Abstract
The marine bacterium Pseudoalteromonas tunicata produces an antibacterial and autolytic protein, AlpP, which causes death of a subpopulation of cells during biofilm formation and mediates differentiation, dispersal, and phenotypic variation among dispersal cells. The AlpP homologue (LodA) in the marine bacterium Marinomonas mediterranea was recently identified as a lysine oxidase which mediates cell death through the production of hydrogen peroxide. Here we show that AlpP in P. tunicata also acts as a lysine oxidase and that the hydrogen peroxide generated is responsible for cell death within microcolonies during biofilm development in both M. mediterranea and P. tunicata. LodA-mediated biofilm cell death is shown to be linked to the generation of phenotypic variation in growth and biofilm formation among M. mediterranea biofilm dispersal cells. Moreover, AlpP homologues also occur in several other gram-negative bacteria from diverse environments. Our results show that subpopulations of cells in microcolonies also die during biofilm formation in two of these organisms, Chromobacterium violaceum and Caulobacter crescentus. In all organisms, hydrogen peroxide was implicated in biofilm cell death, because it could be detected at the same time as the killing occurred, and the addition of catalase significantly reduced biofilm killing. In C. violaceum the AlpP-homologue was clearly linked to biofilm cell death events since an isogenic mutant (CVMUR1) does not undergo biofilm cell death. We propose that biofilm killing through hydrogen peroxide can be linked to AlpP homologue activity and plays an important role in dispersal and colonization across a range of gram-negative bacteria.
Biofilm formation in microorganisms generally involves differentiation processes leading to the formation of multicellular three-dimensional structures. Most biofilms consist of microcolonies encased by an organic polymer matrix. Previously, it has been shown that subpopulations of cells die during the normal course of biofilm development. This phenomenon has been observed for several organisms, including monospecies biofilms of Pseudomonas aeruginosa (52), Serratia marcescens (K. W. Lam, S. A. Rice, and S. Kjelleberg, unpublished data), Caulobacter crescentus (13), and Pseudoalteromonas tunicata (34), as well as mixed-species communities, including dental (4, 27) and river (30a) biofilm communities. Biofilm killing in P. aeruginosa has been linked to the activity of a prophage (52), and killing in P. tunicata is mediated by a large, autolytic protein (AlpP) (34). In both organisms, cell death is localized to the center of microcolonies and is controlled by specific regulatory determinants, such as RpoN (52) and quorum sensing in P. aeruginosa (J. S. Webb and S. Kjelleberg, unpublished data) and a ToxR-like regulator, WmpR, in P. tunicata (11). Mutants deficient in the production of AlpP show no cell death despite the formation of similar biofilm architecture (10, 11, 34). Because it is not necessary for survival per se, it may be suggested that cell death events represent an evolved capacity of importance to the biofilm development in the organism.
AlpP-mediated biofilm cell death in P. tunicata has been linked to the generation of a metabolically active and phenotypically diverse dispersal population (35). A major dispersal event occurs in the wild-type strain after cell death events are observed within the biofilm. It has been shown, using fluorescent staining for metabolic activity, that the dispersal population of the P. tunicata wild type had higher activity than the dispersal population of the ΔAlpP mutant, which does not show cell death (35). Moreover, P. tunicata wild-type dispersal cells displayed a larger variation in motility, growth, and biofilm formation than the ΔAlpP mutant. A metabolically active dispersal population with high variation in traits important to the spread and colonization ability of the organism is hypothesized to be advantageous under changing environmental conditions.
Since the first report of the antibacterial and autolytic protein in P. tunicata (28), a similar protein (LodA, previously marinocine) has been reported to occur in the melanogenic, marine bacterium Marinomonas mediterranea (32). Both proteins show activity against both gram-negative and gram-positive bacteria from a diverse range of environments, as well as displaying autotoxic activity. The antibacterial activity of LodA has recently been shown to be due to the generation of hydrogen peroxide from l-lysine. LodA (EC 1.4.3.20) catalyzes the following reaction: l-lysine + O2 + H2O → 2-aminoadipate 6-semialdehyde + NH3 + H2O2 (17).
In the present study we show that the P. tunicata AlpP produces hydrogen peroxide from l-lysine, and evidence is provided that hydrogen peroxide is responsible for cell death in biofilms. Furthermore, it is demonstrated that the AlpP-homologue, LodA, in M. mediterranea has a similar ecological function to AlpP during biofilm development. LodA production leads to cell death of a subpopulation of cells within microcolonies of M. mediterranea biofilms, which is linked to the generation of a phenotypically diverse dispersal population. Moreover, it is demonstrated that the AlpP-homologues in Chromobacterium violaceum (accession number NP_902938) and in part in C. crescentus (accession number NP_419374) are implicated in similar cell death events during biofilm formation. Our findings suggest that AlpP-mediated autotoxic events may play an important role in biofilm development and differentiation in a range of gram-negative bacterial groups.
MATERIALS AND METHODS
Strains and culture conditions.
All of the bacterial strains and the respective culture media used are shown in Table 1. P. tunicata and M. mediterranea were grown at 25°C and C. violaceum and C. crescentus were grown at 30°C.
TABLE 1.
Bacterial strains and culture media
| Strain | Culture mediuma | Minimal biofilm medium | Source or reference |
|---|---|---|---|
| P. tunicata | VNSS (36) | 3M supplemented with 0.01% trehalose (43) | 26 |
| P. tunicata ΔAlpP mutant Smr Kmr | VNSS + Sm at 100 μg ml−1 + Km at 50 μg ml−1 | 3M supplemented with 0.01% trehalose (43) | 34 |
| M. mediterranea MMB-1 | Marine medium 2216 (Difco) | Marine minimal medium MN (25) | 48 |
| M. mediterranea SB1 lodA mutant Kmr | Marine medium 2216 + Km at 50 μg ml−1 (Difco) | Marine minimal medium MN (25) | 31 |
| C. violaceum | Luria-Bertani broth (Difco) | M9 (39) | UNSW culture collection (accession no. 040100) |
| C. violaceum CVMUR1 Kmr ΔNP_902938 | Luria-Bertani broth + Km at 40 μg ml−1 | M9 (39) | This study |
| C. crescentus CB15 | PYE: peptone, 2 g liter−1; yeast extract, 1 g liter−1; MgSO4·7H2O, 0.2 g liter−1 | M2 supplemented with 0.2% xylose (12) | ACMa (accession no. ACM 5171) |
| C. crescentus CAUMUR1 Smr ΔNP_419374 | PYE + Sm at 5 μg ml−1 | M2 supplemented with 0.2% xylose (12) | This study |
| E. coli DH5α | Luria-Bertani broth | 20 |
Sm, streptomycin; Km, kanamycin.
bACM, Australian Collection of Microorganisms.
