that of wggR was slightly increased (2.7 times).
The effect of disrupting both wggR and mucR (wggR mucR) was an increased level of expression of both wgeA and wggR (∼50%) compared to that in a wggR+ mucR background but not for wgaA or wgdA. This result suggests that in the absence of MucR, WggR appears to function as a negative regulator, as was reported previously (1). With the addition of expR to a wggR mucR background, we observed the same high levels of expression of wgaA as in the expR+ wggR+ mucR+ and expR+ wggR+ mucR backgrounds, an even higher level of expression of wgeA (30% higher than levels of expression in a wggR mucR background), no significant change in the levels of expression of wgdA, and a slight increase (1.7 times) in the level of expression of wggR, implying that in the absence of MucR, WggR is not required for the positive effect of ExpR.
DISCUSSION
Previous studies have shown that the Sin quorum-sensing system in S. meliloti consists of SinI, the AHL synthase, and SinR, a LuxR-type regulator (33). The genes for both proteins are located at the sinR-sinI locus, in which sinI is located downstream of sinR, separated by a 157-bp intergenic region. This intergenic region contains a binding site for ExpR, another LuxR-type regulator, and a so-called lux box, a 16-bp sequence identified on the basis of its homology with the binding sites of LuxR-type regulators from Vibrio fischeri (9) and Agrobacterium tumefaciens (3, 52). lux boxes are typically located upstream of the promoters of its target genes and serve as recognition sites for the LuxR-type regulators upon activation by an AHL (12, 46). In this investigation, we confirm that the expression of sinI is positively regulated by expR in the presence of Sin AHLs and sinR, as was previously reported, and that ExpR does not bind to the lux box but rather binds to a 24-bp binding site located 65 bp upstream of the lux box and 100 bp upstream of sinI (3). A DNA binding site for SinR has not yet been identified, but it is possible that the lux box serves as the binding site for SinR in regulating sinI expression. We have identified the sinI transcription start and show that the −35 region of the sinI promoter is covered by the lux box, consistent with the idea that the lux box is important for the initiation of sinI transcription. However, the exact mode of interaction between the lux box, SinR, and ExpR in the activation of sinI expression remains to be elucidated.
We also show that the activation of sinI is absolutely dependent on sinR, since the expression of sinI was not detectable in the sinR mutant strains independent of the presence or absence of expR or Sin AHLs. Interestingly, sinR promoted sinI expression to a moderate constitutive level even in the absence of expR or sinI. This finding is intriguing because it indicates that either SinR can be activated by another S. meliloti quorum-sensing system or SinR does not require AHLs for this moderate induction of sinI transcription. Perhaps this aspect of the Sin system serves to guarantee a basic Sin AHL-independent expression of sinI at low cell densities that is required for a prompt response to quorum-sensing induction. We also note that expR and Sin AHLs, together with sinR, mediate a high level of expression of sinI. Thus, while the Sin system is not dependent on expR, it is probable that the AHL-activated ExpR serves to ensure the appropriate Sin quorum-sensing response to increasing cell density and that in the absence of ExpR, the Sin system is restricted to a weaker and slower quorum-sensing response.