P. tunicata AlpP purification.
AlpP was purified by using a method adapted from James et al. (28). Briefly, P. tunicata was grown in 3 M for 48 h with shaking at room temperature. Cells were then harvested by centrifugation at 15,000 × g for 15 min at 22°C. The cell pellet was resuspended in fresh medium (0.5 ml/2-g pellet) and incubated for a further 24 h under static conditions. After centrifugation for 1.5 h at 23,700 × g and 4°C, the supernatant was filtered through a 0.22-μm-pore-size membrane. The filtrate was then dialyzed (12,000-kDa cutoff; Sigma, Castle Hill, Australia) overnight against 2 liters of 20 mM Tris (pH 7.4). A strong anionic ion-exchange matrix (High Q strong anion-exchanger; Bio-Rad, Hercules, CA) was used to purify AlpP from the P. tunicata supernatant. The protein was eluted between 250 and 350 mM NaCl in 20 mM Tris-HCl (pH 7.4). The eluate was further purified by ultrafiltration with a 100-kDa molecular cutoff filter (YM100; Diaflo; Amicon, Lexington, MA), and the retentate was saved. The purity of the final sample was checked by running an 8% sodium dodecyl sulfate polyacrylamide protein gel stained with Coomassie blue, yielding only the two bands (60 and 80 kDa) previously identified as AlpP.
The total protein concentration was determined by using the BCA assay (Sigma) according to the manufacturer's instructions.
M. mediterranea LodA purification.
LodA purification from M. mediterranea cultures was performed as previously described (32). Briefly, M. mediterranea was inoculated at an optical density at 600 nm of 0.05 in MN media (25). After 48 h of culture, the supernatant was separated by centrifugation, and 2 volumes of ethanol were added to precipitate LodA by incubation overnight at 4°C. The compound was then recovered by centrifugation at 19,000 × g at 4°C for 20 min. The pellet was allowed to air dry and suspended in 0.1 M sodium phosphate buffer (pH 7). Insoluble material was removed by centrifugation (13,000 × g at 4°C for 20 min). The activity of the extract obtained was calculated by measuring the halos formed in the antibiogram inhibition test against Escherichia coli DH5α (32). Other proteins present in the sample were shown to have no antibacterial activity (data not shown).
Lysine oxidase activity fluorometric measurements.
The Amplex Red hydrogen peroxide/peroxidase assay kit (A22188; Molecular Probes, Eugene, OR) assay was used to test the production of hydrogen peroxide from purified AlpP. In the presence of horseradish peroxidase (HRP), the Amplex red reagent reacts with hydrogen peroxide in a 1:1 stoichiometry to produce the red fluorescent oxidation product resuforin. Resuforin fluorescence was measured at excitation of 550 nm and an emission of 590 nm. Hydrogen peroxide (Univar, Kirkland, WA) concentrations ranging from 2 to 12 μM were used as standard. l-Lysine (50 mM; Sigma) was used as a substrate for AlpP. The ability of AlpP to generate hydrogen peroxide from l-lysine was tested with AlpP concentrations ranging from 0.4 to 4 ng. In a negative control catalase (0.1 mg ml−1; Sigma, St. Louis, MO) was added to the reaction of AlpP and l-lysine.
Lysine detection in biofilm effluent.
Lysine was detected in biofilm effluent samples of P. tunicata and M. mediterranea at 72 and 144 h of biofilm development by using high-performance liquid chromatography (HPLC). Samples were freeze-dried at −50°C overnight. The freeze-dried samples were resuspended in 100 μl of 0.1 mM methionine sulfone in 0.1 M HCl, and 50 μl of each concentrated sample was used for lysine analysis. The sample was derivatized with phenylisothiocyanate according to the protocol from Waters (Rydalmere, Australia). Lysine HPLC analysis of the sample was carried out on HP1100 HPLC unit (Hewlett-Packard, Australia) with an Agilent LC Chemstation (Agilent Technologies, Inc.). A 1/20 volume of the sample was separated in the free amino acids column by following the binary mobile-phase gradient table from Waters (Rydalmere) at 46°C. The derivatized lysine was detected at 254 nm and quantified by comparison to standard solutions.
Determination of the substrate affinity (Km) of AlpP for lysine.
Purified AlpP was used to perform fluorimetric measurements by using an Amplex Red hydrogen peroxide/peroxidase assay kit (A22188; Molecular Probes) to detect hydrogen peroxide production with d- or l-lysine as substrates for the reaction. The assays were carried out in a fluorimetric enzyme-linked immunosorbent assay reader, using an excitation filter of 550 nm and an emission filter of 590 nm. All assays were performed in duplicate in 96-well plates. Background fluorescence in the absence of lysine was subtracted to all data to minimize the effect of other compounds that might be present in the extract. The Michaelis constant (Km) of each reaction was calculated from a Lineweaver-Burk plot.
Flow cell experiments.
All strains were grown in continuous-culture flow cells (channel dimensions 1 by 4 by 40 mm) as previously described (41). Channels were inoculated with 0.5 ml of early-stationary-phase cultures containing approximately 109 cells ml−1 and incubated without flow for 1 h at 25°C. Flow was then started with a mean flow velocity in the flow cells of 0.2 mm s−1, corresponding to laminar flow with a Reynolds number of 0.02. Biofilms were run at 25°C for P. tunicata and M. mediterranea and at 30°C for C. violaceum and C. crescentus.
Biofilm staining.
To investigate cell death during biofilm development, biofilms were stained by using a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes). The two stock solutions of the stain (SYTO 9 and propidium iodide [PI]) were diluted to 3 μl ml−1 in biofilm medium and injected into the flow channels. Live SYTO 9-stained cells and dead PI-stained cells were visualized with a confocal laser scanning microscope (CLSM; Olympus) using fluorescein isothiocyanate and tetramethyl rhodamine isocyanate optical filters, respectively.
In order to provide a quantitative analysis of cell death within the biofilms, all microcolonies (with or without dead cells in the center) were counted in at least 10 fields of view in three replicate flow cells. A percentage value of microcolonies containing dead cells was then determined.
To visualize hydrogen peroxide production during biofilm development, biofilms were stained with Amplex Red. HRP (0.14 U ml−1) and Amplex Red (25 μM) were diluted in the biofilm media and injected into the flow cell. Immediately after staining, biofilms were visualized with an epifluorescence microscope (Leica Microsystem, Wetzlar, Germany). Amplex Red fluorescence was observed at an excitation of 515 to 560 nm and an emission of 590 nm. Bright-field images were taken from the same field of view.
Removal of hydrogen peroxide from biofilms.
To remove hydrogen peroxide from biofilms, catalase (Sigma) was added to the biofilm media. All biofilms were allowed to establish for 24 h before catalase was added at a final concentration of 100 μM. This catalase concentration did not affect the planktonic growth rate of the strains (data not shown). After 3 days of incubation, the biofilm was stained with Amplex Red and observed with an epifluorescence microscope (see above).
Add back of LodA protein to SB1 mutant biofilms.
M. mediterranea LodA was prepared as described above, and approximately 60 μg was added back to each flow cell containing M. mediterranea SB1 mutant biofilms. The add-back experiment was performed as described previously for P. tunicata (34). Briefly, LodA was injected into the flow cells by using a syringe needle. Silicone tubing at either side of the flow cell was then blocked off by using tubing clamps. As a control, 0.1 M phosphate buffer (pH 7.0) was inoculated into separate flow cell channels. Biofilms were incubated at 25°C for 3 h before staining with the LIVE/DEAD kit and visualization with the CLSM.
Add-back of hydrogen peroxide to ΔAlpP mutant biofilms.
ΔAlpP mutant biofilms were grown for 72 h before 1 mM hydrogen peroxide was injected into the flow cells by using a syringe needle in a similar manner to that of the AlpP add-back experiment (34). Silicone tubing at either side of the flow cell was then blocked off by using tubing clamps. As a control, NSS buffer was inoculated into separate flow cell channels. Biofilms were incubated at 25°C for 1 h before staining with the LIVE/DEAD kit and visualization with the CLSM.
Phenotypic variation of M. mediterranea dispersal cells.
To investigate the hypothesis that cell lysis within microcolonies correlates with phenotypic variation in M. mediterranea, effluent was spread plated onto marine agar (Difco/Becton Dickinson) at three time points during biofilm formation: 24 h (before the onset of cell death), 72 h (shortly after the onset of cell death), and 144 h (when cell death was more extended throughout the biofilm). Twenty colonies derived from M. mediterranea wild-type and SB1 biofilms were randomly picked from marine agar and screened for growth and biofilm formation. Portions (15 μl) of overnight cultures of the 20 colonies were inoculated into 1.5 ml of fresh media in 24-well tissue culture plates. Plates were incubated at 25°C with agitation (130 rpm). The optical density at 600 nm was measured after 24 h as an indicator for growth ability. To measure biofilm-forming ability, the wells of the tissue culture plates were stained with crystal violet for 20 min. After the wells were washed twice, crystal violet was extracted in 95% ethanol, and the absorbance was read at 600 nm. The variation coefficient was calculated as the standard deviation of all measurements divided by the mean between all samples.
Creation of alpP/lodA homologue deletion strains in C. crescentus and C. violaceum.
Mutant strains with deletion of the genes homologous to M. mediterranea lodA gene in C. crescentus, locus NP_419374, and C. violaceum, locus NP_902938, were created via homologous recombination using the suicide vectors pNPTS138 (M. R. Alley, unpublished results) and pFSVKCV, respectively. The C. crescentus lodA deletion was created by a two-step recombination method based on sacB counterselection (15). Two fragments of approximately 500 bp upstream and downstream of the gene in C. crescentus were PCR amplified from genomic DNA using the primers CAUDIR1pst and CAUREV1mlu for the upstream fragment and CAUDIR2mlu and CAUREV2sal for the downstream fragment (Table 2). Both fragments were cloned into pNPTS138, placing between them a streptomycin resistance cassette taken from pBSL299 (1). The construct was mobilized into C. crescentus CB15 by electroporation, and strains with integration of the vector by single recombination were selected by growth on peptone-yeast extract with kanamycin at 25 μg/ml before counterselection with streptomycin at 5 μg/ml and sucrose at 3% to select for double-crossover recombinants. Sucrose- and streptomycin-resistant colonies were screened for kanamycin sensitivity, and gene insertion confirmed by PCR with the primers CAUDIR4-CAUREV5 (which gives a 670-bp fragment in wild-type and first recombination mutants maintaining locus NP_419374) and CAUDIR3-CAUREV5 (which gives a 2.3-kb fragment in WT and a 2.1-kb fragment in CAUMUR1 as a result of the substitution of locus NP_419374 with the smaller streptomycin resistance marker). Deletion of the C. violaceum alpP/lodA homologue was achieved by using a previously described method (31). Briefly, a 500-bp region of the gene containing the fourth and fifth conserved domains, out of a total of eight in the homologous proteins, was amplified by PCR using the primers CVDIR2nco and CVREV1 (Table 2). This PCR fragment was cloned into a suicide vector containing a kanamycin resistance cassette to generate pFSVKCV. The plasmid was then transferred to C. violaceum via conjugation with E. coli S17-1λpir. Recombinants were selected on nutrient agar with kanamycin at 40 μg/ml, and gene insertion was confirmed by PCR using the primers CVDIR1 and CVREV1 to amplify a 1-kb fragment from domain 3 to domain 7 present in the wild-type strain and absent in the mutated strain (CVMUR1) and the primers CVDIR1 and KmREV to amplify a 1.4-kb fragment only in the mutant strain from the third conserved domain of the truncated gene to the kanamycin resistance marker in the integrated plasmid (Table 2).
TABLE 2.
Primer design for the creation of C. violaceum CVMUR1 and C. crescentus CAUMUR1 strains
| Primer | Nucleotide sequence (5′-3′) |
|---|---|
| CVDIR2nco | GAACGACTCCATGGACACCC |
| CVREV1 | CCGACGCAGTTGTCCAGC |
| CVDIR1 | CACCTGGCCAACAAGAAGC |
| KmREV | GTAACATCATTGGCAACG |
| CAUDIR1pst | GATCGAGTCTGCAGAGTGGCGCAC |
| CAUREV1mlu | CAGGATGTCGACGCGTAGCTTGG |
| CAUDIR2mlu | CGGTGCGATGACGCGTTGATCG |
| CAUREV2sal | GCAGACCAGTCGACCGCTCTCG |
| CAUDIR4 | GTCCAGGCCAGCCAGCC |
| CAUREV5 | AGGATCAGACGAATGTCTGG |
| CAUDIR3 | GCGTGCAGATGGCCAAGC |
RESULTS
AlpP in P. tunicata produces hydrogen peroxide via lysine oxidase activity.
To investigate whether AlpP from P. tunicata produces hydrogen peroxide from l-lysine, the fluorometric Amplex Red assay was used. High fluorescence intensity (390,000 relative counts) was detected after 6 min when AlpP (0.13 mg ml−1) was incubated with the substrate l-lysine and Amplex Red (Fig. 1). Similar fluorescence intensity was measured in the positive control containing Amplex Red and hydrogen peroxide (12 μM). However, no fluorescence was detected in the negative control (no AlpP) or when catalase was added to the reaction of AlpP and l-lysine, thereby removing the hydrogen peroxide produced (Fig. 1). These data strongly suggest that AlpP has lysine oxidase activity.
FIG. 1.