The Sin system has been shown to be involved in the regulation of galactoglucan production. The disruption of the autoinducer synthase gene sinI abolished galactoglucan production, resulting in a nonmucoid phenotype on phosphate-rich medium (32). Furthermore, the levels of expression of several of the galactoglucan biosynthesis genes, including wgcA, wgeB (expE2), and wgdA, were dramatically reduced during log-phase growth in low-phosphate medium upon the disruption of sinI but were restored by the addition of AHLs from extracts from either the wild-type strain or one of the previously identified Sin-specified AHLs, synthetic palmitoleyl HL (C16:1-HL) (32, 33). In addition to the Sin system, ExpR is also involved in the regulation of galactoglucan production. Several studies have shown that the Sin effect on galactoglucan production requires a functional ExpR (32, 38). However, the details of the regulation of galactoglucan production by the Sin-ExpR system have remained an enigma. Although both genome and proteome approaches have shown the Sin-ExpR system to be involved in the regulation of a multitude of other genes throughout the genome (17, 23), the question of whether ExpR is acting directly or indirectly on its target genes has been unanswered and was one of the specific questions addressed in this investigation. As mentioned above, the single ExpR binding site upstream of sinI was described previously (3). However, while the effect of an active ExpR results in an increase in the level of expression of sinI, such an effect alone is insufficient to explain the dramatic increase in the level of galactoglucan production and the genome-wide changes in gene expression. We postulated that either ExpR binds directly to the promoters of its target genes or some other regulatory intermediate controlled by the Sin-ExpR system is binding to sites within the promoters. The discovery of additional AHL-dependent ExpR binding sites in the promoters of the galactoglucan biosynthesis operons in this investigation confirms our assumptions and allows a better understanding as to precisely how the Sin-ExpR system regulates galactoglucan production. Furthermore, a characterization of the ExpR binding site upstream of sinI, together with a comparison of this site to binding sites in the wgaA and wgeA promoters, allows a homology-based search for other ExpR binding sites that show Sin-ExpR-dependent regulation, as revealed by previous studies involving transcription profiles (23). Accordingly, the detection of ExpR binding sites in the promoter regions of two additional genes, exoI and exsH, demonstrates the success of using this approach. exoI codes for a predicted periplasmic protein of unknown function that is not required for succinoglycan production (5, 20), and exsH codes for an endo-1,3-1,4-β-glycanase which depolymerizes high-molecular-weight succinoglycan (49) to produce symbiotically active low-molecular-weight succinoglycan (48). Both were previously shown to be upregulated in response to Sin-ExpR regulation (19, 23).
The binding of His6-ExpR to the promoter regions of sinI, wgaA, wgeA, exoI, and exsH is specific, since under the same conditions, His6-ExpR did not bind to the wgdA, wggR, or wgcA promoter regions, nor did it bind to several other promoters that contained some homology to the ExpR consensus binding sequence. The low level of transcription activity measured from the wgdA, wggR, and putative wgcA promoters appeared to support this conclusion. Interestingly, a comparison of the ExpR binding sites revealed a consensus sequence unlike the lux box observed in V. fischeri (45, 46) or the tra box in A. tumefaciens (15, 30, 52). The ExpR binding site upstream of sinI contains a low level of similarity with the sites in the wgaA and wgeA promoters and an even lower level of similarity with those in the exoI and exsH promoters, suggesting that ExpR operates quite differently from the other LuxR-type regulators. This observation is also consistent with the solubility and stability of ExpR in the absence of AHLs, in contrast to other LuxR-type regulators such as TraR and LuxR (40, 47, 52).
We have also noted that ExpR is capable of various levels of binding to its target sites in the absence of AHLs in the gel shift experiments, particularly for the exsH and exoI promoter regions. Although the possibility that this is due to suboptimal conditions for the gel shift assays cannot be ruled out, another possibility is that this phenomenon occurs in vivo and plays a role in the regulation of the expression of these promoters. It appears that the interaction between the AHL and ExpR not only enhances the strength of binding of ExpR to the DNA site, as was observed previously (3), but also provides the necessary configuration of an ExpR-DNA complex to allow the RNA polymerase to bind, since the positive regulation of these promoters by ExpR occurs only in the presence of AHLs. However, ExpR binding to a subset of its target sites in the absence of AHLs may negatively regulate the transcription of the downstream gene, serving as another level of control of transcription at low cell densities. In support of this concept, succinoglycan production was recently shown to be reduced in a sinI expR+ strain but not in any of the sinI+ expR+, sinI+ expR, or sinI expR strains, consistent with a positive regulation by Sin-ExpR and a negative regulatory effect by ExpR in the absence of the Sin AHLs (19).
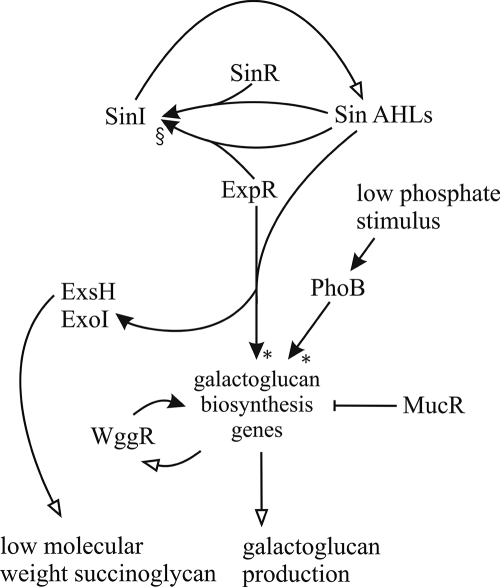
Knowledge of the regulation of galactoglucan production in S. meliloti was recently updated (1). The promoters of the wga, wgd, and wge operons were characterized with respect to transcription starts and the effect of three regulators, PhoB, WggR, and MucR, on the transcription of these promoters during growth in media containing both low and high phosphate concentrations (1). A model illustrating the complex regulation of galactoglucan production by these transcription regulators together with their DNA binding sites within the promoter regions and the distal and proximal transcription starts was presented. The Sin-ExpR quorum-sensing effect presented in this study can be added as yet another level of regulation of galactoglucan production, as shown in Fig. 6. Interestingly, the location of the ExpR binding sites within the wgaA and wgeA promoters almost immediately downstream of the proposed PhoB and WggR binding sites and possibly overlapping the MucR binding sites suggests some sort of cooperation and/or competition among the four regulators. Activation by ExpR is almost completely dependent on the presence of WggR when MucR is present, but in the absence of MucR, WggR is no longer required for the activation by ExpR. This WggR-dependent effect is also seen with PhoB-dependent induction under conditions of phosphate starvation (1) and suggests that WggR functions as a general mediator of galactoglucan biosynthesis gene expression for at least two regulators under different conditions. The close proximity of the PhoB, WggR, MucR, and ExpR binding sites suggests that there exists a complex regulation of the galactoglucan biosynthesis genes involving protein-protein interactions and, possibly, some competition for the DNA target sites. Furthermore, the ExpR binding sites overlap the distal transcription starts of wgaA and wgeA, suggesting that ExpR regulates both wgaA and wgeA by inhibiting the distal promoters and activating the proximal promoters, although this has not been confirmed.
FIG. 6.
Model of regulation of galactoglucan production in S. meliloti. Empty arrows indicate synthesis. Filled arrows indicate regulation, with “↑” and “⟂︁” indicating positive and negative regulation, respectively. Dependency in regulation is indicated beside the filled arrows: “§” indicates a dependency on SinR, where the positive regulation of sinI by ExpR and Sin AHLs is dependent on the presence of SinR, and “*” indicates a dependency on WggR, where the positive regulation of the galactoglucan biosynthesis genes by ExpR and Sin AHLs and by PhoB is dependent on the presence of WggR (1).
No binding between the wggR promoter region and ExpR was observed in the gel shift assays despite the presence of a low level of activation of expression from the wggR promoter due to the presence of expR. One possibility is that our gel shift assay conditions are not optimal enough to detect a weak interaction between the wggR promoter and ExpR, although we consider this to be unlikely, since, as mentioned above, no homology to the known ExpR binding sites upstream of wggR was detected. Perhaps more likely is that the low level of activation of wggR expression by ExpR is indirect, i.e., through another transcription regulator that is yet to be discovered. Yet another possibility is that the binding strength of ExpR is dependent on variations in the structure of the AHL molecule, as was demonstrated previously for the interaction between ExpR and its binding site upstream of sinI (3). That study found that ExpR was activated by a range of AHLs and that, depending on the structure of the acyl chain, the strength of binding between ExpR and DNA varied. The most critical attribute was the number of carbons in the acyl chain, where more than 12 carbons resulted in a dramatic increase in ExpR activation. In this study, we have used one of the functional Sin AHLs, oxo-C14-HL, which was previously shown to cause a maximal induction of sinI expression in S. meliloti (29). It remains to be seen how the other AHLs affect the strength of binding between ExpR and its target promoter regions and the resulting transcription activity from these promoters.