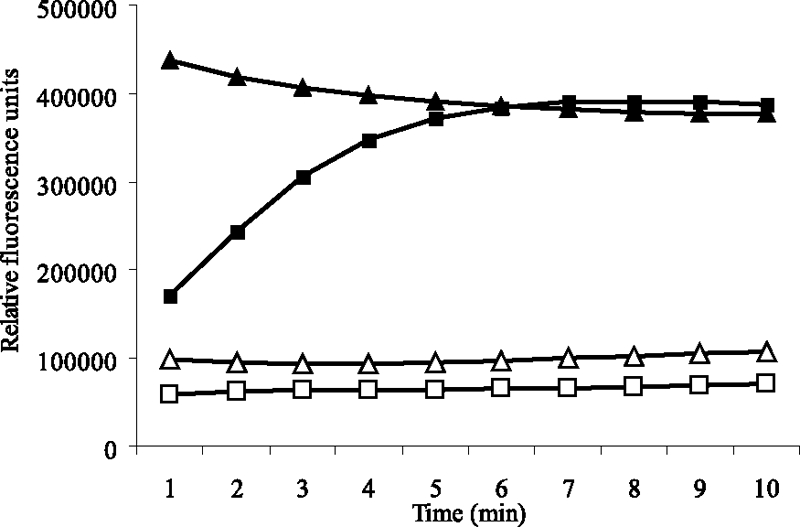
AlpP produces H2O2 from l-lysine. The Amplex Red reagent reacts with H2O2 in the presence of peroxidase (HRP) to produce the red fluorescent oxidation product resuforin. High fluorescence (390,000 counts) was detected in the presence of the H2O2 standard (12 μM) (▴). A similar fluorescence intensity was detected when AlpP (0.13 mg ml−1) was added to the substrate l-lysine (50 mM) (▪) in the presence of Amplex Red. However, no fluorescence was detected in the presence of catalase (0.1 mg ml−1), AlpP (0.13 mg ml−1), and l-lysine (50 mM) (□) and when no AlpP was added to the reaction (▵).
In addition, the substrate affinity of AlpP to lysine was studied by using the Amplex Red assay. AlpP showed a remarkable stereospecificity toward the l-isomer of lysine, showing a very high affinity for this substrate (Km = 24.91 ± 0.48 μM) compared to the d-isomer (Km = 2.08 mM). No lysine oxidase activity was also observed using other compounds (morpholinepropanesulfonic acid, trehalose, and tricine) at the concentration used in the media (data not shown).
Cell death occurs during biofilm development of M. mediterranea.
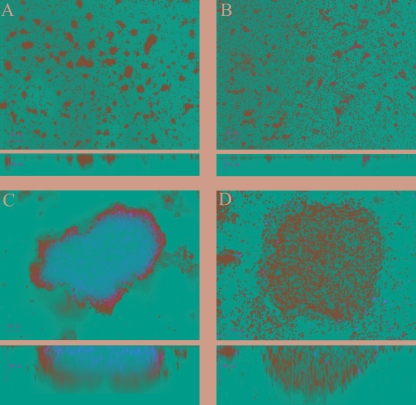
The ability of M. mediterranea to form biofilms was investigated. Particularly, it was explored whether the lysine oxidase, LodA, in M. mediterranea is implicated in cell death events during biofilm formation. Biofilms were allowed to form in continuous culture flow cells and stained with the BacLight LIVE/DEAD kit before visualization with a CLSM. Single cells attached to the substratum and small microcolonies were observed 24 h after inoculation (Fig. 2A). After 48 h, larger microcolonies were formed, consisting only of viable cells. However, at 3 days after inoculation, cell death started to occur within microcolonies, and subpopulations of dead cells were observed in 95% of the microcolonies (Fig. 2C and Table 3). Cell death extended throughout the biofilms, before the biofilm structure started to disrupt and detach, suggesting that, similar to P. tunicata, cell death plays a role during biofilm development and subsequent dispersal in M. mediterranea.
FIG. 2.
Cell death occurs during biofilm development of M. mediterranea wild-type but not the SB1 mutant. Biofilms were stained with the BacLight LIVE/DEAD viability kit. (A) Wild-type, 24 h; (B) SB1 mutant, 24 h; (C) wild-type, 72 h; (D) SB1 mutant, 72 h.
TABLE 3.
H2O2 can be detected in biofilms at the onset of cell death, and the addition of catalase prevents biofilm cell death
| Biofilma | Fluorescence type | Scoreb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| P. tunicata wild type | ΔAlpP mutant | M. mediterranea wild type | SB1 mutant | C. violaceum wild type | CVMUR1 | C. crescentus wild type | CAUMUR1 | ||
| In minimal medium | PI fluorescence (dead cells) | +++ | - | +++ | - | +++ | - | +++ | ++ |
| AR fluorescence (H2O2) | +++ | - | +++ | + | +++ | - | ++ | ++ | |
| In minimal medium plus catalase | PI fluorescence (dead cells) | + | - | - | - | - | - | + | - |
| AR fluorescence (H2O2) | - | - | + | + | - | - | - | - | |
Two sets of triplicate biofilms were inoculated for each strain, one set in minimal medium and the other set in minimal medium with the addition of catalase (100 μg ml−1) after 24 h. Biofilms were allowed to establish for 4 days before staining with the BacLight LIVE/DEAD viability kit to detect cell death and with the Amplex Red (AR) reagent to localize hydrogen peroxide, as indicated in column 2.
The fluorescence intensity was scored as follows: +++, high; ++, medium; +, low; -, none.
The SB1 mutant strain does not show cell death during biofilm development.
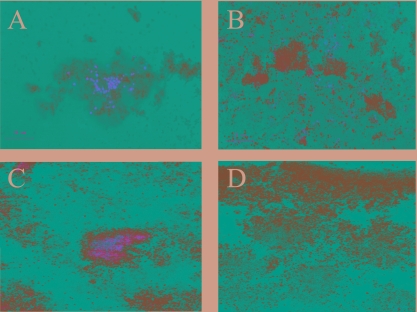
To assess whether biofilm cell death in M. mediterranea is mediated by LodA, biofilm development of a lodA mutant strain (SB1) (31) was investigated. The SB1 mutant also formed a biofilm with microcolony-based architecture. However, similar to the P. tunicata ΔAlpP mutant major cell death events did not occur during any stage of biofilm development (Fig. 2B and D and Table 3), and only a few individual dead cells were observed in the SB1 mutant biofilm. To further support the hypothesis that LodA causes cell death in M. mediterranea, purified LodA was added back to mature SB1 mutant biofilms. The add-back of purified LodA to mature SB1 mutant biofilms induced cell death (Fig. 3A) in 78% of the microcolonies, while the buffer control still showed only viable cells, suggesting that LodA mediates cell death in M. mediterranea biofilms (Fig. 3B).
FIG. 3.
Add-back of purified LodA to M. mediterranea SB1 mutant and add-back of hydrogen peroxide to ΔAlpP mutant biofilms can induce cell death. Biofilms were stained with the BacLight LIVE/DEAD kit. (A) Add-back of LodA to 72-h SB1 mutant biofilms; (B) 72-h SB1 mutant biofilm plus buffer control, (C) add-back of 1 mM H2O2 to 72-h ΔAlpP mutant biofilms; (D) 72-h ΔAlpP mutant biofilm plus buffer control.
Hydrogen peroxide can be detected in biofilms of P. tunicata and M. mediterranea.