As noted above, galactoglucan can be produced either upon a “low-phosphate” stimulation of the galactoglucan biosynthesis genes or through the Sin-ExpR quorum-sensing system. However, only quorum-sensing stimulation results in low-molecular-weight galactoglucan that is important for symbiosis (34, 38, 43, 51). Under phosphate-limiting conditions, the wga, wgd, and wge operons were all stimulated at comparable levels through positive regulation by WggR and PhoB (1). Under quorum-sensing regulation, however, only those of wgaA and wgeA were dramatically upregulated in the presence of both ExpR and WggR. One possibility is that this strong upregulation of one or more of the products from the wga and/or wge operon is responsible for the production of low-molecular-weight galactoglucan. Alternatively, ExpR may directly regulate the transcription of a gene encoding a hydrolase that results in the generation of low-molecular-weight galactoglucan. The detection of an ExpR binding site upstream of exsH, a succinoglycan glucanase, leading to the generation of low-molecular-weight succinoglycan (19), lends support to this hypothesis. As yet another alternative, ExpR or one or more of the products from the open reading frames within the S. meliloti genome that are predicted to encode LuxR homologs (SMc00658, SMc00877, SMc00878, and SMc04032) are responsible for the production of symbiotically active galactoglucan by the regulation of other genes via DNA binding sites that remain to be identified. Further investigations into the role of quorum sensing in the regulation of various cell processes in S. meliloti, including the production of exopolysaccharides, will allow a greater understanding of the complex formation of symbiosis.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft in the framework of Collaborative Research Center SFB613 and by the Freiburg Initiative for Systems Biology, supported by the Bundesministerium für Bildung und Forschung.
We thank Juan E. González for providing the sinI mutant. We thank Patrik Plattner for supplying oxo-C14-HL.
Footnotes
Published ahead of print on 30 May 2008.
REFERENCES
- 1.Bahlawane, C., B. Baumgarth, J. Serrania, S. Rüberg, and A. Becker. 2008. Fine-tuning of galactoglucan biosynthesis in Sinorhizobium meliloti by differential WggR (ExpG)-, PhoB-, and MucR-dependent regulation of two promoters. J. Bacteriol. 1903456-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartels, F. W., B. Baumgarth, D. Anselmetti, R. Ros, and A. Becker. 2003. Specific binding of the regulatory protein ExpG to promoter regions of the galactoglucan biosynthesis gene cluster of Sinorhizobium meliloti—a combined molecular biology and force spectroscopy investigation. J. Struct. Biol. 143145-152. [DOI] [PubMed] [Google Scholar]
- 3.Bartels, F. W., M. McIntosh, A. Fuhrmann, C. Metzendorf, P. Plattner, N. Sewald, D. Anselmetti, R. Ros, and A. Becker. 2007. Effector-stimulated single molecule protein-DNA interactions of a quorum sensing system in Sinorhizobium meliloti. Biophys. J. 924391-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgarth, B., F. W. Bartels, D. Anselmetti, A. A. Becker, and R. Ros. 2005. Detailed studies of the binding mechanism of the Sinorhizobium meliloti transcriptional activator ExpG to DNA. Microbiology 151259-268. [DOI] [PubMed] [Google Scholar]
- 5.Becker, A., A. Kleickmann, H. Kuster, M. Keller, W. Arnold, and A. Pühler. 1993. Analysis of the Rhizobium meliloti genes exoU, exoV, exoW, exoT, and exoI involved in exopolysaccharide biosynthesis and nodule invasion: exoU and exoW probably encode glucosyltransferases. Mol. Plant-Microbe Interact. 6735-744. [DOI] [PubMed] [Google Scholar]
- 6.Becker, A., S. Rüberg, H. Küster, A. A. Roxlau, M. Keller, T. Ivashina, H. P. Cheng, G. G. Walker, and A. Pühler. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriol. 1791375-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84188-198. [DOI] [PubMed] [Google Scholar]
- 8.Bertram-Drogatz, P. A., I. Quester, A. Becker, and A. Pühler. 1998. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 257433-441. [DOI] [PubMed] [Google Scholar]
- 9.Callahan, S. M., and P. V. Dunlap. 2000. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 1822811-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casse, F., C. Boucher, J. S. Julliot, M. Michel, and J. Denarie. 1979. Identification and characterization of large plasmids in Rhizobium meliloti using agarose-gel electrophoresis. J. Gen. Microbiol. 113229-242. [Google Scholar]
- 11.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 1805183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol. Microbiol. 311197-1204. [DOI] [PubMed] [Google Scholar]
- 13.Flanco, A. G., M. Sola, F. X. Gomis-Rüth, and M. Coll. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10701-713. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua, C. M., R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35439-468. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 1762796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorhölter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293668-672. [DOI] [PubMed] [Google Scholar]
- 17.Gao, M., H. Chen, A. Eberhard, M. R. Gronquist, J. B. Robinson, B. G. Rolfe, and W. D. Bauer. 2005. sinI- and expR-dependent quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 1877931-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glazebrook, J., and G. C. Walker. 1989. A novel exopolysaccharide can function in place of the Calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56661-672. [DOI] [PubMed] [Google Scholar]
- 19.Glenn, S. A., N. Gurich, M. A. Feeney, and J. E. González. 2007. The ExpR/Sin quorum-sensing system controls succinoglycan production in Sinorhizobium meliloti. J. Bacteriol. 1897077-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glucksmann, M. A., T. L. Reuber, and G. C. Walker. 1993. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J. Bacteriol. 1757045-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González, J. E., B. L. Reuhs, and G. C. Walker. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. USA 938636-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 874645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoang, H., A. Becker, and J. E. González. 2004. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 1865460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8528-535. [PubMed] [Google Scholar]
- 25.Hübner, P., J. C. Willison, P. M. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 1732993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller, M., A. Roxlau, W. M. Weng, M. Schmidt, J. Quandt, N. Karsten, D. Jording, W. Arnold, and A. Pühler. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant-Microbe Interact. 8267-277. [DOI] [PubMed] [Google Scholar]
- 27.Leigh, J. A., and G. C. Walker. 1994. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1063-67. [DOI] [PubMed] [Google Scholar]
- 28.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 826231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llamas, I., N. Keshavan, and J. E. González. 2004. Use of Sinorhizobium meliloti as an indicator for specific detection of long-chain N-acyl homoserine lactones. Appl. Environ. Microbiol. 703715-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, Z. Q., and S. K. Farrand. 1999. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc. Natl. Acad. Sci. USA 969009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marketon, M. M., and J. E. González. 2002. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J. Bacteriol. 1843466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marketon, M. M., S. A. Glenn, A. Eberhard, and J. E. González. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 185325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marketon, M. M., M. R. Groquist, A. Eberhard, and J. E. González. 2002. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 1845686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendrygal, K. E., and J. E. González. 2000. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227197-203. [DOI] [PubMed] [Google Scholar]
- 37.Pellock, B. J., H.-P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 1824310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellock, B. J., M. Teplitski, R. P. Boinay, W. D. Bauer, and G. C. Walker. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 1845067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pobigaylo, N., D. Wetter, S. Szymczak, U. Schiller, S. Kurtz, F. Meyer, T. W. Nattkemper, and A. Becker. 2006. Construction of a large signature-tagged mini-Tn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti. Appl. Environ. Microbiol. 724329-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin, Y., Z. Luo, A. J. Smyth, P. Gao, S. B. von Bodman, and S. K. Farrand. 2000. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 195212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reuhs, B. L., R. W. Carlson, and J. S. Kim. 1993. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J. Bacteriol. 1753570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes, F., M. D. Roldan, W. Klipp, F. Castillo, and C. Moreno-Vivian. 1996. Isolation of periplasmic nitrate reductase genes from Rhodobacter sphaeroides DSM 158: structural and functional differences among prokaryotic nitrate reductases. Mol. Microbiol. 191307-1318. [DOI] [PubMed] [Google Scholar]
- 43.Rüberg, S., A. Pühler, and A. Becker. 1999. Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 145603-611. [DOI] [PubMed] [Google Scholar]
- 44.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1784-791. [Google Scholar]
- 45.Stevens, A. M., and E. P. Greenberg. 1997. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J. Bacteriol. 179557-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevens, A. M., K. M. Dolan, and E. P. Greenberg. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 9112619-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urbanowski, M. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urzainqui, A., and G. C. Walker. 1992. Exogenous suppression of the symbiotic deficiencies of Rhizobium meliloti exo mutants. J. Bacteriol. 1743403-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.York, G. M., and G. C. Walker. 1997. The Rhizobium meliloti exoK gene and prsD/prsE/exsH genes are components of independent degradative pathways which contribute to production of low-molecular-weight succinoglycan. Mol. Microbiol. 25117-134. [DOI] [PubMed] [Google Scholar]
- 50.Yuan, Z. C., R. Zaheer, R. Morton, and T. M. Finan. 2006. Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res. 342686-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhan, H. J., C. C. Lee, and J. A. Leigh. 1991. Induction of the second exopolysaccharide (EPSb) in Rhizobium meliloti SU47 by low phosphate concentrations. J. Bacteriol. 1737391-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 964832-48378. [DOI] [PMC free article] [PubMed] [Google Scholar]