Because of the finding that AlpP and LodA mediate cell death and produce hydrogen peroxide, we hypothesized that hydrogen peroxide can be detected in biofilms at the onset of killing. Amplex Red staining was used to visualize hydrogen peroxide in biofilms of P. tunicata and M. mediterranea. High red fluorescence was observed in wild-type biofilms at the time of killing (Table 3), indicating the presence of hydrogen peroxide in the biofilms. However, little or no fluorescence was detected in the ΔAlpP mutant of P. tunicata or the SB1 mutant of M. mediterranea, where no cell death occurs, supporting the hypothesis that hydrogen peroxide is involved in biofilm killing. To provide further evidence for this model, catalase was added to the biofilm media to remove hydrogen peroxide. Biofilm cell death was almost entirely prevented by the addition of catalase, and little or no hydrogen peroxide was detected within the biofilms after the catalase treatment (Table 3).
To further strengthen the notion that hydrogen peroxide is involved in biofilm killing in organisms containing AlpP homologues, ΔAlpP mutant biofilms were exposed to hydrogen peroxide to induce cell death. The add-back of hydrogen peroxide led to cell death within the center of the majority of microcolonies (78%) in a fashion similar to when the purified protein was added to the mutant biofilms (34) (Fig. 3C).
Lysine is produced in P. tunicata and M. mediterranea biofilms.
Because the biofilm medium used in the present study does not contain lysine, it was investigated whether endogenous lysine is produced by the cells in the biofilm to serve as a substrate for AlpP or LodA, respectively. Lysine was detected in the biofilm effluent of P. tunicata and M. mediterranea at 72 and 144 h of biofilm development by using HPLC analysis. The lysine concentration in the biofilm effluent at 72 h was 313.2 μM for P. tunicata and 165.5 μM for M. mediterranea. Similar lysine levels were measured at 144 h with 333.6 μM in P. tunicata and 126.4 μM in M. mediterranea biofilm effluent. Because these concentrations are at least 1 order of magnitude above the Km's of AlpP (see above) and LodA (17), we expect to have maximum catalytic activity in the biofilm.
LodA-mediated cell death in M. mediterranea is linked to phenotypic variation.
In P. tunicata, AlpP-mediated cell death has been linked to the generation of a metabolically active and phenotypically diverse dispersal population (35). Detached cells of the ΔAlpP mutant showed significantly lower variation in motility, growth, and biofilm formation. In the present study, we investigated whether LodA-mediated cell death in M. mediterranea is also implicated in the generation of phenotypic variation among biofilm dispersal cells. M. mediterranea wild-type and SB1 mutant (which does not show cell death during biofilm formation) dispersal cells were tested for variation in growth and biofilm formation at three different time points during biofilm development. The M. mediterranea wild type showed higher variation in growth ability at all time points investigated. Variation in growth among the 20 randomly picked colonies was 19% in the wild-type compared to only 5% in the SB1 mutant strain. Variation in biofilm formation was highest in the wild-type at the 144-h time point when cell death had occurred extensively throughout the biofilm. At this time point variation was 45% in the wild-type compared to only 25% in the mutant strain (Fig. 4). These results suggest that LodA-mediated biofilm cell death in M. mediterranea is linked to the generation of a phenotypically diverse dispersal population.
FIG. 4.
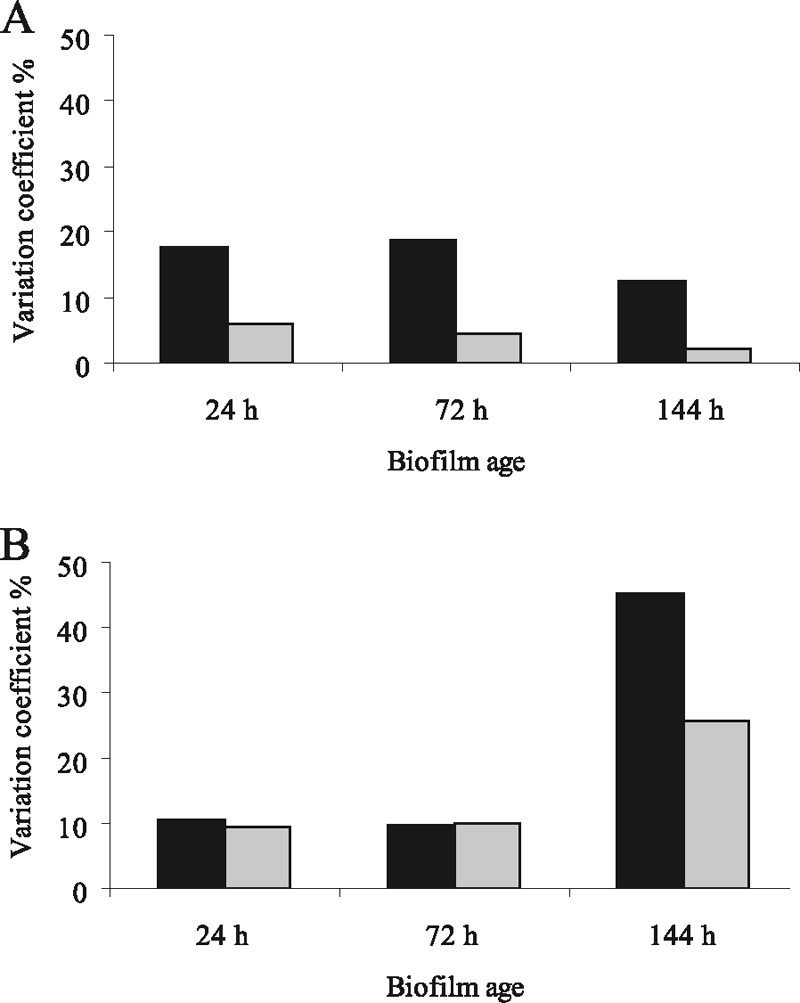
M. mediterranea wild-type dispersal cells show a higher variation than M. mediterranea SB1 mutant. Variation coefficient of M. mediterranea wild-type (▪) and SB1 mutant ( ) biofilm dispersal cells in growth (A) and biofilm formation (B). Variation coefficient (%) calculated for 20 colonies for time points 24, 72, and 144 h after biofilm inoculation.
) biofilm dispersal cells in growth (A) and biofilm formation (B). Variation coefficient (%) calculated for 20 colonies for time points 24, 72, and 144 h after biofilm inoculation.
Hydrogen peroxide mediates cell death in biofilms of C. violaceum and C. crescentus.
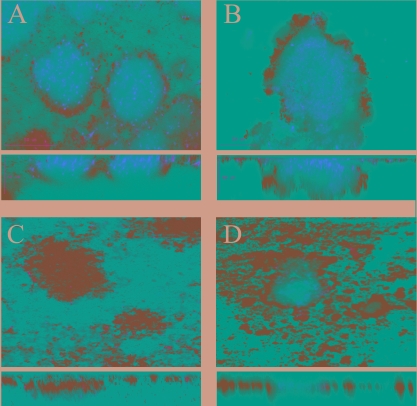
Because AlpP homologues were also identified in other gram-negative bacteria, including C. violaceum (32% identity) and C. crescentus (27% identity), we investigated whether biofilm cell death also occurs in these two organisms. C. violaceum and C. crescentus formed a microcolony-based biofilm, and cell death occurred within the center of mature microcolonies at 3 days postinoculation (Fig. 5A and B). Ninety-three and ninety-five percent of the microcolonies of C. violaceum and C. crescentus, respectively, showed dead cells within the microcolony center. Similar to the observation for P. tunicata and M. mediterranea, hydrogen peroxide was detected at the time of killing associated with C. violaceum and C. crescentus microcolonies using Amplex Red staining. Moreover, the addition of catalase removed hydrogen peroxide and also prevented cell death in biofilms of C. violaceum and C. crescentus, indicating that hydrogen peroxide is involved in the killing (Table 3).
FIG. 5.
Cell death also occurs in C. violaceum and C. crescentus biofilms. Biofilms were stained with the BacLight LIVE/DEAD kit at 4 days postinoculation. (A) C. violaceum wild-type; (B) C. crescentus wild-type; (C) C. violaceum CVMUR1; (D) C. crescentus CAUMUR1.
To investigate whether hydrogen peroxide in C. violaceum and C. crescentus biofilms is a result of the lysine oxidase activity of the AlpP-homologues, isogenic mutants (C. violaceum CVMUR1 and C. crescentus CAUMUR1) were created and tested for the occurrence of biofilm cell death and hydrogen peroxide. Both mutants formed a biofilm with a similar architecture than the wild types consisting of microcolonies. The CVMUR1 strain did not show cell death during biofilm development, and no hydrogen peroxide could be detected, indicating that its AlpP homologue is responsible for biofilm killing in the wild type. In comparing the wild-type and the C. crescentus CAUMUR1 strains, we noted that cell death occurred in many but not all microcolonies of the mutant strain (81%). Moreover, hydrogen peroxide was detected localized into the microcolonies in CAUMUR1 biofilms, suggesting that additional processes leading to hydrogen peroxide-mediated cell death occur in this organism.
DISCUSSION
Selective lysis of cell subpopulations appears to be a highly evolved and widespread mechanism among prokaryotes (3, 16), and several roles have been suggested for this process. It was shown that killing of siblings in Streptococcus pneumoniae leads to the release of a key virulence factor (Ply) and other cytoplasmic contents during colonization in humans (16, 19). Moreover, it was recently demonstrated that the suicide gene spxB mediates cell death via hydrogen peroxide and confers a survival advantage in colonization in S. pneumoniae (46). In B. subtilis, sporulating cells can trigger the lysis of sibling cells and feed on subsequently released nutrients, thereby delaying the energy-costly sporulation process (18). The death of cell subpopulations is also required during differentiation processes in Streptomyces antibioticus (14, 38) and during the formation of multicellular fruiting bodies of myxobacteria (53). In classically nondifferentiating bacteria cell death occurs during biofilm formation (4, 13, 27, 34, 52). The occurrence of cell death events of subpopulations of cells in different microorganisms and within diverse processes suggests a role for cell death for enhanced survival of the remaining cells as seen, for example, by the generation of phenotypically different biofilm dispersal variants.
AlpP and its homologues mediate cell death via the production of hydrogen peroxide.
The present study shows that AlpP can produce hydrogen peroxide from the oxidation of l-lysine (Fig. 1). Such a mode of action was first discovered for the AlpP homologue LodA produced by M. mediterranea (31). Interestingly, our results show that the substrate lysine is produced endogenously by the biofilm bacteria and consequently allows for AlpP/LodA activity. The fact that the lysine concentration detected in the biofilm effluent is significantly higher than the Km determined for AlpP (see results above) and LodA (17) strongly suggests that there is enough substrate for the enzyme to have maximum activity. A similar finding has recently been reported in the sea hare Aplysia californica, where a lysine oxidase and the amino acid are released from the cells at the same time (29). Interestingly, endogenous production of hydrogen peroxide was also shown in the human bacterial pathogen S. pneumoniae, where the gene responsible for hydrogen peroxide production induces an apoptosis-like death that confers a survival advantage in colonization (46).
In eukaryotes l-lysine oxidases were first isolated from the fungus Trichoderma sp. (30). Recently, an l-amino acid oxidase with lysine oxidase activity was detected in the ink of the sea hare A. californica (54). These enzymes have since become a focus of research because of their unique properties, which include cytotoxic, antitumor, antimetastatic, antiinvasive, antiviral, and antibacterial activities (5, 33). These characteristics are suggested to be due to a decrease in the essential amino acid l-lysine and the production of hydrogen peroxide. The antibacterial activity of lysine oxidases was first demonstrated using a rec mutant of Bacillus subtilis as a target strain. The rec mutant was more sensitive to the antibacterial effect of the lysine oxidase than the B. subtilis wild-type (30), suggesting that the damaging effect of hydrogen peroxide on DNA is responsible for the antibacterial activity, rather than the decrease of l-lysine. Further evidence for this resulted from the ability of catalase to protect the cells by removing hydrogen peroxide (30).
The AlpP homologue LodA also mediates cell death in M. mediterranea biofilms, leading to increased phenotypic variation among biofilm dispersal cells.
M. mediterranea strain MMB-1 is a melanogenic marine bacterium originally isolated from a water column sample from the Mediterranean Sea (48). Recently, however, new strains have been isolated from the microbial communities associated with the sea grass Posidonia oceanica in different Mediterranean areas (E. Marco-Noales, unpublished data; E. Espinosa et al., unpublished data), suggesting that this could be the microhabitat it occupies. The results of the present study demonstrated, for the first time, that M. mediterranea is able to form biofilms which may contribute to its colonization ability and establishment of M. mediterranea on the plant surface. Similarly to P. tunicata, subpopulations of cells die during its biofilm formation. Furthermore, it was demonstrated that cell death in M. mediterranea biofilms is mediated by its lysine oxidase LodA and that this process plays a role in the dispersal of the organism.
Similar to the P. tunicata biofilm life cycle, cell death seems to play a role in generating a phenotypically diverse dispersal population from M. mediterranea biofilms. The present study shows that the onset of LodA-mediated killing correlates with the generation of high variation among dispersal cells of M. mediterranea biofilms. However, because some variation also occurs before the onset of cell death in wild-type dispersal cells, it is possible that while LodA may be produced at low undetectable levels at early stages of biofilm formation, these concentrations are sufficient to induce variation before the onset of cell death. Variation in biofilm formation also increased with biofilm age among dispersal cells of the SB1 mutant; hence, other factors may play a role in inducing variation in M. mediterranea biofilms. Many organisms display variation in cells growing in biofilms, including P. aeruginosa (7, 9, 23, 51), Staphylococcus aureus (45, 47), Staphylococcus epidermidis (8, 21), Vibrio cholerae (2, 37), Listeria monocytogenes (42), and P. tunicata (35). Diverse mechanisms that may lead to an increased genetic and phenotypic variation in biofilm-forming bacteria have been investigated. These mechanisms include phase variation (9), adaptive mutation (6, 44), enhanced gene transfer through conjugation, transformation (22, 24, 40), phage induction (51), and self-induced lysis (35). The process of generating variation is hypothesized to be beneficial to the population as the chances of thriving under different environmental conditions are enhanced.
AlpP homologues are common proteins and may have similar roles during biofilm development.
Several AlpP homologues exist in a range of gram-negative organisms, including Marinomonas sp. strain MWYL1, M. mediterranea MMB-1, C. violaceum, Magnetococcus sp. strain MC-1, C. crescentus CB15, Shewanella woodyi ATCC 51908, Hahella chejuensis KCTC 2396, Roseovarius sp. strain HTCC2601, Burkholderia ambifaria IOP40-10, Herpetosiphon aurantiacus ATCC 23779, Marinomonas sp. strain MED121, Delftia acidovorans SPH-1, Rhodopseudomonas palustris CGA009, Synechococcus sp. strain WH 7805, Rhodopirellula baltica SH 1, Alphaproteobacterium strain BAL199, Ralstonia solanacearum UW551, Kordia algicida OT-1, Nitrobacter hamburgensis X14, Plesiocystis pacifica SIR-1, Saccharophagus degradans 2-40, Nitrobacter sp. strain Nb-311A, and Laccaria bicolor S238NH82, suggesting that it is a common protein among bacteria, where it may play a similar role to the one described here.
Apart from P. tunicata and M. mediterranea, a possible role for AlpP homologues in biofilm formation in two organisms—C. violaceum and C. crescentus—was investigated. To our knowledge, the present study is the first report describing biofilm development and architecture of C. violaceum. We find that C. violaceum also forms a differentiated, microcolony-based biofilm, and cell death occurs in the center of microcolonies during the normal course of biofilm development. The AlpP homologue was clearly linked to these cell death events because the isogenic mutant C. violaceum CVMUR1 did not undergo biofilm killing. Previously, biofilm development of C. crescentus has been investigated, and the formation of microcolonies was observed. Moreover, the occurrence of cell death within the center of microcolonies was described and suggested to be due to senescence (13). Interestingly, biofilm cell death was still observed in the C. crescentus CAUMUR1 mutant, although to a lesser extent than in the wild type. Although our results show that hydrogen peroxide causes cell death in C. crescentus biofilms (because it can be detected at the time of killing and catalase significantly reduces cell death), it remains to be elucidated what processes, apart from a possible lysine oxidation by the AlpP homologue, are involved in generating hydrogen peroxide. Because C. crescentus is an obligate aerobe bacterium and several oxidases have been identified in its genome, alternative ways for generating hydrogen peroxide during biofilm formation are feasible.
Hydrogen peroxide plays a role in biofilm cell death of P. tunicata. M. mediterranea, C. violaceum, and C. crescentus.
Hydrogen peroxide is a low-molecular-weight compound that acts on a wide range of molecules and is likely to facilitate cell death and dispersal even in complex microbial communities such as those on living marine surfaces. Hydrogen peroxide was detected in biofilms of P. tunicata, M. mediterranea, C. violaceum, and C. crescentus at the time when cell death occurs (Table 3). Although the exact mechanism of hydrogen peroxide-mediated cell death remains to be elucidated, cells in the center of microcolonies appear to be more susceptible because the add-back of hydrogen peroxide leads to cell death within the center of microcolonies. In the add-back experiment a molar concentration of hydrogen peroxide three times higher than the apparent lysine concentration was required to induce cell death in the majority of microcolonies in the ΔAlpP mutant biofilms. It was previously shown that high catalase activity can prevent penetration of hydrogen peroxide into biofilms of P. aeruginosa at a concentration of 50 mM (49). The P. tunicata genome also possesses three catalase genes (T. Thomas et al., unpublished results), which could be responsible for preventing penetration of exogenously added hydrogen peroxide, as well as playing a role in different susceptibility of the cells to hydrogen peroxide.
In summary, the present study suggests that endogenous hydrogen peroxide-mediated lysis of a subpopulation of cells occurs in P. tunicata, M. mediterranea, C. violaceum, and C. crescentus. AlpP homologue-mediated hydrogen peroxide appears to be fully responsible for autolysis of biofilm cells in P. tunicata, M. mediterranea, and C. violaceum. We propose that this process is an important mechanism for facilitating dispersal and generating diversity. It is suggested that hydrogen peroxide allows (directly or indirectly) both killing of a subpopulation of cells (30) and a possible increase in DNA damage and mutation frequency (50) of the remaining live cells, which may lead to the observed phenotypic variation in the dispersal cell population.
Acknowledgments
We acknowledge the assistance by Geoff Kornfeld and Abraham Choy for the lysine analysis. We also thank Antonio A. Iniesta for the plasmid pNPTS138 and helpful advice on the generation of the C. crescentus CAUMUR1 mutant strain.
This research was supported by grants from the Australian Research Council and the Centre for Marine Bio-Innovation at the University of New South Wales (UNSW), Sydney, Australia. P.L.-E. was the recipient of a grant from the Consejería de Educación y Cultura of the Comunidad Autónoma de la Región de Murcia, Murcia, Spain, which allowed her to visit UNSW.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. New mini-Tn5 derivatives for insertion mutagenesis and genetic engineering in gram-negative bacteria. Can. J. Microbiol. 411053-1055. [DOI] [PubMed] [Google Scholar]
- 2.Ali, A., M. H. Rashid, and D. K. Karaolis. 2002. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl. Environ. Microbiol. 685773-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ameisen, J. C. 2002. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 9367-393. [DOI] [PubMed] [Google Scholar]
- 4.Auschill, T. M., N. B. Arweiler, L. Netuschil, M. Brecx, E. Reich, A. Sculean, and N. B. Artweiler. 2001. Spatial distribution of vital and dead microorganisms in dental biofilms. Arch. Oral Biol. 46471-476. [DOI] [PubMed] [Google Scholar]
- 5.Barsby, T. 2006. Drug discovery and sea hares: bigger is better. Trends Biotechnol. 241-3. [DOI] [PubMed] [Google Scholar]
- 6.Bjedov, I., O. Tenaillon, B. Gerard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 3001404-1409. [DOI] [PubMed] [Google Scholar]
- 7.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 10116630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 1866208-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416740-743. [DOI] [PubMed] [Google Scholar]
- 10.Egan, S., S. James, C. Holmstrom, and S. Kjelleberg. 2002. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ. Microbiol. 4433-442. [DOI] [PubMed] [Google Scholar]
- 11.Egan, S., S. James, and S. Kjelleberg. 2002. Identification and characterization of a putative transcriptional regulator controlling the expression of fouling inhibitors in Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 68372-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204372-384. [DOI] [PubMed] [Google Scholar]
- 13.Entcheva-Dimitrov, P., and A. M. Spormann. 2004. Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J. Bacteriol. 1868254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez, M., and J. Sanchez. 2002. Nuclease activities and cell death processes associated with the development of surface cultures of Streptomyces antibioticus ETH 7451. Microbiology 148405-412. [DOI] [PubMed] [Google Scholar]
- 15.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore, M. S., and W. Haas. 2005. The selective advantage of microbial fratricide. Proc. Natl. Acad. Sci. USA 1028401-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez, D., P. Lucas-Elio, A. Sanchez-Amat, and F. Solano. 2006. A novel type of lysine oxidase: l-lysine-epsilon-oxidase. Biochim. Biophys. Acta 17641577-1585. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301510-513. [DOI] [PubMed] [Google Scholar]
- 19.Guiral, S., T. J. Mitchell, B. Martin, and J. P. Claverys. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. USA 1028710-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 21.Handke, L. D., K. M. Conlon, S. R. Slater, S. Elbaruni, F. Fitzpatrick, H. Humphreys, W. P. Giles, M. E. Rupp, P. D. Fey, and J. P. O'Gara. 2004. Genetic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J. Med. Microbiol. 53367-374. [DOI] [PubMed] [Google Scholar]
- 22.Hausner, M., and S. Wuertz. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 653710-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haussler, S. 2004. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ. Microbiol. 6546-551. [DOI] [PubMed] [Google Scholar]
- 24.Hendrickx, L., M. Hausner, and S. Wuertz. 2003. Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl. Environ. Microbiol. 691721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez-Romero, D., P. Lucas-Elio, D. Lopez-Serrano, F. Solano, and A. Sanchez-Amat. 2003. Marinomonas mediterranea is a lysogenic bacterium that synthesizes R-bodies. Microbiology 1492679-2686. [DOI] [PubMed] [Google Scholar]
- 26.Holmström, C., S. James, B. A. Neilan, D. C. White, and S. Kjelleberg. 1998. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 481205-1212. [DOI] [PubMed] [Google Scholar]
- 27.Hope, C. K., D. Clements, and M. Wilson. 2002. Determining the spatial distribution of viable and nonviable bacteria in hydrated microcosm dental plaques by viability profiling. J. Appl. Microbiol. 93448-455. [DOI] [PubMed] [Google Scholar]
- 28.James, S. G., C. Holmström, and S. Kjelleberg. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 622783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, P. M., C. E. Kicklighter, M. Schmidt, M. Kamio, H. Yang, D. Elkin, W. C. Michel, P. C. Tai, and C. D. Derby. 2006. Packaging of chemicals in the defensive secretory glands of the sea hare Aplysia californica. J. Exp. Biol. 20978-88. [DOI] [PubMed] [Google Scholar]
- 30.Kusakabe, H., K. Kodama, A. Kuninaka, H. Yoshino, and K. Soda. 1979. Antibacterial activity of l-lysine α-oxidase against rec+ and rec− strains of Bacillus subtilis. Agric. Biol. Chem. 431371-1373. [Google Scholar]
- 30a.Lawrence, J. R., M. R. Chenier, R. Roy, D. Beaumier, N. Fortin, G. D. Swerhone, T. R. Neu, and C. W. Greer. 2004. Microscale and molecular assessment of impacts of nickel, nutrients, and oxygen level on structure and function of river biofilm communities. Appl. Environ. Microbiol. 704326-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucas-Elío, P., D. Gómez, F. Solano, and A. Sanchez-Amat. 2006. The antimicrobial activity of marinocine, synthesized by Marinomonas mediterranea, is due to the hydrogen peroxide generated by its lysine oxidase activity. J. Bacteriol. 1882493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas-Elio, P., P. Hernandez, A. Sanchez-Amat, and F. Solano. 2005. Purification and partial characterization of marinocine, a new broad-spectrum antibacterial protein produced by Marinomonas mediterranea. Biochim. Biophys. Acta 1721193-203. [DOI] [PubMed] [Google Scholar]
- 33.Lukasheva, E. V., and T. T. Berezov. 2002. l-Lysine alpha-oxidase: physicochemical and biological properties. Biochemistry 671152-1158. [DOI] [PubMed] [Google Scholar]
- 34.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 703232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mai-Prochnow, A., J. S. Webb, B. C. Ferrari, and S. Kjelleberg. 2006. Ecological advantages of autolysis during the development and dispersal of Pseudoalteromonas tunicata biofilms. Appl. Environ. Microbiol. 725414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marden, P., A. Tunlid, K. Malmcrona-Friberg, G. Odham, and S. Kjelleberg. 1985. Physiological and morphological changes during short term starvation of marine bacterial isolates. Arch. Microbiol. 142326-332. [Google Scholar]
- 37.Matz, C., D. McDougald, A. M. Moreno, P. Y. Yung, F. H. Yildiz, and S. Kjelleberg. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 10216819-16824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miguelez, E. M., C. Hardisson, and M. B. Manzanal. 1999. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. J. Cell Biol. 145515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York, NY.
- 40.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilization of the biofilm structure. Curr. Opin. Biotechnol. 14255-261. [DOI] [PubMed] [Google Scholar]
- 41.Moller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monk, I. R., G. M. Cook, B. C. Monk, and P. J. Bremer. 2004. Morphotypic conversion in Listeria monocytogenes biofilm formation: biological significance of rough colony isolates. Appl. Environ. Microbiol. 706686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2881251-1254. [DOI] [PubMed] [Google Scholar]
- 45.Proctor, R. A., P. van Langevelde, M. Kristjansson, J. N. Maslow, and R. D. Arbeit. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 2095-102. [DOI] [PubMed] [Google Scholar]
- 46.Regev-Yochay, G., K. Trzcinski, C. M. Thompson, M. Lipsitch, and R. Malley. 2007. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J. Bacteriol. 1896532-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadowska, B., A. Bonar, C. von Eiff, R. A. Proctor, M. Chmiela, W. Rudnicka, and B. Rozalska. 2002. Characteristics of Staphylococcus aureus, isolated from airways of cystic fibrosis patients, and their small colony variants. FEMS Immunol. Med. Microbiol. 32191-197. [DOI] [PubMed] [Google Scholar]
- 48.Solano, F., E. Garcia, E. Perez De Egea, and A. Sanchez-Amat. 1997. Isolation and characterization of strain MMB-1 (CECT 4803), a novel melanogenic marine bacterium Appl. Environ. Microbiol. 633499-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart, P. S., F. Roe, J. Rayner, J. G. Elkins, Z. Lewandowski, U. A. Ochsner, and D. J. Hassett. 2000. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 66836-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Termini, J. 2000. Hydroperoxide-induced DNA damage and mutations. Mutat. Res. 450107-124. [DOI] [PubMed] [Google Scholar]
- 51.Webb, J. S., M. Lau, and S. Kjelleberg. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1868066-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1854585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wireman, J. W., and M. Dworkin. 1977. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J. Bacteriol. 129798-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, H., P. M. Johnson, K. C. Ko, M. Kamio, M. W. Germann, C. D. Derby, and P. C. Tai. 2005. Cloning, characterization, and expression of escapin, a broadly antimicrobial FAD-containing l-amino acid oxidase from ink of the sea hare Aplysia californica. J. Exp. Biol. 2083609-3622. [DOI] [PubMed] [Google Scholar]